The KASH proteins Klar and MSP-300 cooperate to promote even myonuclear spacing by linking the MSP-300 nuclear ring to the astral microtubule network.
Abstract
Striated muscle fibers are characterized by their tightly organized cytoplasm. Here, we show that the Drosophila melanogaster KASH proteins Klarsicht (Klar) and MSP-300 cooperate in promoting even myonuclear spacing by mediating a tight link between a newly discovered MSP-300 nuclear ring and a polarized network of astral microtubules (aMTs). In either klar or msp-300ΔKASH, or in klar and msp-300 double heterozygous mutants, the MSP-300 nuclear ring and the aMTs retracted from the nuclear envelope, abrogating this even nuclear spacing. Anchoring of the myonuclei to the core acto-myosin fibrillar compartment was mediated exclusively by MSP-300. This protein was also essential for promoting even distribution of the mitochondria and ER within the muscle fiber. Larval locomotion is impaired in both msp-300 and klar mutants, and the klar mutants were rescued by muscle-specific expression of Klar. Thus, our results describe a novel mechanism of nuclear spacing in striated muscles controlled by the cooperative activity of MSP-300, Klar, and astral MTs, and demonstrate its physiological significance.
Introduction
Striated muscle fibers are large multinucleated cells with highly ordered cytoplasmic organization (Squire, 1997; Sparrow and Schöck, 2009). Cellular organelles such as mitochondria and myonuclei are evenly spaced along the entire muscle fiber and are separated from the bulk of the cytoplasm, which contains densely arranged myofibrils. The mechanisms establishing and maintaining this highly ordered distribution of organelles and its relevance to muscle function has not yet been elucidated.
In a variety of cell types, a family of KASH domain proteins promote positioning of nuclei as well as mitochondria (Crisp et al., 2006; Fridkin et al., 2009; Starr and Fridolfsson, 2010; Mellad et al., 2011; Razafsky et al., 2011; Starr, 2011). KASH domains insert into the outer nuclear membrane and associate across the perinuclear space with SUN domain proteins in the inner nuclear membrane. This pairing brings about a mechanical linkage between the nucleoskeleton and the cytoskeleton (LINC complex; Crisp et al., 2006; Wilhelmsen et al., 2006). KASH proteins interact with actin, microtubule (MT) motor proteins, and/or intermediate filaments through their N-terminal domains (Morris, 2000; Tran et al., 2001; Starr and Han, 2002; Zhen et al., 2002; Padmakumar et al., 2004; Wilhelmsen et al., 2005; Roux et al., 2009), whereas SUN proteins associate with nuclear lamins via their C-terminal region (Crisp et al., 2006; Haque et al., 2010).
Previous studies in C. elegans and mice have suggested additional important roles of KASH and SUN proteins in muscles, particularly in the positioning of myonuclei and mitochondria (Starr and Han, 2002; Zhang et al., 2007b). For example, mice lacking the KASH domains of both Nesprin/syne-1 and Nesprin/syne-2 exhibit lethality shortly after birth. Nevertheless, the primary cause of their death appeared to be associated with aberrant alveolus sac morphology, resulting in respiratory failure; this lung phenotype complicates the interpretation of the contribution of these proteins to muscle function in a later stage (Zhang et al., 2007b). Double SUN-1 and SUN-2 knockout mice also die soon after birth; however, their lethality could be rescued by neuronal-specific expression of SUN-1 (Lei et al., 2009), casting doubt on the contribution of the KASH-containing isoforms to muscle function. However, because muscle performance was not tested directly in these mice, specific muscle defects cannot be excluded. It also appears that the genetic background of knockout mice is important for the penetrance of the phenotype in muscles, as in a later study, knockout of the Nesprin-1 KASH domain on a different genetic background resulted in 50% lethality, and the surviving mice exhibited progressive muscle wasting and abnormal gait (Puckelwartz et al., 2009). Similarly, knockout mice lacking the C-terminal spectrin repeat region of Nesprin-1 showed 60% lethality and muscle abnormalities (Zhang et al., 2010), which suggests a critical function of KASH proteins in proper muscle function. Thus, although mispositioning of nuclei and mitochondria in muscles has been described, the contribution of this phenotype to muscle function has not been established (Starr and Han, 2002). In all, the distinct contribution of KASH proteins as well as their possible cooperation, and the biological significance of their activities for muscle function, are not clear, primarily because of the combinatorial coexpression of KASH proteins in muscles, as well as in nonmuscle tissues.
Recent genetic analyses have implicated Nesprins in various human myopathies. For example, Nesprin/Syne-1 or Nesprin/Syne-2 are associated with Emery-Dreifuss muscular dystrophy (EDMD; Puckelwartz et al., 2009) and other myopathies (Zhang et al., 2007a; Attali et al., 2009) as well as severe cardiomyopathies (Puckelwartz et al., 2010). It is therefore critical to elucidate the specific function and mechanism of action of Nesprins in muscle tissue.
Drosophila melanogaster express two known KASH domain proteins, MSP-300 and Klarsicht (Klar). Klar is expressed in a wide range of tissues and has been shown to be essential for positioning of photoreceptor and cone cell nuclei in the developing eye and for the transport of lipid droplets in early embryos (Welte et al., 1998, 2005; Fischer et al., 2004; Patterson et al., 2004; Guo et al., 2005; Xie and Fischer, 2008); MSP-300 is not required for these processes (Xie and Fischer, 2008). Both proteins are expressed in striated muscles, but their contribution to organelle distribution and muscle function has not been previously reported. In larval stages, MSP-300 is specifically expressed in muscles and not in the nervous system; thus, its impact on muscle performance can be directly studied. Klar is expressed in other tissues, but its muscle-specific function can, in principle, be deduced by muscle-specific rescue experiments. Therefore, unlike mammalian KASH proteins, it is possible to dissect the function and individual contribution of MSP-300 and Klar to muscle performance in great detail.
In this paper, we show that MSP-300 and Klar act together to promote even spacing of the myonuclei at the periphery of striated muscle fibers by mediating a tight association between a nuclear ring structure of MSP-300 and the plus ends of a unique astral MT network. In the absence of Klar, or in an MSP-300 mutant lacking only the KASH domain, both the MSP-300 nuclear ring and the astral MT dissociate from the myonuclei; these nuclei remain anchored to the acto-myosin myofibrils, but their even spacing is lost. Nuclear anchoring to the core myofibril compartment is mediated solely by MSP-300 KASH-independent isoforms. Similarly, MSP-300 but not Klar promotes the anchoring and spacing of the mitochondria and ER within the muscle fiber. Significantly, we show that larval locomotion is impaired in both msp-300 and klar mutants, and the latter can be rescued by muscle-specific Klar expression. These results provide a mechanistic explanation for myonuclear positioning in striated muscles and demonstrate that this mechanism differs from that operating at earlier developmental stages in myotubes before the development of muscle sarcomeric architecture.
Results
Differential localization of MSP-300 and Klar in muscle fibers
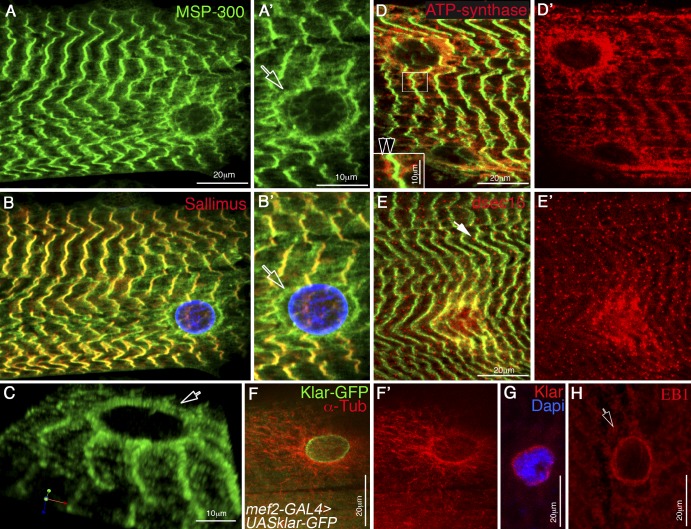
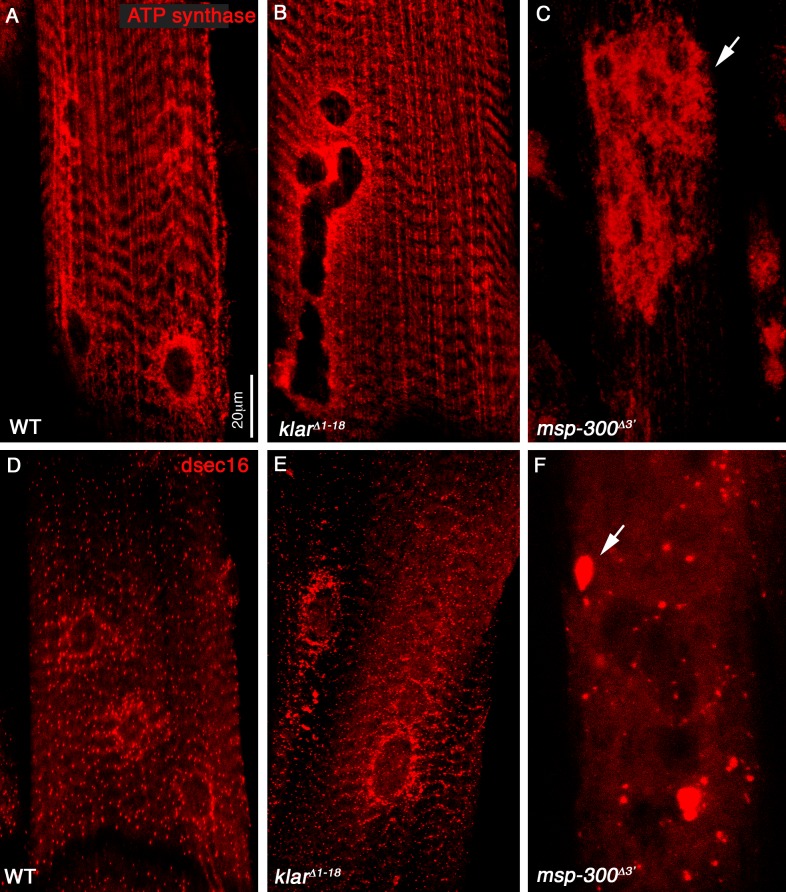
MSP-300 is highly expressed in striated muscles of third instar larvae (Volk, 1992). We used flat open larvae to reveal its fine subcellular distribution in muscles by colabeling MSP-300 with various organelle markers. MSP-300 showed extensive overlap with Sallimus/d-Titin along the Z-discs (Fig. 1 B). Notably, MSP-300 was also detected in a ring-like structure that surrounds each myonucleus and is located close to the sarcolemma (Fig. 1, A′, B′, and C). Co-labeling of MSP-300 with the mitochondrial marker ATP-synthase showed that the mitochondria were adjacent to the Z-discs, and in addition, were enriched around each nucleus (Fig. 1, D and D′). Similarly, a marker for the ER exit sites (anti-dsec16) showed a specific dotted pattern observed along the Z-discs, as well as around each nucleus, but did not fully overlap with MSP-300 (Fig. 1, E and E′). In contrast, staining with anti-Klar (Fig. 1 G) or labeling with Klar-GFP (Fig. 1 F) showed a single subcellular distribution that overlapped the nuclear envelope. Remarkably, a MT astral network surrounded each nucleus (Fig. 1 F), with their plus ends (labeled with anti-EB1) facing the nuclear membrane (Fig. 1 H).
Figure 1.
MSP-300 distribution in striated muscle fiber differs from that of Klar. (A and B) Striated muscles from third instar wild-type larvae labeled with MSP-300 (green; A), and with Sallimus (red) and lamin (blue; B). A′ and B′ represent twofold magnification of the nuclear region shown in A and B. Note the colocalization of MSP-300 and Sallimus in the Z-discs. (C) 3D image of the MSP-300 nuclear ring. Arrows in A′, B′, and C point to the MSP-300 nuclear ring. (D) Striated larval muscles stained with MSP-300 (green) and anti–ATP-synthase (red), labeling mitochondria. D′ shows the corresponding red channel. Note the accumulation of the mitochondria around the nucleus and along the Z-discs. The inset in D shows twofold magnification of the Z-disc region (boxed region), demonstrating the close proximity between the mitochondria and MSP-300. Arrowheads indicate the Z-band (green) and the mitochondria (red). (E) Striated larval muscles stained with MSP-300 (green) and with anti-dsec16 (red), which labels ER exit sites. E′ shows the red channel. Note the accumulation of dsec16 around the nucleus as well as along MSP-300 in Z-discs (arrow in E). (F) Striated larval muscles expressing UAS-Klar-GFP driven by a muscle-specific mef2-GAL4 driver (GFP-green), and labeled with anti–α-Tubulin (anti–α-Tub; red). F′ shows only the red channel of α-Tub. Notice the astral organization of the MT around the myonuclei. (G) Labeling of larval muscle nucleus with anti-Klar (red) and DAPI (blue). (H) Labeling of larval muscle nucleus with anti-EB1, a marker for MT plus ends demonstrating staining close to the nucleus. The arrow indicates the nuclear localization of EB1. Bars: (A, D, E, F, G, and H) 20 µm; (A’, C, D, inset) 10 µm.
In summary, the two Drosophila KASH proteins exhibit unique subcellular distributions: MSP-300 is associated with both the nuclear membrane and Z-discs, whereas Klar is located around the nuclei.
Both MSP-300 and Klar are essential for myonuclear spacing but differ in their contribution to anchoring of nuclei to the myofibrils
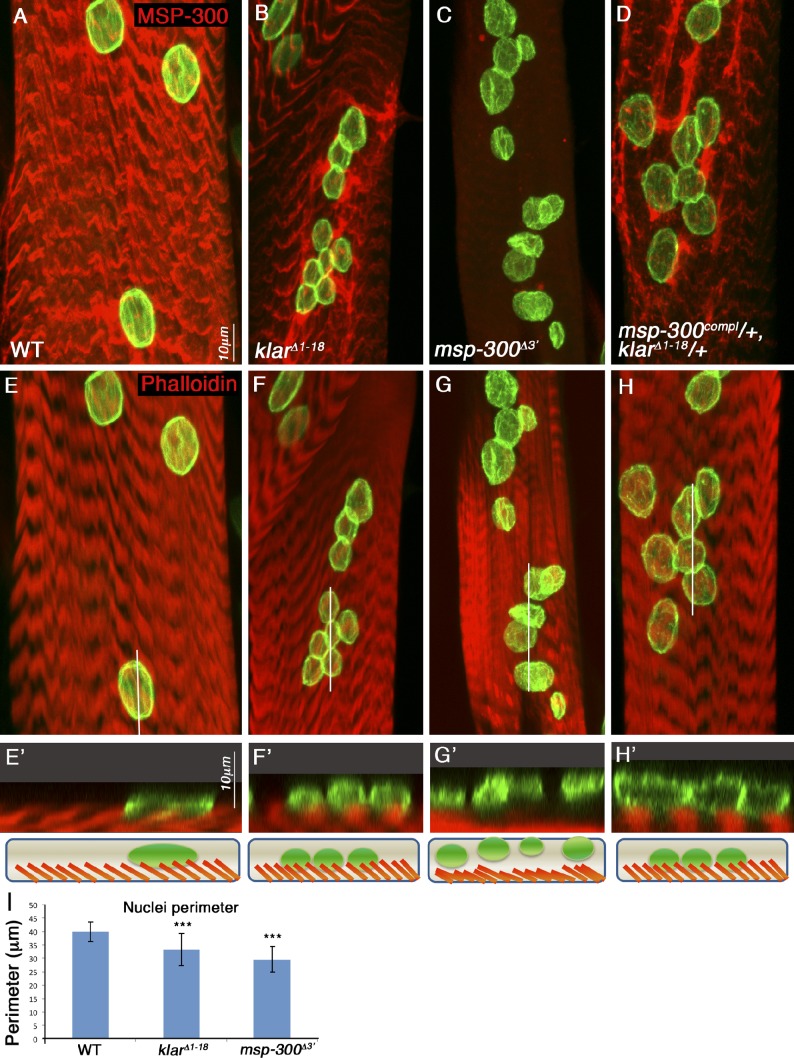
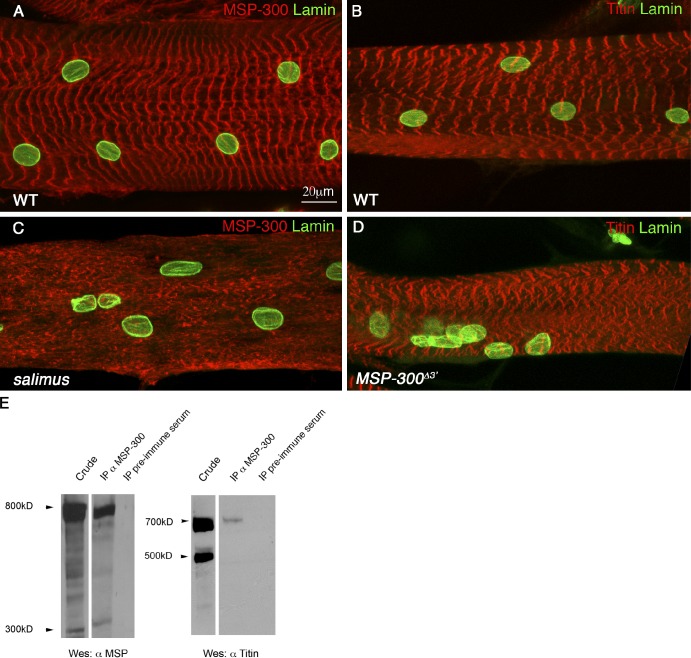
Next, we examined the contribution of MSP-300 and Klar to myonuclei positioning. We used the alleles klarΔ1–18, which deletes the entire klar locus, and msp-300Δ3′, which deletes roughly half of the 3′ region of the msp-300 locus (for molecular details of these alleles, see Fig. S1). We found that both klar and msp-300 mutant larvae exhibited aggregation of the myonuclei and aberrant nuclear shape, as well as variable nuclear size (Fig. 2, B, C, F, G, and I). Orthogonal confocal optical sections showed that in contrast to wild-type and klar mutant muscles, in which the nuclei were tightly associated with the myofibril compartment (Fig. 2, E′, F′, and H′), in msp-300Δ3′ mutant muscles the myonuclei dissociated from the core acto-myosin compartment and floated above it (Fig. 2 G′). Interestingly, when we simultaneously reduced the dosage of both Klar and MSP-300 (using heterozygous flies for a complete deletion of MSP-300 [msp-300coml] and klarΔ1–18), myonuclei were aberrantly positioned, but still associated with the acto-myosin network. Reduction of the expression of either Klar or MSP-300 alone had no nuclear positioning phenotype (unpublished data). Thus, there is a significant genetic interaction between the KASH proteins in promoting nuclear spacing but not in mediating nuclear anchoring.
Figure 2.
Both MSP-300 and Klar are essential for nuclear spacing, but MSP-300 uniquely contributes to nuclear anchoring. (A–H) Striated muscle of third instar larvae taken from wild-type (A, E, and E′), klarΔ1–18 mutant (B, F, and F′), msp-300Δ3′ mutant (C, G, and G′), or double heterozygous msp-300complete/+;klarΔ1–18/+ labeled with MSP-300 (red; A–D) and lamin (green), or with phalloidin (red, marks F-actin; E, E′, F, F′, G, G′, H, and H′) and lamin (green). Nuclear aggregation and variable nuclear sizes are observed in all mutant combinations. (I) Measurements of the nuclear perimeter in wild-type, klar, and msp-300 mutant muscles shows that in both mutants the nuclear size is significantly smaller (***, P < 0.0001). Orthogonal confocal cross sections of the region labeled by the white line in the projection images presented in E, F, G, and H are shown in E′. F′, G′, and H′ are shown at twofold magnification. Note that nuclear detachment from the acto-myosin compartment labeled with phalloidin is observed uniquely in msp-300Δ3′ mutant muscle. Corresponding schemes of the orthogonal sections of wild-type or mutant muscles are shown in the lower panels. Error bars indicate ±SE. Bars, 10 µm.
These results indicate that MSP-300 and Klar collaborate to promote even spacing between myonuclei; yet, MSP-300 uniquely directs myonuclei anchoring to the myofibrillar domain.
The KASH domains of both MSP-300 and Klar are essential for promoting the association of the MSP-300 nuclear ring with the nuclear membrane
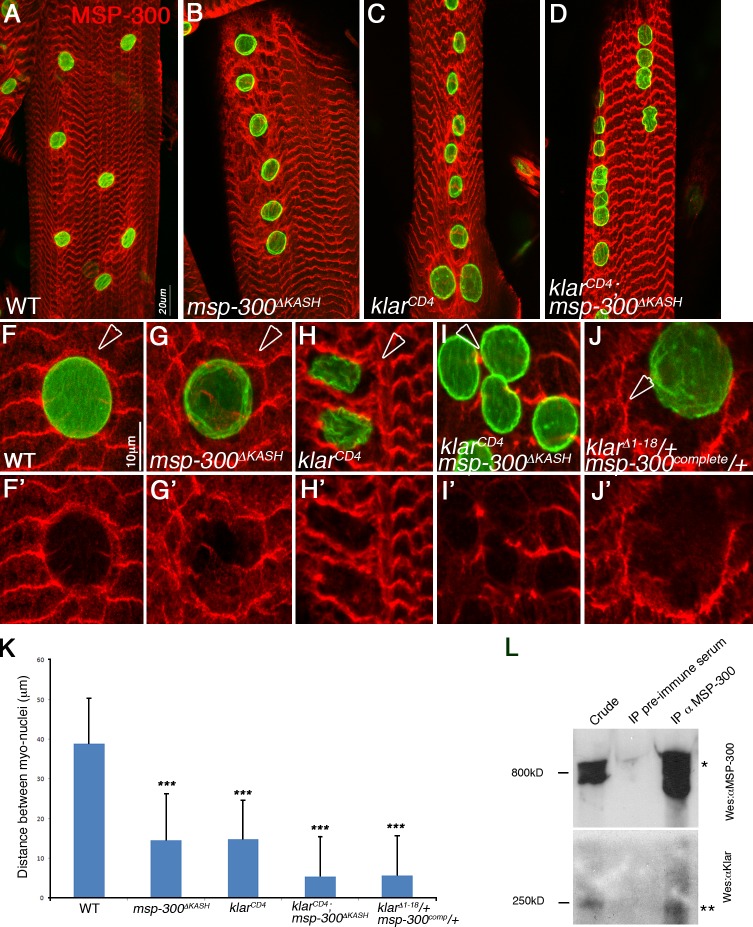
Both klar and msp-300 genes produce KASH as well as non-KASH protein isoforms (according to genome databases; Guo et al., 2005). To determine the importance of the KASH domains for myonuclear positioning, we analyzed MSP-300ΔKASH and/or klarmCD4, two alleles that specifically lack the KASH domains of MSP-300 and Klar, respectively. Larvae homozygous mutant for msp-300ΔKASH exhibited a milder phenotype relative to msp-300Δ3′ (compare Fig. 2 C and Fig. 3 B), and the nuclei remained connected to the myofibrillar domain (not depicted). Homozygous mutant larvae for klarCD4 exhibited a phenotype that was slightly weaker than that of klarΔ1–18 (compare Fig. 2 B to Fig. 3 C). Larvae double mutant for both klarcd4 and msp-300ΔKASH exhibited a significantly more severe nuclear mispositioning phenotype than each of these single mutants (Fig. 3 K), but in each of these cases the nuclei remained connected to the myofibrillar domain (not depicted), which suggests either parallel or cooperative function of both proteins in directing nuclear spacing.
Figure 3.
MSP-300 and Klar cooperate to connect the MSP-300 nuclear ring to the nuclear envelope, a process required for nuclear spacing. (A–K) Striated larval muscles of wild type (WT; A, F, and F′), msp-300ΔKASH (B, G, and G′), klarcd4 (C, H, and H′), klarcd4;msp-300ΔKASH double mutant (D, I, and I′), or klarΔ1–18/+;msp-300Δ3′/+ double heterozygous mutant (J and J′). Arrowheads mark the MSP-300 nuclear ring. Muscles were labeled with anti–MSP-300 (red) and anti-lamin (green). Note that the MSP-300 nuclear ring dissociates from the nuclear envelope in msp-300 or in klar mutants lacking only the KASH domain, as well as in the double heterozygous klar;msp-300 mutants, whereas the ring disappears in the double klar;msp-300 KASH knockout mutants. Dissociation of the MSP-300 nuclear ring correlates with the severity of the nuclear spacing phenotype, summarized in K. ***, P < 0.0001. Error bars indicate ±SE. (L) Immunoprecipitation experiment with anti–MSP-300 antibodies. The left lane shows the crude extract of larval muscles, the middle lane shows immunoprecipitation with preimmune serum, and the right lane shows the immunoprecipitated material with anti–MSP-300 antibody, reacted by Western blotting with anti–MSP-300 (top) or with anti–Klar-M (bottom). The immunoprecipitated MSP-300 is detected around the 800 kD (*) and Klar is detected at 250 kD (**). Bars: (A–D) 20 µm; (F–J) 10 µm.
We noticed that the MSP-300 nuclear ring dissociated from the nuclear envelope in both msp-300ΔKASH and in klarmCD4 homozygous mutants (compare Fig. 3, G and H, to Fig. 3 F). Because the nuclear ring is still detected by the antibody (raised against the spectrin repeats domain) in msp-300ΔKASH mutants, we assumed that the ring itself is comprised of non-KASH sequences. Interestingly, the ring structure was entirely lost in the double msp-300ΔKASH;klarCD4 mutants (Fig. 3 I′), which suggests that Klar as well as MSP-300 KASH-containing isoforms function to attach the ring to the nuclear envelope. In support of a cooperative activity between both KASH proteins in linking MSP-300 nuclear ring to the nuclear envelope, muscles from double heterozygous msp-300complete/+;klarcd4/+ animals exhibited dissociation of the MSP-300 nuclear ring (Fig. 3, I and J) in parallel to the aberrant nuclear spacing shown in Fig. 2 D. Moreover, in an attempt to evaluate whether MSP-300 and Klar associate in a protein complex, we performed an immunoprecipitation experiment with anti–MSP-300 antibodies from larval muscles and showed that Klar coimmunoprecipitated with MSP-300 (Fig. 3 L).
These results indicate that the MSP-300 nuclear ring is anchored to the nuclear envelope through the cooperative activity of both MSP-300 and Klar KASH domains, and suggest that this ring represents a key structure in promoting nuclear spacing at the periphery of the muscle fiber.
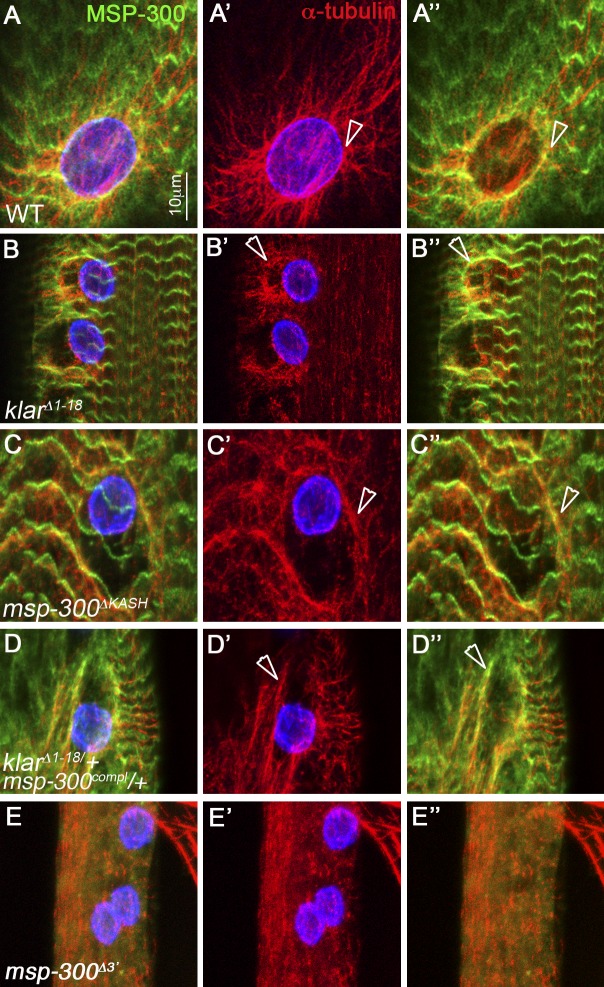
MSP-300 and Klar mediate the attachment of the astral MT to the nuclear membrane
The peripheral localization of both the MSP-300 nuclear ring and astral MT prompted us to examine possible links between the two structures. In wild-type larval muscles, we found that the MSP-300 nuclear ring often overlapped with α-tubulin staining, which showed accumulation around the nucleus (Fig. 4, A–A″). In klarΔ1–18, the network of the astral MT dissociated from the nucleus and often overlapped the MSP-300 nuclear ring (Fig. 4, B–B″). Similarly, in msp-300ΔKASH, we detected dissociation of the MT from the nuclear envelope that overlapped with the MSP-300 nuclear ring (Fig. 4, C–C″), as well as in msp-300compl/+;klarΔ1–18/+ double heterozygotes (Fig. 4, D–D″). Thus, the association of the astral MT with the nuclear membrane depends on the cooperative function of both MSP-300 and Klar. Interestingly, we detected a partial overlap between the dissociated MT and the MSP-300 nuclear ring (arrowheads in Fig. 4, B″, C″, and D″) observed in the various mutants. We therefore suggest that in the wild type, the MTs associate with MSP-300 in a KASH-independent manner; Klar as well as MSP-300 KASH domains connect both the MT and the MSP-300 ring to the nuclear envelope. Deletion of most of the spectrin repeats domain of MSP-300 (as in msp-300Δ3′) leads to complete disruption of MT organization at the periphery of the muscle fiber (Fig. 4, E–E″). Collectively, these results support a cooperative function of MSP-300 and Klar KASH domains in mediating the astral organization and association of the MTs to each myonucleus.
Figure 4.
The astral MTs detach from the myonuclei in msp-300 and klar mutants and overlap the MSP-300 nuclear ring. Striated larval muscles of wild type (A, A′, and A″), klarΔ1–18 (B, B′, and B″), msp-300ΔKASH (C, C′, and C″), msp-300compl/+;klarΔ1-18/+ double heterozygous (D, D′, and D″), and msp-300Δ3′ (E, E′, and E″) mutants labeled with MSP-300 (green; A, A″, B, B″, C, C″, D, D″, E, and E″), α-Tubulin (red; all panels), and lamin (blue; A, A′, B, B′, C, C′, D, D′, E, and E′). Arrowheads in all panels indicate the overlap between α-Tub staining and the MSP-300 nuclear ring. Note the complete disruption of the astral MT in msp-300Δ3′ mutant. Bar, 10 µm.
MSP-300 is essential for anchoring the mitochondria and ER to the Z-discs, whereas Klar is dispensable
To further elucidate the role of MSP-300 and Klar in the proper distribution of the mitochondria and ER within the muscle fiber, we used anti–ATP-synthase and anti–ER exit site–specific protein (dsec16). In wild-type larvae, the mitochondria accumulate around each nucleus, as well as along the Z-discs (Fig. 1, D and D′), and the staining for the ER exit site also appears along the Z-discs and surrounding each nucleus (Fig. 1, E and E′). Muscles from larvae homozygous mutant for msp-300Δ3′ showed mitochondrial as well as ER aggregation (Fig. 5, C and F), whereas klarΔ1–18 mutants (Fig. 5, B and E), as well as msp-300ΔKASH (not depicted) displayed normal distribution of both organelles.
Figure 5.
MSP-300 is uniquely required for the correct distribution of mitochondria and ER in striated muscle. Striated larval muscles of wild type (A and D), klarΔ1–18 (B and E), and msp-300Δ3′ (C and F) mutants labeled with anti–ATP-synthase (red; A–C) or with anti-dsec16 (red; D–F). Arrows in C and F indicate severe aggregation of the mitochondria and the ER exit sites, observed only in msp-300 mutant muscles. Bar, 20 µm.
These results suggested a major functional difference between MSP-300 and Klar in promoting organelle distribution; MSP-300 is required for their correct distribution, whereas Klar is dispensable for these processes.
d-Titin/Sallimus recruits the 800-kD isoform of MSP-300 to the Z-discs
The results so far suggest a critical and unique function for MSP-300 in anchoring nuclei, mitochondria, and ER structures to the Z-discs, in addition to its cooperative activity with Klar in nuclear spacing. To address the mechanism recruiting MSP-300 to the Z-discs, we immunoprecipitated MSP-300 from larval muscles and screened for the presence of Z-disc–specific proteins in the precipitated material. We found that a specific 700-kD isoform of d-Titin/Sallimus coprecipitated with MSP-300 (Fig. 6 E). The relatively low amount of d-Titin in the precipitated sample suggests that only a small fraction of this protein interacts with MSP-300.
Figure 6.
d-Titin recruits MSP-300 to the Z-discs. (A and C) Dorsal muscles of wild-type (A) or d-titin/sallimus mutant (C) larvae stained with MSP-300 (red) and lamin (green). (B and D) Dorsal muscles of wild-type (B) and msp-300 mutant (D) larvae stained with D-Titin (red) and lamin (green). Note the dissociation of MSP-300 from the Z-discs observed in the d-titin/sallimus mutant (C). (E) Immunoprecipitation of MSP-300 from wild-type larval extracts using anti–MSP-300 antibodies. The membrane was first reacted with anti-Titin antibodies (right) and then with anti–MSP-300 antibodies (left). Immunoprecipitation with preimmune serum served as a control. Bar, 20 µm.
This interaction is functionally important. MSP-300 localization along the Z-discs was significantly abrogated in sallimus mutant muscles, and consistent with these results, myonuclei were not properly distributed (compare Fig. 6 C to Fig. 6 A). In contrast, D-Titin/Sallimus localization along the Z-discs of msp-300Δ3′ mutant muscles was only slightly affected (compare Fig. 6 D to Fig. 6 B), which suggests that MSP-300 depends on D-Titin/Sallimus for its proper localization at the Z-discs, and not vice versa. Significantly, the sarcomeric organization of the msp-300Δ3′ mutant muscles was retained, as shown by Sallimus staining. Thus, the aggregation of the muscle organelles in msp-300Δ3′ mutant is not secondary to sarcomeric disorganization, which supports the direct involvement of MSP-300 in promoting correct distribution of these organelles.
Proper myonuclear positioning is essential for correct larval locomotion
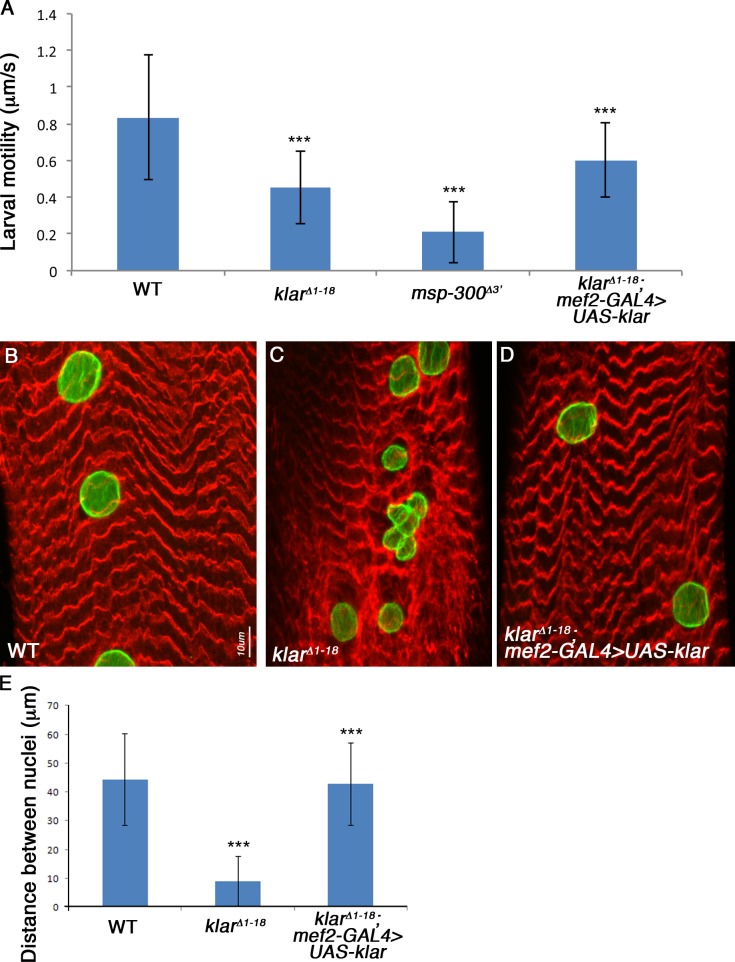
To address whether proper myonuclei positioning contributes to muscle function, we developed an assay to measure larval locomotion using video cameras that followed larval movements in a given time period (see Materials and methods). First we compared wild-type larvae with klarΔ1–18 mutant larvae and found that the homozygous mutants exhibited ∼50% reduction in their motility relative to wild-type larvae (Fig. 7 A), which supports the notion that even nuclear positioning is essential for correct motility. Remarkably, MSP-300Δ3′ mutants showed 70% reduction in their motility relative to wild-type larvae (Fig. 7 A), presumably reflecting the combined activity of this protein on mitochondria, ER, and myonuclear positioning. Assuming that the reduced motility of the larvae reflects aberrant muscle function, these experiments demonstrate that nuclear positioning is essential for correct muscle function, and that additional displacement of the mitochondria and ER worsen larval locomotion. Consistent with these results, we also found that adult flies homozygous for msp-300Δ3′ or for klarΔ1–18 are unable to fly.
Figure 7.
Larval motility is impaired in msp-300 and klar mutants, and can be rescued by muscle-specific expression of Klar. (A) Summary of larval motility measurements performed by video imaging analysis on wild type (n = 5), klarΔ1–18 (n = 8), msp-300Δ3′ (n = 6), or klar mutant rescued by mef2-GAL–driven expression of Klar (n = 10). ***, P < 0.0001. (B) Nuclear spacing rescued by mef2-GAL–driven expression of Klar (D), compared with wild type (A), and to klar mutant (C) muscles. Muscles were labeled with MSP-300 (red) and with lamin (green). Bar, 10 µm. (E) Quantification of nuclear spacing in wild-type (n = 36), klar (n = 30), and klar mutant muscles rescued by muscle-specific Klar expression (n = 21). ***, P < 0.0001 when comparing klar mutant to wild type, or rescued larvae compared with klar mutant. Error bars indicate ±SE.
Because Klar is expressed in various tissues, it is crucial to evaluate whether its effect on larval locomotion is muscle-specific. To this end, we rescued klarΔ1–18 mutant larvae with muscle-specific expression of Klar-GFP (using a mef2-GAL4 driver). Myonuclei positioning in these larvae was fully rescued (compare Fig. 7, B–D, and Fig. 7 E). Significantly, larval locomotion was restored in these larvae to wild-type levels (Fig. 7 A). We therefore concluded that in Drosophila larvae, proper myonuclei positioning is absolutely required for correct muscle function. However, when additional organelles such as mitochondria and ER are mispositioned (as is the case in MSP-300Δ3′ mutant larvae), the degree of disruption of muscle function is significantly extended.
Nuclear positioning in nonstriated myotubes depends solely on Klar activity
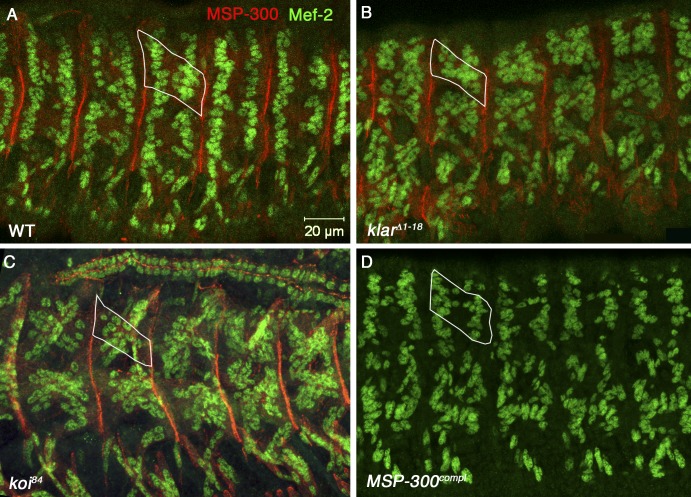
Drosophila muscles acquire their stereotypic sarcomeric architecture in which the acto-myosin network is arranged in striated fashion only toward the end of embryogenesis; however, a typical organization of the myonuclei is evident several hours earlier, shortly after the formation of the myotendinous junction (MTJ; Fig. S2). We analyzed the pattern of myonuclei in the four dorsal muscles DA1, DO1, DA2, and DO2 at stage 16 of embryonic development before muscle sarcomeric organization, as well as in third instar larvae in which sarcomeric organization has been established. The nuclei underwent significant rearrangement after the establishment of sarcomeric organization (Fig. S2). This suggested that the mechanism promoting nuclear positioning in muscles before and after sarcomerization might be different. We therefore further dissected the position of myonuclei in msp-300, klar, and klaroid mutants in embryonic muscles. Indeed, we found that myonuclei positioning is aberrant in klarΔ1–18 homozygous mutant embryos (Fig. 8 B) but not in msp-300complete homozygous mutants (Fig. 8 D). Interestingly, a similar myonuclei phenotype was also observed in homozygous mutants for the Drosophila SUN domain protein klaroid (koi) (Fig. 8 C). Thus, before muscle sarcomeric architecture had been established, Klar was the only KASH protein responsible for myonuclear organization, and nuclear organization in various states of muscle differentiation was promoted by distinct mechanisms.
Figure 8.
Klar and Koi but not MSP-300 are required for muscle nuclear organization in embryonic muscles. Wild-type (A), klarD1–18-null (B), koi84-null (C), and MSP-300compl-null (D) mutant embryos double labeled with MEF2 (green) and MSP-300 (red; lateral dorsal view of the embryo). The white outline in each panel represents a single dorsal muscle cell. The two nuclear rows are characteristic of wild-type dorsal muscle disappearing in klar and koi mutant muscles. Note the lack of staining with anti–MSP-300 of the MSP-300complt mutant. Bar, 20 µm.
Discussion
In this paper, we show for the first time cooperative, as well as unique, activities of the two Drosophila KASH proteins MSP-300 and Klar in promoting even spacing and anchoring of myonuclei in striated muscle fibers. A novel MSP-300 nuclear ring assembles and anchors the MTs to the nuclear envelope in a Klar- and MSP-300 KASH–dependent manner, mediating MT astral organization around each nucleus. We suggest that the astral MT associated with each myonucleus, forming a basic unit, which in the steady-state holds each nucleus in place. However, during muscle fiber growth, each unit might change its relative position as a result of MT growth so that the distance between the myonuclei is maintained. Anchoring of the myonuclei to the core acto-myosin fibrillar compartment is mediated exclusively by MSP-300, which maintains physical continuity between the nuclear ring and the Z-discs, presumably through dimerization of the spectrin repeats capable of forming filaments.
Recent results (Metzger et al., 2012) suggested that reducing KASH proteins from the nuclear membrane (by overexpressing the KASH domain) did not affect nuclear positioning in C2C12 cells. It is possible that residual KASH-dependent activity was present in these cells, capable of rescuing nuclei position.
Our physiological measurements demonstrate the critical contribution of nuclear spacing to larval locomotion and muscle activity, which is consistent with the results of Metzger et al. (2012). Significantly, our studies further demonstrate that the contribution of MSP-300 to muscle function is more critical relative to that of Klar (as msp-300Δ3′ mutant larvae were significantly slower than klarΔ1–18 mutant larvae), presumably because MSP-300 affects the positioning of the myonuclei as well as that of the mitochondria and ER.
The MT network appears to possess the dynamic properties required for myonuclear organization. Consistently, recent data show the active involvement of opposing MT motors in driving robust nuclear dynamics in differentiating CTC12 cells (Wilson and Holzbaur, 2012). During embryogenesis, MT arrangement in myotubes is polarized with their plus ends close to the MTJ (Clark et al., 1997). However, after muscle sarcomerization, the MTs undergo a significant rearrangement into astral organization with their plus ends facing the nuclei (shown by staining with EB1; Fig. 1). This suggests that nuclear positioning and spacing differs mechanistically between early embryonic and larval stages, i.e., before and after the establishment of sarcomeric architecture. In support of this model, we show that: (a) Klar is the only KASH protein required for nuclear positioning in embryonic myotubes, whereas MSP-300 is dispensable (Fig. 8). (b) The polarity and distribution of the MT network changes during development: in embryos (stage 16), the plus ends are close to the MTJ (Clark et al., 1997), whereas in striated muscle fibers, the MT forms astral structures surrounding each nucleus with their plus ends facing the nuclear envelope (Fig. 1). (c) The nuclei are arranged in distinct patterns at each stage; e.g., in the embryonic dorsal acute and oblique muscles, the nuclei are positioned in a typical half circle close to the MTJ, whereas in third instar larvae, the nuclei of the same muscles rearrange and are distributed evenly along the entire muscle fiber. Thus, we suggest that sarcomerization involves a significant rearrangement of the MT network and the nuclei, at the end of which the nuclei are spaced evenly along the muscle fiber and attached to the Z-discs.
The mechanism regulating this rearrangement has not been elucidated. We suggest that as part of this rearrangement, MSP-300 is translocated from the MTJ, where it accumulates during embryonic stages (Volk 1992 and unpublished data) into the Z-discs at the end of embryogenesis. Presumably, during this latter stage MSP-300 isoforms containing KASH are produced, and collaborate with Klar to promote the attachment of the astral MT network to the nuclear envelope. Klar function has been previously linked to MTs as a regulator of the MT motors dynein (associated with MT minus ends) and/or kinesin (associated with MT plus ends; Welte et al., 1998; Mosley-Bishop et al., 1999; Fischer et al., 2004; Patterson et al., 2004; Welte, 2004). Therefore, Klar function in promoting the association of the MSP-300 nuclear ring and MT plus ends with the nuclear envelope might be mediated through its ability to recruit MT plus-end motors. Nesprin 1 and 2 were also shown to interact with dynein/dynactin and kinesin-1 in developing mouse neurons (Zhang et al., 2009), and the spectrin repeats region of Nesprin 1 was shown to bind kinesin-2 (Fan and Beck, 2004). Therefore, Klar can potentially mediate an interaction with MT, which is also consistent with the demonstration that kinesin heavy chain is essential for myonuclei positioning in Drosophila larval muscles (Metzger et al., 2012). MSP-300 might be indirectly linked to this activity through its association with Klar.
Because muscle size increases dramatically during larval growth, and the number of nuclei remains constant (Beckett and Baylies, 2006), it is not clear how nuclear spacing is maintained during this growth process. We suggest that similarly to nuclear spacing in preblastoderm embryos (Baker et al., 1993), the minus ends of the MT associate with each other, whereas the plus ends grow so that the distance between the nuclei remains equal. In this manner, nuclei are able to compare their relative position and maintain even spacing in all directions. A prerequisite for this mechanism is the anchoring of the astral MT to the nuclear membrane in a polarized fashion, and this function is provided by the MSP-300 ring, which might associate with MT in a klar-independent manner.
Our experiments suggest a major function for MSP-300 both in promoting nuclear spacing (cooperatively with Klar) and in mediating anchoring of the nuclei as well as other organelles to the muscle core myofibrillar domain. The spectrin repeats domain is critical for this latter function, as most of it is eliminated in msp-300Δ3′. Similarly to other proteins containing the spectrin repeats domain, the multiple (52) spectrin repeats are likely to promote formation of MSP-300 dimers that form elongated filaments capable of connecting between different organelles and the cytoskeleton. In striated muscle, we suggest that such filaments connect between the various organelles and the Z-discs. The association of these filaments with the Z-discs requires the activity of d-Titin/Sallimus, another large protein that associates with the Z-discs (Zhang et al., 2000). The function of the N-terminal domain of MSP-300 containing the calponin-homology (CH) actin-binding motifs is still elusive because of the early lethality of the homozygous mutants.
Our analysis provides a mechanistic explanation for the severity of Nesprin-related diseases. We suggest that as in Drosophila, Nesprin 1 and 2 and a mammalian klar orthologue (yet to be identified) cooperate to promote myonuclei positioning in vertebrate muscle fibers. Thus, elimination of the KASH domains of both nesprin1 and nesprin2 in mutant mice might still leave a putative mammalian Klar orthologue intact, thereby only partially affecting nuclear positioning in the mutant muscles. However, elimination of additional domains of Nesprin described in Zhang et al. (2010) might abrogate MT organization more profoundly so that other Nesprins are incapable of complementing this situation. In addition, the position of other organelles might be disrupted as observed in msp-300Δ3′, leading to a more severe phenotype in mice, and possibly also in humans.
A recent study suggested that the amyotrophic lateral sclerosis (ALS)-associated protein VAPB is secreted from motor neurons and promotes the correct positioning of mitochondria in muscle fibers (Han et al., 2012). It would be interesting to test for a possible functional link between VAPB and MSP-300 activity in promoting mitochondrial positioning in striated muscles.
In summary, our studies provide a mechanistic explanation for the process of myonuclear positioning at distinct developmental stages in striated muscles. These studies may help in the prognosis of the severity of various Nesprin-related diseases in which distinct domains are missing, and provide a basis for future therapeutic approaches.
Materials and methods
Fly stocks
Fly stocks used in this study included w− (WT), mef2-gal4 (Bloomington Drosophila Stock Center), MSP-300ΔKASH, klarmCD4/TM6B (from J.A Fischer, University of Texas, Austin, TX), w1118;Df(2L)msp3003’/CyO, w1118;Df(2L)msp300complete/CyO,kr-GFP (from S. Roth, University of Cologne, Cologne, Germany), klar Δ118/TM6B (generated by us), slsg05/slse4 (from M.C. Beckerle, University of Utah, Salt Lake City, UT), and duf-gal4,UAS-RFP-NLS/FM7 (from B. Shilo, Weizmann Institute, Rehovot, Israel). The upstream activator sequence (UAS) line (produced by us) was GFP-Klar A.
Generation of the klar null allele
To generate the klar null allele, FLP-mediated recombination was used between two PBac elements, PBac[WH] f06799 and PBac{WH}klarf06231, using established strategies (Parks et al., 2004; Thibault et al., 2004). This approach generated a deletion of ∼108 kb of genomic DNA that removes all coding exons of klar as well as the neighboring gene CG13891; the noncoding exon 0 (Guo et al., 2005) remained intact. We refer to this allele as klarΔ1–18 because exons 1–18 were removed.
Expression constructs
To generate the GFP-Klar–expressing flies, GFP was amplified from pEGP-C1 (Takara Bio Inc.) and cloned into the KpnI and NotI sites of pUASp. A tobacco etch virus (TEV) protease site was generated by oligonucleotide synthesis and cloned 3′ to the GFP gene. Full-length klar α was amplified from the cDNA clone klar1 (Mosley-Bishop et al., 1999) and cloned in-frame 3′ to the TEV site. The resulting plasmid was injected into Drosophila embryos by Genetic Services, Inc.
Immunochemical reagents
Primary antibody staining was performed at room temperature for 2 h (for Western blotting) or overnight (for immunostaining), and the secondary staining was performed for 1 h. We used the following primary antibodies: guinea pig anti–MSP-300 (Volk, 1992) was used at 1:200 for immunostaining, and at 1:1,000 for Western blotting. Rat anti–α-Tubulin was used at 1:10 for immunostaining (MCA78S; AbD Serotec). Rabbit anti–Mef-2 (from H. Nguyen, Friedrich-Alexander University of Erlangen-Nuremberg, Erlangen, Germany) was used at 1:200 for immunostaining. Mouse anti-lamin (from Y. Gruenbaum, Hebrew University of Jerusalem, Jerusalem, Israel) was used at 1:10 for immunostaining. Mouse anti-DEB1 raised against Drosophila EB1 protein (J.A Knoblich, Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria.) was used at 1:10 for immunostaining. Rat anti-kettin/d-Titin (Klg16, MAC155; Abcam) was used at 1:200 for immunostaining and 1:500 for Western blotting. Chick anti-GFP (Abcam) was used at 1:500 for immunostaining. Mouse anti-ATP synthase (MitoSciences) was used at 1:500 for immunostaining. Rabbit anti-Sec16 (ER exit site) raised against a GST fusion protein comprising amino acids 655–817 of dSec16 (Ivan et al., 2008) was used at 1:200 for immunostaining. Mouse anti–Klar-C, which recognizes an epitope in exon 18 (Guo et al., 2005), was used at 1:5. Mouse anti–Klar-M, which recognizes an epitope in exon 9, was used at 1:30 for Western blotting (Guo et al., 2005). Secondary antibodies conjugated with Cy3, Cy5, and Cy2 raised against guinea pig, mouse, and rabbit were purchased from Jackson ImmunoResearch Laboratories. Alexa Fluor 488 Phalloidin (Invitrogen) was used at 1:200.
Fixation and immunostaining
Staining of embryos.
Embryos were collected, dechorionated, fixed with a mixture of 3% paraformaldehyde and heptane, devitellinized with a methanol-heptane mixture, washed, and incubated with a blocking solution of 10% BSA in PBT (PBS + 0.1% Triton X-100) for 1 h. Embryos were then incubated in primary antibody overnight, washed, incubated in the secondary antibody for 2 h, and washed and submerged in 80% glycerol, followed by mounting using Immu-Mount solution (Thermo Fisher Scientific).
Staining of larval flat preparations.
Third instar larvae were selected and pinned on Sylgard plates (silicone elastomer-coated Petri plates, prepared by using a cocktail obtained from Dow Corning). The larvae were slit open, cleaned from fat bodies and organs, and fixed in 4% PFA in PBS for 1 h, followed by blocking with 5% BSA in PBS with 0.1% Triton X-100 (PBT) for 15 min. The preparations were stained with primary antibody in PBT (overnight), washed with PBT, reacted with secondary antibody for 1 h, washed with PBT, and mounted using Immu-Mount solution (Thermo Fisher Scientific) on a glass slide such that the somatic muscles faced the coverslip.
Western blotting and immunoprecipitation
Third instar larvae were collected and crushed in RIPA buffer (1% Triton X-100, 0.1% SDS, 0.15 M NaCl, and 0.05 Tris, pH 7) and protease inhibitors (P 8340; Sigma-Aldrich), then incubated on ice for 10 min. For immunoprecipitation, equal amounts of protein lysate were incubated with protein A/G beads (SC-2003, Santa Cruz Biotechnology, Inc.) coupled with guinea pig anti–MSP-300 polyclonal antibody or mouse anti-GFP monoclonal antibody, overnight at 4°C. Beads were washed three times with the RIPA lysis buffer and boiled in protein sample buffer to elute the proteins. Western analysis was performed according to standard procedures, as described previously (Volk, 1992).
High molecular weight protein gels
High molecular weight proteins were analyzed by SDS-PAGE using 2.5% acrylamide gels strengthened with 1.5% agarose (Burkart et al., 2007). The gel solution was prewarmed to 50°C in a water bath in 50-ml Falcon tubes. Tetramethylethylenediamine (Temed) was added last and the gel was poured immediately. The gel was run for 1–2 h at 180 V at room temperature. Proteins were transferred to a nitrocellulose membrane by electrophoresis at 500 mA for 6 h in a buffer containing 0.1% (wt/vol) SDS (Tatsumi and Hattori, 1995).
Larval locomotion assay
Third instar larvae were collected and placed on agar plates (5 cm in diameter) at room temperature. Their movement across the plate was filmed with a video camera (OM-1; Olympus) with known filming velocity. Film frames spanning 20–70 s, showing continuous movement, were used for analysis of distance and movement velocity. Quantification was performed using ImageJ software.
Confocal microscopy
Immunofluorescence microscopy was performed with a confocal system (LSM710; Carl Zeiss) mounted on a microscope (Axiovert; Carl Zeiss) at room temperature. A C-Apochromat 40×, NA 1.20 water immersion objective lens was used. The digital fluorescent images were processed using LSM software (ZEN 2010) and Photoshop CS (Adobe). 3D images were processed using Volocity software (PerkinElmer).
Online supplemental material
Fig. S1 shows a scheme of MSP-300 and klar gene loci and the various alleles used in the present study. Fig. S2 shows confocal scans of muscle nuclei in dorsal muscles of embryonic and larval stages, demonstrating the rearrangement of the nuclei in muscles prior to and following sarcomerization. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201204102/DC1.
Supplementary Material
Acknowledgments
We thank J.A. Fischer, B. Bullard, S. Roth, M.C. Beckerle, J.A. Knoblich, H.N. Nguyen, C. Rabouille, and Y. Gruenbaum for fly stocks and/or various antibodies. We also thank the Bloomington Stock Center for various fly lines, B. Bullard for providing the immunoprecipitation and Western protocols for large proteins, and for Flybase. We thank Slava Kalchenko from the Weizmann Imaging Center for setting up the larval locomotion assay.
This study was supported by a grant from Israel Science Foundation (ISF) to T. Volk, by National Institute of General Medical Sciences (NIGMS) grant GM64687, and by start-up support from the University of Rochester to M. Welte.
Footnotes
Abbreviations used in this paper:
- Klar
- Klarsicht
- MT
- microtubule
- MTJ
- myotendinous junction
References
- Attali R., Warwar N., Israel A., Gurt I., McNally E., Puckelwartz M., Glick B., Nevo Y., Ben-Neriah Z., Melki J. 2009. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum. Mol. Genet. 18:3462–3469 10.1093/hmg/ddp290 [DOI] [PubMed] [Google Scholar]
- Baker J., Theurkauf W.E., Schubiger G. 1993. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J. Cell Biol. 122:113–121 10.1083/jcb.122.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett K., Baylies M.K. 2006. The development of the Drosophila larval body wall muscles. Int. Rev. Neurobiol. 75:55–70 10.1016/S0074-7742(06)75003-6 [DOI] [PubMed] [Google Scholar]
- Burkart C., Qiu F., Brendel S., Benes V., Hååg P., Labeit S., Leonard K., Bullard B. 2007. Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J. Mol. Biol. 367:953–969 10.1016/j.jmb.2007.01.059 [DOI] [PubMed] [Google Scholar]
- Clark I.E., Jan L.Y., Jan Y.N. 1997. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 124:461–470 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Beck K.A. 2004. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 117:619–629 10.1242/jcs.00892 [DOI] [PubMed] [Google Scholar]
- Fischer J.A., Acosta S., Kenny A., Cater C., Robinson C., Hook J. 2004. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 168:1385–1393 10.1534/genetics.104.028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A., Penkner A., Jantsch V., Gruenbaum Y. 2009. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell. Mol. Life Sci. 66:1518–1533 10.1007/s00018-008-8713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Jangi S., Welte M.A. 2005. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol. Biol. Cell. 16:1406–1416 10.1091/mbc.E04-10-0920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.M., Tsuda H., Yang Y., Vibbert J., Cottee P., Lee S.J., Winek J., Haueter C., Bellen H.J., Miller M.A. 2012. Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev. Cell. 22:348–362 10.1016/j.devcel.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Mazzeo D., Patel J.T., Smallwood D.T., Ellis J.A., Shanahan C.M., Shackleton S. 2010. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 285:3487–3498 10.1074/jbc.M109.071910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V., de Voer G., Xanthakis D., Spoorendonk K.M., Kondylis V., Rabouille C. 2008. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell. 19:4352–4365 10.1091/mbc.E08-03-0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K., Zhang X., Ding X., Guo X., Chen M., Zhu B., Xu T., Zhuang Y., Xu R., Han M. 2009. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. USA. 106:10207–10212 10.1073/pnas.0812037106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellad J.A., Warren D.T., Shanahan C.M. 2011. Nesprins LINC the nucleus and cytoskeleton. Curr. Opin. Cell Biol. 23:47–54 10.1016/j.ceb.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., Baylies M.K. 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 484:120–124 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N.R. 2000. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148:1097–1101 10.1083/jcb.148.6.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop K.L., Li Q., Patterson L., Fischer J.A. 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9:1211–1220 10.1016/S0960-9822(99)80501-6 [DOI] [PubMed] [Google Scholar]
- Padmakumar V.C., Abraham S., Braune S., Noegel A.A., Tunggal B., Karakesisoglou I., Korenbaum E. 2004. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 295:330–339 10.1016/j.yexcr.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Patterson K., Molofsky A.B., Robinson C., Acosta S., Cater C., Fischer J.A. 2004. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell. 15:600–610 10.1091/mbc.E03-06-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz M.J., Kessler E., Zhang Y., Hodzic D., Randles K.N., Morris G., Earley J.U., Hadhazy M., Holaska J.M., Mewborn S.K., et al. 2009. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 18:607–620 10.1093/hmg/ddn386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz M.J., Kessler E.J., Kim G., Dewitt M.M., Zhang Y., Earley J.U., Depreux F.F., Holaska J., Mewborn S.K., Pytel P., McNally E.M. 2010. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol. 48:600–608 10.1016/j.yjmcc.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D., Zang S., Hodzic D. 2011. UnLINCing the nuclear envelope: towards an understanding of the physiological significance of nuclear positioning. Biochem. Soc. Trans. 39:1790–1794 10.1042/BST20110660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., Burke B. 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 106:2194–2199 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J.C., Schöck F. 2009. The initial steps of myofibril assembly: integrins pave the way. Nat. Rev. Mol. Cell Biol. 10:293–298 10.1038/nrm2634 [DOI] [PubMed] [Google Scholar]
- Squire J.M. 1997. Architecture and function in the muscle sarcomere. Curr. Opin. Struct. Biol. 7:247–257 10.1016/S0959-440X(97)80033-4 [DOI] [PubMed] [Google Scholar]
- Starr D.A. 2011. KASH and SUN proteins. Curr. Biol. 21:R414–R415 10.1016/j.cub.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Fridolfsson H.N. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi R., Hattori A. 1995. Detection of giant myofibrillar proteins connectin and nebulin by electrophoresis in 2% polyacrylamide slab gels strengthened with agarose. Anal. Biochem. 224:28–31 10.1006/abio.1995.1004 [DOI] [PubMed] [Google Scholar]
- Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Tran P.T., Marsh L., Doye V., Inoué S., Chang F. 2001. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153:397–411 10.1083/jcb.153.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T. 1992. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 116:721–730 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2004. Bidirectional transport along microtubules. Curr. Biol. 14:R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Welte M.A., Gross S.P., Postner M., Block S.M., Wieschaus E.F. 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 92:547–557 10.1016/S0092-8674(00)80947-2 [DOI] [PubMed] [Google Scholar]
- Welte M.A., Cermelli S., Griner J., Viera A., Guo Y., Kim D.H., Gindhart J.G., Gross S.P. 2005. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr. Biol. 15:1266–1275 10.1016/j.cub.2005.06.062 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K., Ketema M., Truong H., Sonnenberg A. 2006. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119:5021–5029 10.1242/jcs.03295 [DOI] [PubMed] [Google Scholar]
- Wilson M.H., Holzbaur E.L. 2012. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Fischer J.A. 2008. On the roles of the Drosophila KASH domain proteins Msp-300 and Klarsicht. Fly (Austin). 2:74–81 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Featherstone D., Davis W., Rushton E., Broadie K. 2000. Drosophila D-titin is required for myoblast fusion and skeletal muscle striation. J. Cell Sci. 113:3103–3115 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T., et al. 2007a. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 16:2816–2833 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., Han M. 2007b. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 134:901–908 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Felder A., Liu Y., Guo L.T., Lange S., Dalton N.D., Gu Y., Peterson K.L., Mizisin A.P., Shelton G.D., et al. 2010. Nesprin 1 is critical for nuclear positioning and anchorage. Hum. Mol. Genet. 19:329–341 10.1093/hmg/ddp499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y.Y., Libotte T., Munck M., Noegel A.A., Korenbaum E. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207–3222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.