Six1 in satellite cells is important for muscle regeneration and homeostasis of the stem cell niche by regulating MyoD, Myogenin, and Dusp6-ERK signaling.
Abstract
Satellite cells (SCs) are stem cells that mediate skeletal muscle growth and regeneration. Here, we observe that adult quiescent SCs and their activated descendants expressed the homeodomain transcription factor Six1. Genetic disruption of Six1 specifically in adult SCs impaired myogenic cell differentiation, impaired myofiber repair during regeneration, and perturbed homeostasis of the stem cell niche, as indicated by an increase in SC self-renewal. Six1 regulated the expression of the myogenic regulatory factors MyoD and Myogenin, but not Myf5, which suggests that Six1 acts on divergent genetic networks in the embryo and in the adult. Moreover, we demonstrate that Six1 regulates the extracellular signal-regulated kinase 1/2 (ERK1/2) pathway during regeneration via direct control of Dusp6 transcription. Muscles lacking Dusp6 were able to regenerate properly but showed a marked increase in SC number after regeneration. We conclude that Six1 homeoproteins act as a rheostat system to ensure proper regeneration of the tissue and replenishment of the stem cell pool during the events that follow skeletal muscle trauma.
Introduction
Adult skeletal muscles possess a remarkable capacity for regeneration. The muscle tissue is composed of terminally differentiated multinucleated cells called myofibers, which contain densely packed myofibrils that assemble the sarcomeres, forming the basic machinery necessary for muscle contraction. Mature myofibers cannot regenerate, and reconstruction of the skeletal muscle tissue after damage, such as physical trauma, repeated exercise, or as a result of disease, relies on the stem cell potential of resident satellite cells (SCs; Le Grand and Rudnicki, 2007). SCs express the paired-box transcription factor Pax7 and are indispensable for adult skeletal muscle regeneration, as conditional ablation of SCs results in a failure to regenerate skeletal muscle up to 2 mo after ablation and muscle injury (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). Under resting conditions, SCs are quiescent and located in small depressions between the sarcolemma of their host myofibers and the basal lamina (Mauro, 1961). After damage to the myofibers, SCs will activate, proliferate, and give rise to a population of transient-amplifying myogenic cells called myoblasts expressing the myogenic regulatory factors (MRFs) MyoD and/or Myf5. Myoblasts subsequently express the MRF Myogenin, commit to terminal differentiation, and fuse to reconstruct their host fibers or to generate new myofibers and repair the damaged tissue (Tedesco et al., 2010).
The renewal of the SC population is crucial for sustained muscle regeneration during the life span of the individual. Transplantation experiments of either intact myofibers with their associated SCs (Collins et al., 2005) or single SCs (Sacco et al., 2008) demonstrated that a subpopulation of SCs are capable of both extensive contribution to muscle regeneration and self-renewal by giving rise to new SCs within the transplanted host muscle. In vivo, SCs undergo self-renewal to replenish the stem cell pool around the newly formed myofibers during regeneration of the tissue. Strikingly, the restored SC population is equivalent to the initial one (Le Grand et al., 2009; Shea et al., 2010). However, the mechanisms by which a subset of SCs or their progeny bypass cues to differentiate and instead return to quiescence to replenish the quiescent adult muscle stem cell pool remain poorly understood.
In multiple stem cell systems, the niche environment is responsible for the induction or inhibition of stem cell differentiation, based on the size and the composition of the niche. During regeneration, Wnt7a signaling through the planar cell polarity (PCP) pathway regulates the homeostatic level of satellite stem cells within the tissue. Wnt7a stimulation allows dividing SCs to maintain contact with the basal lamina and thus preserve their orientation relative to the niche (Le Grand et al., 2009). In parallel, recent data showed that cells located in the SC neighborhood (smooth muscle cells, fibroblasts) regulate self-renewal by controlling the return to quiescence of a subset of SCs. The paracrine effect of neighboring cells is mediated by Angiopoietin-1 (Ang1)/Tie2 signaling, through the extracellular signal-regulated kinases 1/2 (ERK1/2) pathway (Abou-Khalil et al., 2009). Still, the molecular pathways coordinated by signals from the niche to the SCs remain largely undefined.
During embryonic myogenesis in the body, a stem/progenitor population that expresses Pax3 and Pax7 arises from the somatic paraxial mesoderm and is maintained throughout embryogenesis within the developing skeletal muscles. Pax3/7+ progenitors continuously generate MRF-expressing myoblasts that will fuse to generate myofibers. Late in fetal development, the resident stem/progenitor population generates cells in a satellite position around myofibers (Gros et al., 2005; Kassar-Duchossoy et al., 2005; Relaix et al., 2005). Interestingly, our previous work demonstrated that the homeodomain transcription factors Six1 and Six4 are major regulators of embryonic myogenesis (Laclef et al., 2003; Grifone et al., 2005). Six1/4 sequentially control every step of embryonic myogenesis, and Six1 has been shown to lie genetically upstream of Pax3, Myf5, and Myogenin during limb myogenesis (Spitz et al., 1998; Giordani et al., 2007; Grifone et al., 2007). We thus hypothesized that Six1 might have an important role in adult regenerative myogenesis.
In this study, we analyzed the role of Six1 in SC physiology. We found that Six1 is expressed in all adult SCs, and that conditional Six1 gene disruption within the adult Pax7 lineage does not perturb SC quiescence. In contrast, Six1 is required for SC myogenic commitment ex vivo, and for proper skeletal muscle regeneration in vivo, via the control of MyoD and Myogenin but not Myf5 expression. Moreover, we show that Six1 is a critical regulator of SC self-renewal, in part via the regulation of Dusp6-ERK1/2 signaling. These data define a novel role for Six1 in governing muscle stem cell niche homeostasis in vivo.
Results
Six1 is expressed by SCs, but is not required for quiescence or activation
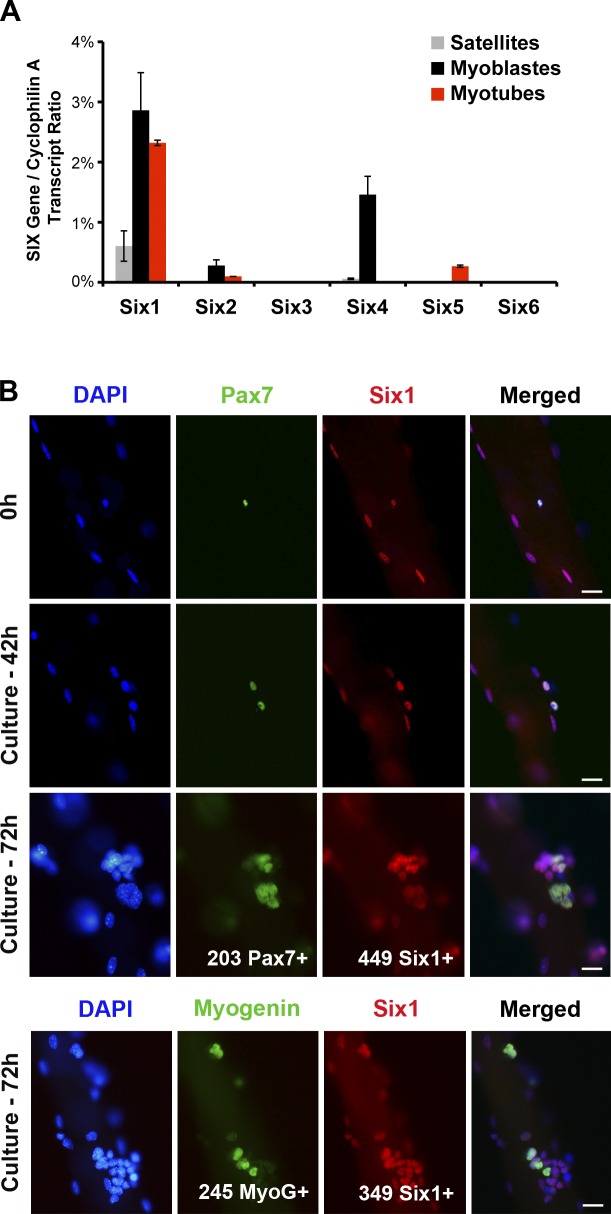
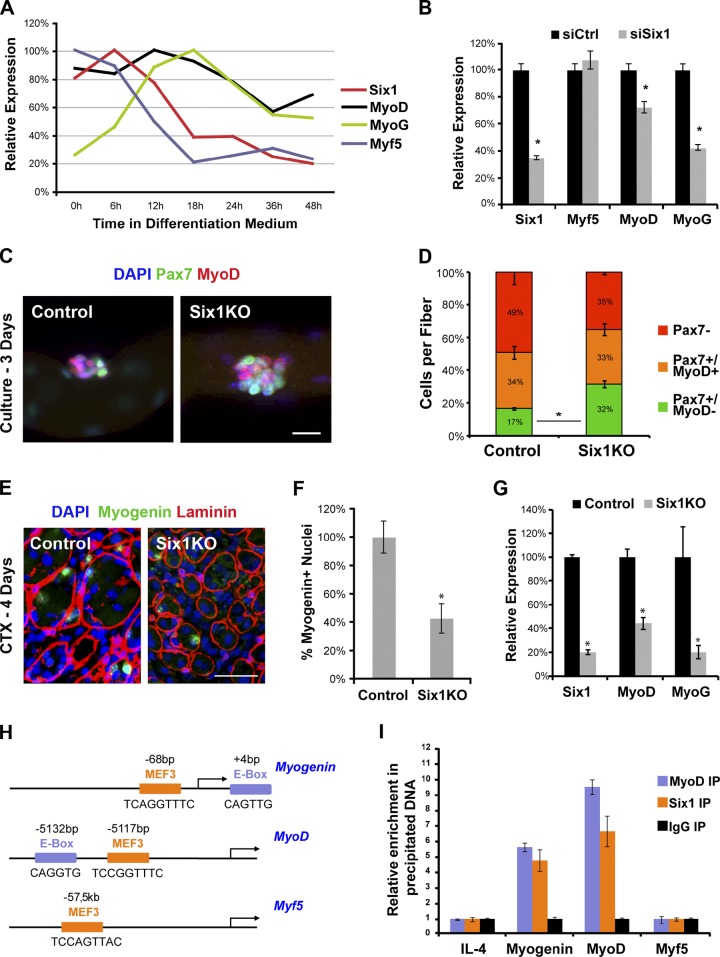
Recent studies highlighted that SIX homeoproteins are expressed by SCs isolated from adult skeletal muscle (Pallafacchina et al., 2010; Yajima et al., 2010). We performed quantitative RT-PCR (qRT-PCR) analysis for SIX family transcripts expression by freshly sorted SCs (Satellites) and by the same cells cultured ex vivo in growth conditions for 3 d (Myoblasts) or differentiated in multinucleated cells for 4 d (Myotubes). We found that Six1 is the main SIX gene to be expressed by adult myogenic progenitors at all stages analyzed, with myoblasts expressing relatively higher levels of Six1 transcripts compared with myotubes and SCs. Low Six4 expression was detected in quiescent SCs and myoblasts compared with Six1. Marginal relative amounts of Six2 and Six5 transcripts were also detected in proliferating and differentiated cells, respectively. Six3 and Six6 transcripts were not detected above the cut-off threshold of 30 cycles of amplification (Fig. 1 A).
Figure 1.
Satellite cells express Six1. (A) qRT-PCR analysis indicated expression of SIX family transcripts by freshly FACS-sorted SCs (Satellites), myogenic cells cultured in growth medium (Myoblasts), or induced to differentiate by serum removal for 3 d (Myotubes). Error bars indicate standard deviations. (B) Single myofibers isolated from EDL muscles of C57BL/6 mice. Myofibers were cultured in floating conditions and immunolocalized for Six1 and Pax7 or Myogenin proteins at different times after isolation. All quiescent, dividing, or differentiating SCs expressed Six1. Bars, 10 µm.
Immunocytochemical analysis of myofibers isolated from extensor digitorum longus (EDL), soleus, and plantaris muscles demonstrated that all quiescent SCs expressed Six1 proteins (n = 6 mice, >200 cells/mouse), independently of muscle fiber type composition (Fig. 1 B and not depicted). We then observed that all doublets of dividing SCs on cultured EDL myofibers were positive for Six1 expression when scored 42 h after isolation. Similarly, differentiating (Myogenin+) and proliferating (Pax7+) SC descendants on cultured myofibers all expressed Six1 proteins after 3 d ex vivo (n = 3 mice, >200 cells/mouse; Fig. 1 B).
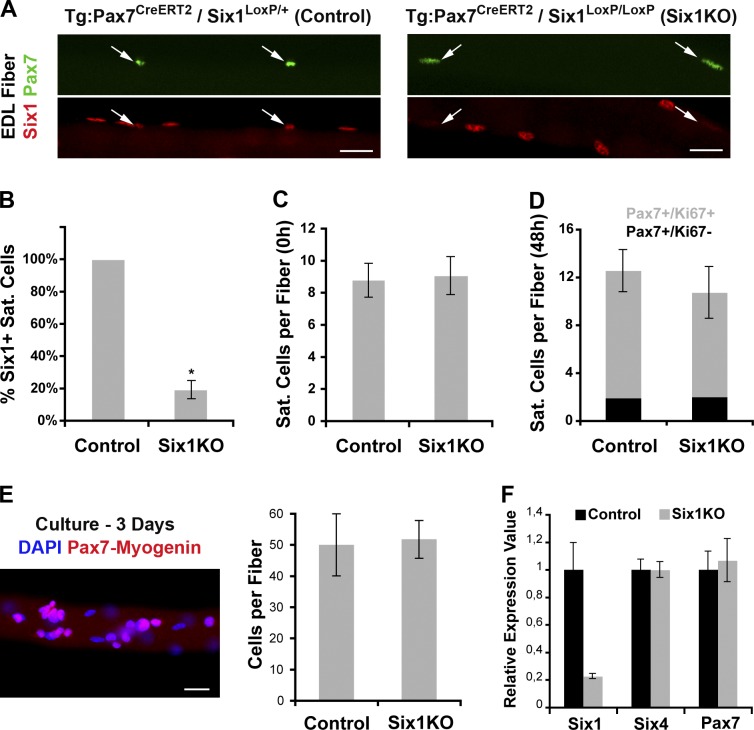
To investigate the role of Six1 in adult SC biology, we generated a conditional Six1flox allele, which after Cre-mediated recombination becomes the null allele Six1Δ (Fig. S1). To inactivate Six1 specifically in the adult SCs, we used transgenic Tg:Pax7-CreERT2 mice, which express a tamoxifen (TM)-inducible Cre recombinase-estrogen receptor fusion protein in cells that express Pax7 (Mourikis et al., 2012). We first assayed for inducible Cre activity by crossing Tg:Pax7-CreERT2 mice to Rosa26 reporter (R26R) mice. Efficacy and specificity of TM-dependent CreERT2 activity was validated by 89% co-immunolocalization of β-galactosidase with Pax7 or Syndecan4 on SCs 1 wk after TM administration (n = 3 mice, >100 cells/mouse; unpublished data). We then produced Tg:Pax7-CreERT2::Six1flox/flox mice (termed Six1KO mice) to permanently disrupt Six1 function in adult Pax7+ SCs upon the administration of TM. Tg:Pax7-CreERT2::Six1wt/wt mice treated with TM were used as controls. The loss of Six1 protein expression by SCs on EDL myofibers (Fig. 2 A) 1 wk after TM administration demonstrated that a high degree of recombination was obtained in Six1KO SCs (82% Pax7+/Six1− cells; n = 4 mice, >100 cells/mouse, P = 0.001; Fig. 2 B), whereas a moderate number of SCs have escaped recombination and remained Six1+. Importantly, Six1KO and control mice did not show any significant differences in terms of muscle tissue weights or histology up to 1 yr after TM injection (unpublished data).
Figure 2.
Six1 gene disruption does not influence SC quiescence, activation, or proliferation. (A) Single myofibers isolated from EDL muscles of control (Tg:Pax7-CreERT2::Six1flox/+) and Six1KO (Tg:Pax7CreERT2/Six1flox/flox) mice 1 wk after TM injection. Six1 protein expression is lost in Six1KO SCs (arrows). (B) The majority of SCs from Six1KO EDL and TA muscles are negative for Six1 expression. (C) Quantification of quiescent sublaminar Pax7+ SCs per EDL myofibers isolated from control and Six1KO mice 6 wk after TM injection. Six1 loss does not perturb SC quiescence in vivo. (D) EDL myofibers from control and Six1KO animals were cultured for 2 d to visualize SC activation (Pax7+/Ki67+). Six1 loss does not perturb SC activation ex vivo. (E) EDL myofibers from control and Six1KO animals were cultured for 3 d. SC descendants were immunolocalized for both Pax7 and Myogenin proteins. Six1 loss does not perturb SC proliferation ex vivo. (F) Primary myoblasts were isolated from control and Six1KO limb muscles. qRT-PCR analysis indicated expression of Six1, Pax7, and Six4 transcripts. Six1 gene disruption does not have an impact on Pax7 and Six4 expression levels. Error bars indicate standard deviations. *, P < 0.001. Bars, 10 µm.
To determine if Six1 has a role in SC quiescence, we injected 2-mo-old mice with TM and analyzed the SC compartment after 6 wk. Conditional Six1 disruption in adult Pax7+ cells did not lead to any loss of SCs associated with EDL myofibers (n = 3 mice, >200 cells/mouse; Fig. 2 C), which indicates that Six1 is not required for their maintenance. Furthermore, conditional mutant cells maintained SC characteristics as shown by CD34 and α7-Integrin expressions in Pax7+/Six1− SCs (Fig. S2 A). We then cultured single EDL myofibers ex vivo to determine if Six1 disruption has an impact on SC activation and proliferation after exit from quiescence. The numbers of activated (Pax7+/Ki67+) cells after 48 h of culture (n = 3 mice, >150 cells/mouse; Fig. 2 D), and the numbers of myogenic cells (Pax7+ or Myogenin+) after 72 h of culture (n = 3 mice, >400 cells/mouse; Fig. 2 E) were not significantly different between Six1KO and control myofibers. We then derived primary myoblasts from mice leg muscles, maintained them in growth conditions, and observed that both control and Six1KO cells express the myoblasts marker Desmin and similar levels of Six4 transcription factor ex vivo (Fig. S2 B). qRT-PCR analysis demonstrated an efficient disruption of Six1 at the transcription level in Six1KO primary myoblasts, and no compensatory increase in Pax7 and Six4 expression levels (Fig. 2 F).
Collectively, these results demonstrate that Six1 is expressed in quiescent SCs as well as in proliferating and differentiating myogenic cells. Conditional Six1 gene disruption in Pax7-expressing SCs is efficient, and does not induce changes in SC quiescence, activation, and proliferation dynamics.
Six1 is necessary for SCs regenerative potential
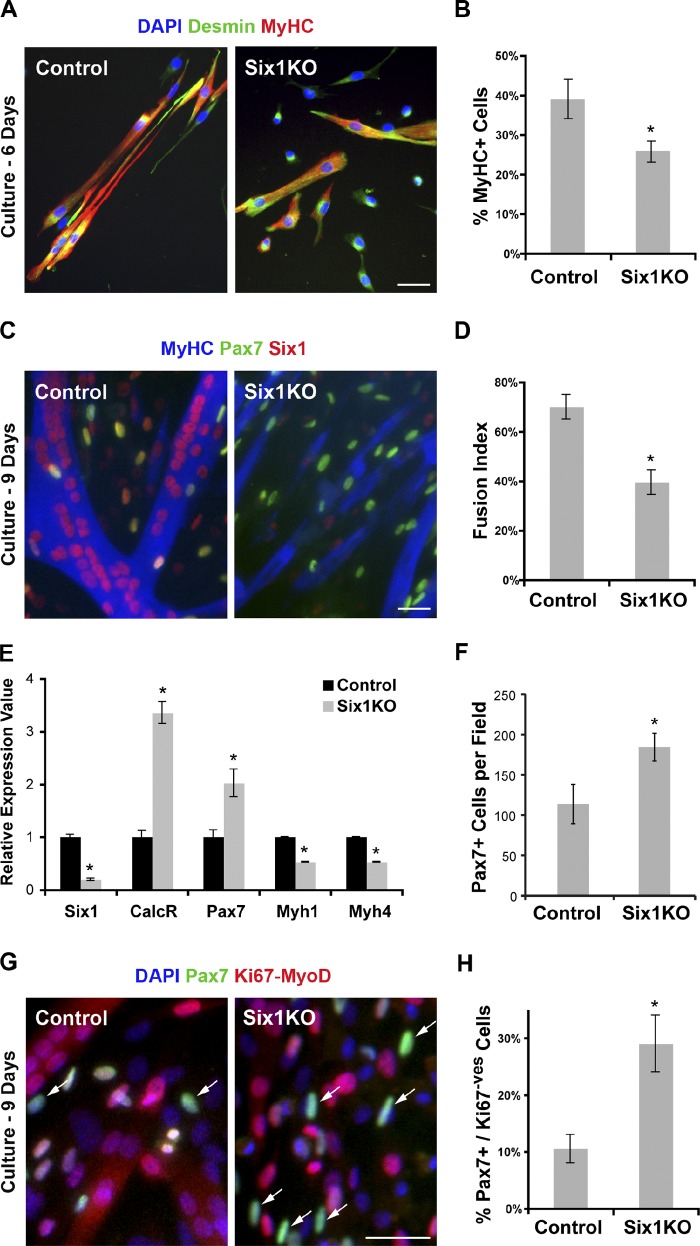
To investigate the role of Six1 in adult SC functions, we plated single myofibers on Matrigel ex vivo to allow SCs to proliferate and differentiate. After 6 d of culture, SC descendants from both control and Six1KO mice fused to form differentiated myotubes (Fig. 3 A), but Six1KO cells committed less efficiently to differentiation compared with control cells (n = 3 cultures, >900 cells scored per culture, P = 0.01; Fig. 3 B). After 9 d ex vivo (Fig. 3 C), control myogenic cells fused extensively and formed long multinucleated myotubes, whereas Six1KO myogenic cells formed smaller myotubes containing fewer nuclei (n = 3 cultures, >1,000 nuclei scored per culture, P = 0.02; Fig. 3 D). qRT-PCR analysis demonstrated that cells in Six1KO cultures expressed higher levels of the SC markers Pax7 and Calcitonin Receptor, and lower levels of the differentiation markers Myosin Heavy Chain 1 and 4 (Fig. 3 E). In parallel, more cells remained Pax7+ and mononuclear in Six1KO cultures compared with control cultures (Fig. 3 F). Examination of the Pax7+/MyoD−/Ki67− “reserve” cell population (Fig. 3 G) showed that Six1KO cell cultures contained a high proportion of cells that escaped the differentiation program (n = 3 cultures, >300 cells scored per culture, P = 0.01; Fig. 3 H).
Figure 3.
Six1 gene disruption perturbs myogenic differentiation of SC descendants ex vivo. 1 wk after TM treatment, EDL myofibers from control and Six1KO mice were plated on Matrigel, and cultures were analyzed after 6 and 9 d of culture ex vivo. (A) Myogenic cells grown for 6 d were immunolocalized for Desmin (myoblast marker) and MyHC (differentiation marker) proteins. (B) Six1KO cells exhibit limited differentiation potential ex vivo compared with control cells. (C) Myogenic cells grown for 9 d were immunolocalized for Six1, Pax7 (undifferentiated state marker), and MyHC (differentiated state marker) proteins. (D) Six1KO cells fuse less efficiently and form smaller myotubes compared with control cells. (E) qRT-PCR analysis indicated expression of Six1, CalcR, Pax7 (SC markers), and Myh1, Myh4 (differentiation markers) transcripts by differentiated myogenic cells. (F) Six1KO cell cultures generate more Pax7+ cells compared with control cells. (G) SC-derived myogenic cells grown for 9 d were immunolocalized for Pax7 and both MyoD and Ki67 proteins. (H) Six1KO cells generate more “reserve” cells (Pax7+/MyoD−/Ki67−; arrows) compared with control cells. Error bars indicate standard deviations. *, P < 0.02. Bars, 10 µm.
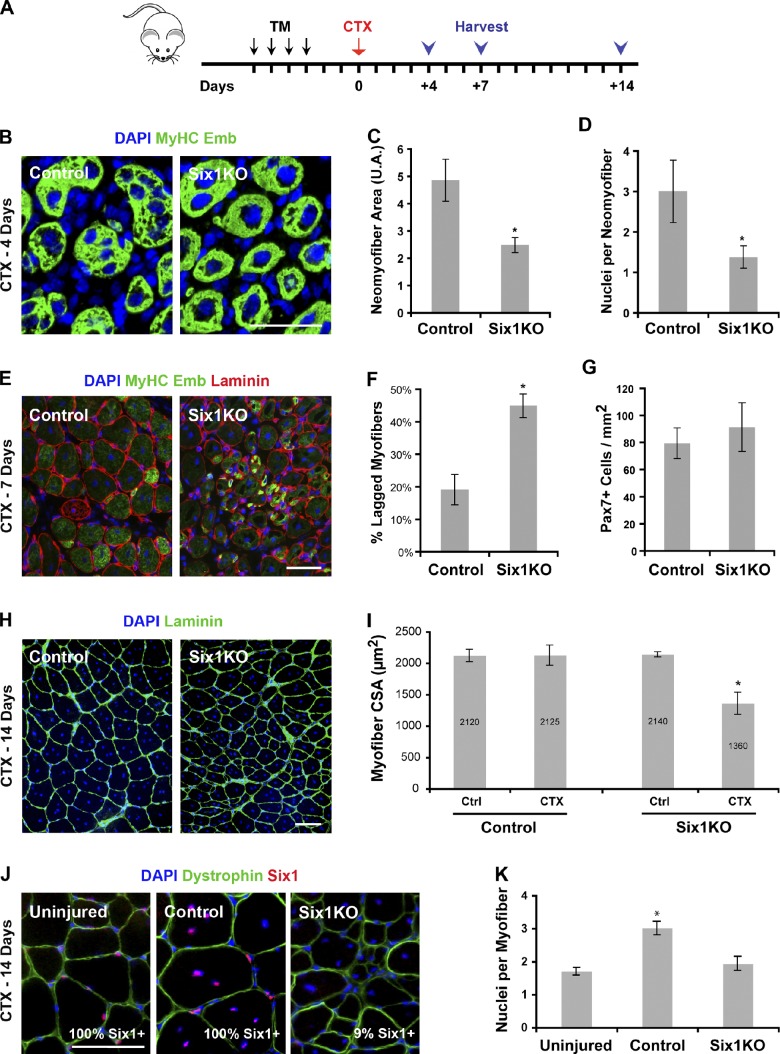
To confirm a role for Six1 in controlling SC myogenic potential, we used in vivo regeneration assays. Tibialis anterior (TA) muscles of TM-treated control and Six1KO animals were subjected to a single cardiotoxin (CTX) injury and then allowed to recover for 4–14 d before analysis of the regenerated tissue (Fig. 4 A). During the acute phase of regeneration (4 d after injury), SC descendants fuse to form new myofibers expressing embryonic (emb) myosin heavy chain (MyHC emb; Fig. 4 B). Six1KO regenerating muscles were composed of smaller newly formed myofibers (Fig. 4 C), containing fewer myonuclei compared with control muscles (n = 3 animals, >300 fibers scored per sample, P < 0.001; Fig. 4 D). At 7 d after injury, the muscle tissue is composed of regenerated myofibers that have down-regulated MyHC emb expression (Fig. 4 E). In contrast, Six1KO muscles contained numerous “lagged” fibers of small caliber expressing embryonic MyHC (n = 4 animals, >500 fibers scored per sample, P = 0.0001; Fig. 4 F). The lack of differentiated cells in Six1KO muscle does not appear to arise from a defect in SC proliferation because the number of Pax7+ cells is equivalent between control and Six1KO animals at this time point (Fig. 4 G).
Figure 4.
Six1 expression by SCs is necessary for proper skeletal muscle regeneration. (A) 3 d after TM treatment, TA muscles of control and Six1KO mice were injured by a single CTX injection and analyzed at various times during the regeneration process. (B) Cryosections of 4-d regenerating TA muscles. Immunolocalization of MyHC emb proteins marks the newly formed myofibers. (C) Regenerating myofibers of Six1KO animals are smaller compared with controls. (D) Regenerating myofibers of Six1KO animals contain fewer nuclei compared with controls. (E) Cryosections of 7-d regenerating TA muscles. Laminin staining shows basal lamina of myofibers. Strong MyHC emb staining marks a population of small, delayed myofibers. (F) 7-d regenerating muscles of Six1KO animals exhibit a significant proportion of lagged myofibers compared with controls. (G) 7-d regenerating control and Six1KO muscles do not contain significantly different amounts of Pax7+ cells. (H) Cryosections of regenerated TA muscles 14 d after CTX injection. Laminin staining shows basal lamina of myofibers. Note the abnormal accumulation of matrix in Six1KO muscles. (I) Quantification of muscle fiber caliber in 14-d regenerated TA muscles. Regenerated Six1KO muscles contain smaller fibers compared with regenerated control muscles. (J) Cryosections of regenerated TA muscles 14 d after CTX injection. Dystrophin staining shows myofibers sarcolemma. Shown is the percentage of Six1+ myonuclei. (K) Regeneration of the muscle tissue results in a higher number of nuclei per myofiber on cross sections in controls but not in Six1KO animals. Error bars indicate standard deviations. *, P < 0. 01. Bars, 50 µm.
2 wk after injury, the TA is composed of neomyofibers with centrally located nuclei (Fig. 4 H). Although the size of the regenerated myofibers is similar to the undamaged myofibers in control muscles, regenerated myofibers cross-sectional area (CSA) was 37% smaller compared with undamaged myofibers in Six1KO muscles (n = 4 animals, >800 fibers scored per sample, P = 0.02; Fig. 4 I). Of note, only 9% of the myonuclei within neomyofibers in Six1KO regenerated muscles expressed Six1, demonstrating that they were mostly formed by the fusion of Six1-null SC descendants (Fig. 4 J). Moreover, the reduction in cell size in regenerated Six1KO muscles was accompanied by a striking reduction in the number of nuclei per neomyofiber (n = 4 animals, >250 fibers scored per sample, P = 0.001; Fig. 4 H).
We further noticed that Six1KO regenerated muscles contained numerous necrotic myofibers (n = 4 animals, P = 0.0005; Fig. S3 A) and displayed aberrant fibrotic tissue formation between the regenerated myofibers of Six1KO muscle (n = 4 animals, P = 0.04; Fig. S3 B). Collectively, these data demonstrate that Six1 expression by SCs is necessary for proper repair of damaged skeletal muscle tissue.
Six1 directly controls MyoD and Myogenin expression by SCs
Previous work established that in early Six1−/− embryos, activation of the MRFs is reduced and delayed in limb buds (Laclef et al., 2003; Giordani et al., 2007). Therefore, we decided to test if Six1 might control MRF genes expressions by adult SCs.
First, we differentiated SC-derived primary myoblasts in low mitogen medium, and extracted RNA every 6 h for the first 2 d of culture after serum removal. qRT-PCR analysis of the expression of Six1, MyoD, Myogenin, and Myf5 showed that the first three genes peaked in transcription during differentiation, before being reduced after 48 h. Notably, Six1 expression was the first to be increased (n = 3 independent cultures; Fig. 5 A). We then reduced Six1 expression level by siRNA transfection 12 h before differentiation, and extracted RNA 12 h after serum removal. We observed that Six1 silencing resulted in a reduction in MyoD and Myogenin, but not Myf5, expression by differentiating myogenic cells (n = 3, P < 0.05; Fig. 5 B).
Figure 5.
Six1 activates MyoD and Myogenin expression by SCs in vivo. (A) qRT-PCR analysis of differentiating myogenic cells shows transient up-regulation of Six1, MyoD, and Myogenin expressions during the first day after serum removal. (B) qRT-PCR analysis of differentiating myogenic cells shows that Six1 silencing decreases MyoD and Myogenin expression but not Myf5 expression ex vivo. (C) EDL single myofibers were cultured for 3 d, and immunolocalized for Pax7 and MyoD protein expression. Representative myogenic cell clusters are shown. (D) Percentage of SC descendants at the surface of cultured myofibers. Loss of Six1 decreases the proportion of committed cells (Pax7−/MyoD+) and increases the proportion of undifferentiated (Pax7+/MyoD−) cells in clusters. (E) Cryosections of 4-d regenerating TA muscles. Laminin staining shows basal lamina of myofibers. Immunolocalization of Myogenin proteins marks differentiating myonuclei. (F) Six1 gene disruption in SCs results in decreased Myogenin+ nuclei numbers during muscle regeneration. (G) qRT-PCR analysis of Six1, MyoD, and Myogenin transcripts levels by 4-d regenerating TA muscles. Six1 gene disruption decreases MyoD and Myogenin expression in vivo. (H) Schematic representations of Myogenin, MyoD, and Myf5 regulatory regions region. Shown are the localization and sequences of E-box (bHLH binding) and MEF3 (SIX binding) sites. (I) qRT-PCR analysis of locus enrichment in ChIP assays from differentiating myogenic cells. MyoD and Six1 proteins are bound to the MyoD and Myogenin upstream regulatory elements, but not on Myf5 enhancer. Error bars indicate standard deviations. *, P < 0.04. Bars: (C) 20 µm; (E) 50 µm.
We then observed that SCs clusters on 3-d cultured Six1KO myofibers (Fig. 5 C) contained a higher proportion of undifferentiated Pax7+/MyoD− cells, and a reduced proportion of differentiated Pax7−/MyoD+ cells compared with control myofibers (n = 3 animals, >250 cells scored per sample, P = 0.04; Fig. 5 D). We then observed that 4-d regenerating Six1KO muscles (Fig. 5 E) contained fewer Myogenin+ nuclei (n = 4 animals, >200 cells scored per sample, P = 0.02; Fig. 5 F) and expressed lower amounts of MyoD and Myogenin transcripts (n = 4 mice; Fig. 5 G) compared with control muscles.
To test if MRF genes are direct targets of Six1 in adult myogenic cells, we analyzed the binding of Six1 proteins to the MEF3 site located within the Myogenin proximal promoter (Spitz et al., 1998), to a novel MEF3 site located within the MyoD distal regulatory region (DRR; required for MyoD expression during muscle regeneration; Tapscott et al., 1992), and to the MEF3 site located within Myf5 limb enhancer region (Fig. 5 H; Giordani et al., 2007). Chromatin immunoprecipitation (ChIP) assays with differentiating myogenic cells demonstrated that Six1 proteins were bound to the Myogenin promoter and to the MyoD DRR, but not to the Myf5 enhancer during myogenic differentiation of SCs (relative to a mock immunoprecipitation and to the control locus IL4 intron). Because MEF3 sites are located in close proximity to E-boxes, we designed PCR primers to encompass both MEF3 and E-box sites for both loci. We then found that MyoD proteins are also bound to the Myogenin promoter and to the MyoD DRR in myogenic cells (n = 2 independent experiments; Fig. 5 I). Collectively, our results indicate that Six1 regulates the entry into the differentiation program of SC descendants during adult regenerative myogenesis via direct control of MyoD and Myogenin expression.
Six1 limits SC self-renewal
During skeletal muscle regeneration, the SC population is maintained by self-renewal. In vivo, self-renewal occurs between 2 and 4 wk after injury; at the end of this period the majority of renewed SCs have returned to quiescence (Abou-Khalil et al., 2009; Le Grand et al., 2009; Shea et al., 2010).
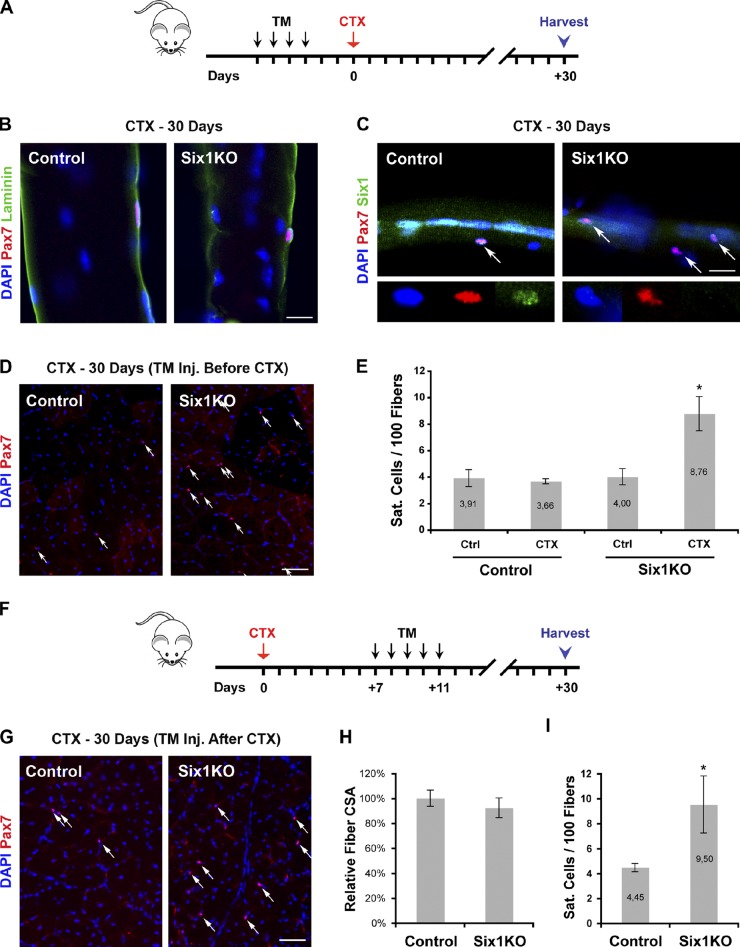
To investigate a possible role for Six1 in SC self-renewal, we analyzed regenerated skeletal muscle tissues from control and Six1KO animals and sampled them 1 mo after CTX injury (Fig. 6 A). We first extracted single myofibers from regenerated EDL muscles, and observed that SCs had relocated under the basal lamina both in control and Six1KO animals (Fig. 6 B). We then validated the finding that 97% of Pax7+ cells within control and Six1KO regenerated muscles were negative for the proliferation marker Ki67 (n = 2 animals, >300 cells scored per sample; Fig. S4 A). However, examination of SCs on single myofibers revealed that although self-renewed SCs in control muscles still expressed Six1 (100% Pax7+/Six1+) and are present in a number similar to the undamaged contralateral muscle, self-renewed SCs in Six1KO muscles did not express Six1 (95% Pax7+/Six1−; Figs. 6 C and S4 B) and were 2.2-fold more frequent compared with the undamaged contralateral muscle (n = 4 animals, >280 cells scored per sample, P = 0.02; Figs. 6 C and S4 C).
Figure 6.
Six1 limits SC self-renewal in vivo. (A) 3 d after TM treatment, TA muscles of control and Six1KO mice were injured by a single CTX injection and analyzed 30 d after the injury. (B) Single myofibers isolated from 30-d regenerated EDL muscles of control and Six1KO animals. Renewed Pax7+ SCs are located in sublaminar position around host myofibers in both control and Six1KO muscles. (C) Six1 is expressed by centrally located myonuclei and renewed SCs in control myofibers but not in Six1KO myofibers. Six1KO myofibers contain a higher number of renewed SCs (arrows). (D) Cryosections of 30-d regenerated TA muscles. Immunolocalization of Pax7 proteins mark quiescent SCs (arrows). (E) The SC pool is increased 2.4-fold in regenerated Six1KO TA muscles. (F) TA muscles of non-TM treated control and Six1KO mice were injured by a single CTX injection. Mice were then subjected to TM administration between 7 and 11 d after injury. Muscles were analyzed 30 d after the injury. (G) Cryosections of 30-d regenerated TA muscles. Immunolocalization of Pax7 proteins mark quiescent SCs (arrows). (H) Although the size of regenerated myofibers of control and Six1KO animals are similar, the SC pool is increased 2.1-fold in regenerated Six1KO TA muscles when TM was administrated after myogenesis has occurred. Error bars indicate standard deviations. *, P < 0.02. Bars: (B) 20 µm; (C) 10 µm; (D and G) 50 µm.
To validate the finding that disruption of Six1 in SCs lead to an increase in muscle SC niche occupancy after regeneration, we scored the number of quiescent sublaminar Pax7+ SCs on cryosections of 30-d regenerated TA muscles (Fig. 6 D). Loss of Six1 resulted in a 2.4-fold increase in SC niche occupancy in Six1KO regenerated muscles compared with control regenerated muscles or Six1KO contralateral undamaged muscles (n = 4 animals, >200 cells scored per sample, P = 0.0006; Fig. 6 E). Interestingly, a second round of regeneration increased the pool of resident SCs up to 2.8-fold in Six1KO muscle (Fig. S4 D).
We then decided to score the number of Pax7+ cells on TA cryosections during the course of muscle repair. We observed that although the number of Pax7+ cells was not significantly different between Six1KO and control muscles during the early stages of the repair process, Six1KO muscles had already accumulated Pax7+ cells at 14 d after CTX injury (Fig. S4 E). To test the implication of Six1 on restoration of the muscle SC pool in a timely fashion, we injured animals that were not subjected beforehand to TM administration, and subsequently injected TM between 7 and 11 d after injury (Fig. 6 F). We observed that “late” TM injection allowed for Six1KO muscles to regenerate similarly to controls (no significant differences in neomyofiber CSA; Fig. 6 H), but also resulted in a 2.1-fold increase in SC niche occupancy in SixKO muscles compared with control muscles (n = 3 animals, >250 cells scored per sample, P = 0.02; Fig. 6, G and I).
Collectively, our results indicate that Six1 is intrinsically required for proper homeostasis of the SC pool during skeletal muscle regeneration, and that this is independent of the myofiber repair process.
Six1 does not regulate polarity of SC divisions
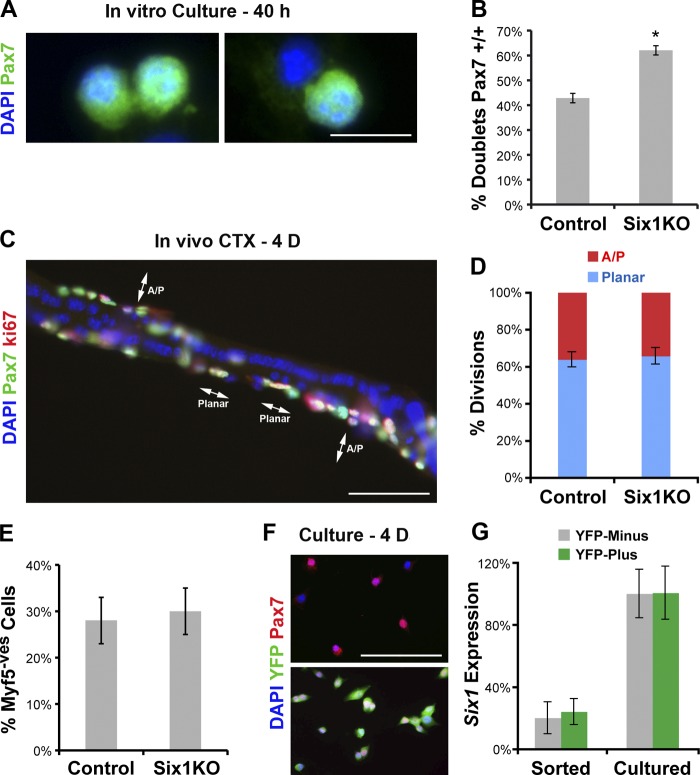
To test if self-renewal is increased in Six1KO SCs, we plated FACS-sorted SCs and analyzed the frequency of Pax7-positive doublets ex vivo (Fig. 7 A). We observed that Six1 gene disruption increased SC self-renewal (Fig. 7 B). To visualize the plane of SC divisions in vivo, we isolated myofibers from the adjacent EDL muscle 4 d after CTX injection into the TA muscle (Fig. 7 C). Examination of doublets of sister SCs beneath the basal lamina of regenerating myofibers did not reveal any differences in the frequency of planar orientations between control and Six1KO myofibers (n = 2 animals, >200 doublets scored per sample; Fig. 7 C). Similarly, symmetric expansion of SCs can be monitored in vivo by counting the number of Pax7+/Myf5− cells (Le Grand et al., 2009). Again, we did not observe any differences in the number of SCs that do not express the Myf5 protein on control and Six1KO myofibers (n = 3 animals, >80 cells scored per sample; Fig. 7 E). Lastly, we prepared cDNAs from SCs isolated by FACS and separated on the basis of Myf5Cre conditional YFP fluorescence (Fig. 7 F). Both quiescent and ex vivo cultured YFP+ and YFP− SCs expressed similar amounts of Six1 transcripts (Fig. 7 G). Together these results indicate that Six1 does not control SC division polarity in vivo, and that increased SC self-renewal observed in Six1KO SCs is not related to an increase in the expansion of Pax7+/Myf5− stem cells.
Figure 7.
Six1 gene disruption increases SCs self-renewal, but does not perturb the orientation of SC divisions. (A) FACS-sorted SCs were plated ex vivo and fixed after the first division. Typical doublets of sister SCs with Pax7+/+ or Pax7+/− gene signature are shown. (B) Six1KO SCs have higher self-renewal potential. (C) Immunolocalization of Pax7 and Ki67 proteins on myofibers separated from 4-d regenerating EDL muscles. (D) Quantification of SC division orientation. Six1 gene disruption does not have an impact on the rate of planar-to-perpendicular divisions. (E) EDL myofibers were separated from 4-d regenerating EDL muscles, and immunolocalized for Pax7 and Myf5 protein expression. Six1 gene disruption does not have an impact on the Myf5-negative satellite stem cell population. (F) FACS-sorted SCs separated on the basis of Myf5-Cre–driven reporter fluorescence and plated ex vivo. Immunolocalization of Pax7 and YFP proteins on YFP− (stem) and YFP+ (committed) myoblasts. (G) qRT-PCR analysis indicated expression of Six1 transcripts by YFP+ and YFP− SCs and myoblasts. Error bars indicate standard deviations. *, P < 0.01. Bars: (A) 10 µm; (C and F) 50 µm.
Six1 dampens ERK signaling via Dusp6
During skeletal muscle regeneration, Ang1/Tie2 signaling, acting through the ERK1/2 pathway, regulates SC return to quiescence (Abou-Khalil et al., 2009). Analysis of previous microarray experiments (Richard et al., 2011) revealed that the expression of the Dual-specificity Phosphatase 6 (Dusp6), a physiological restrainer of ERK1/2 signaling (Maillet et al., 2008), is reduced in embryonic and fetal muscles that lack Six1/4.
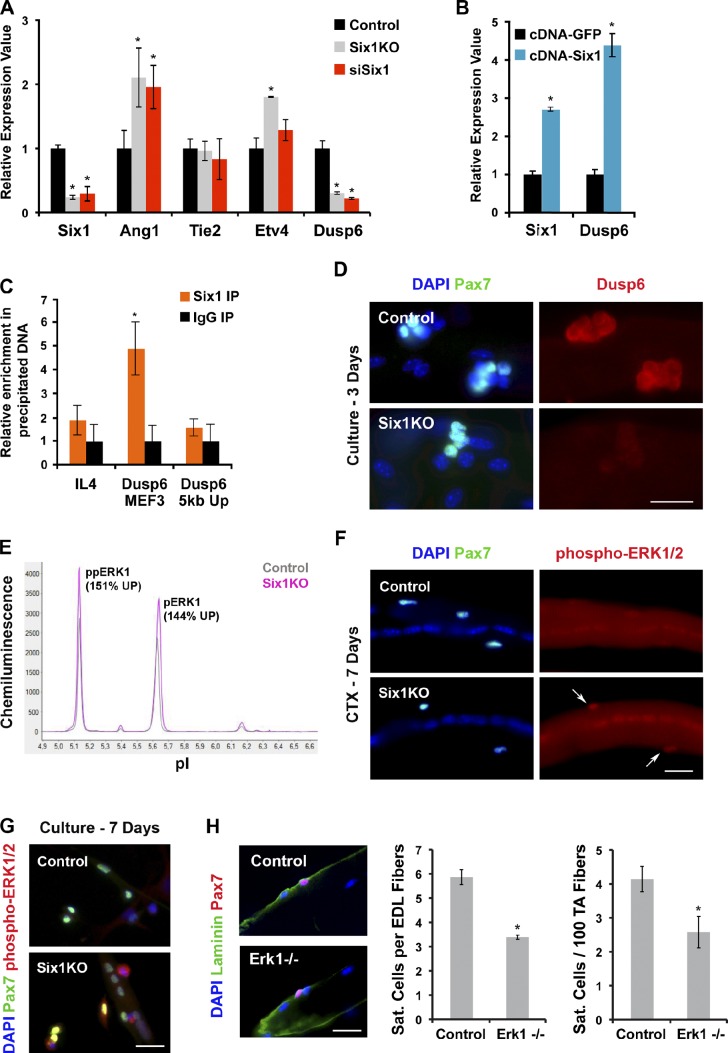
Therefore, to evaluate Ang1/Tie2/ERK1/2 signaling in Six1KO SCs, we analyzed the relative expression levels of Ang1, Tie2, and Dusp6 and the downstream transcription factor Etv4 in SC-derived myoblasts by qRT-PCR. Interestingly, Ang1 and Etv4 transcripts levels were elevated, whereas Dusp6 expression was decreased in myoblasts with reduced Six1 activity compared with control myoblasts (n = 3, P < 0.05; Fig. 8 A). We then overexpressed Six1 by transfection of a CMV-Six1 plasmid in primary myoblasts, and found that a 2.8-fold increase in Six1 expression induced a 4.4-fold increase in Dusp6 expression compared with empty-vector transfected cells (n = 3, P < 0.05; Fig. 8 B). ChIP assays further demonstrated that Six1 proteins were bound to a MEF3 site located 2 kb upstream of the Dusp6 gene (relative to a mock immunoprecipitation and to the control loci) in proliferating myogenic cells (Fig. 8 C). We did not detect Dusp6 proteins in quiescent SCs nor in skeletal myofibers, but we did observe that Dusp6 protein is strongly expressed in all Pax7+ cells on control myofibers cultured for 3 d but not on Six1KO myofibers (n = 3 mice, >100 cells/mouse; Fig. 8 D). Collectively, these results indicate that Six1 directly controls Dusp6 expression in SC, and suggest that ERK1/2 signaling might be elevated in Six1KO cells.
Figure 8.
Six1 negatively regulates ERK signaling in SCs. (A) qRT-PCR analysis of Six1, Ang1, Tie2, Etv4, and Dusp6 expression in proliferating myoblasts. Six1 gene disruption or silencing increases Ang1 and Etv4 transcription levels and decreases Dusp6 expression. (B) qRT-PCR analysis of Six1 and Dusp6 expression in proliferating myoblasts. Six1 overexpression increases Dusp6 transcription. (C) Real-time PCR analysis of locus enrichment in ChIP from proliferating myoblasts. Six1 proteins are bound to the MEF3 element upstream of the Dusp6 gene. (D) Immunolocalization of Pax7 and Dusp6 proteins on 3-d cultured EDL myofibers. Proliferating Six1KO SCs do not express Dusp6. (E) Detection of phosphorylated ERK from control (gray) and Six1KO (fushia) myoblasts. pI values are plotted against signal intensities. Different ERK isoforms and the relative increases in signal intensity of Six1KO over control are indicated (n = 3). Six1KO myoblasts present elevated ERK1 signaling ex vivo. (F) Immunolocalization of Pax7 and phosphorylated ERK1/2 proteins on 7-d regenerating EDL muscles. Six1KO SCs present elevated ERK signaling in vivo. (G) Immunolocalization of Pax7 and phosphorylated ERK1/2 proteins on 6-d cultured EDL myofibers. Six1KO SCs present elevated ERK signaling ex vivo. (H) EDL myofibers from control and Erk1−/− animals. Immunostaining indicated that quiescent SCs express Pax7+ and have a correct sublaminar position in mutant muscles. Pax7+ sublaminar SCs were scored on EDL single fibers (left) and on TA cryosections (right). The SC pool is diminished in muscles from Erk1−/− mice compared with controls. Error bars indicate standard deviations. *, P < 0.05. Bars, 10 µm.
To test if the deficiency in Dusp6 expression induced by Six1 gene disruption in SCs leads to elevated ERK1/2 signaling, we used the capillary-based NanoPro assay to analyze ERK1/2 phosphorylation states in primary myoblasts. We observed that ERK1 (but not ERK2) signaling is increased by 35% in Six1KO myoblasts compared with control cells (Fig. 8 E). We then isolated myofibers from the adjacent EDL muscle 7 d after CTX injection into the TA muscle and observed that Six1KO Pax7+ cells at the surface of regenerated myofibers had elevated levels of phospho-ERK1/2 compared with controls (Fig. 8 F). Likewise, we grew SCs from EDL myofibers on Matrigel, and observed strong levels of phopho-ERK1/2 only in Six1KO cells (Fig. 8 G).
To validate the impact of ERK1 signaling in SC niche occupancy in vivo, we investigated Erk1−/− mice. 2-mo-old Erk1−/− mice did not exhibit visible defects in skeletal muscle tissue. However, evaluation of SC niche occupancy by enumerating sublaminar Pax7+ cell populations on both TA cryosections and EDL myofibers showed that Erk1−/− muscles had 40% less quiescent SCs compared with control littermates (n = 4 mice, >200 cells scored, P = 0.001; Fig. 8 H). Collectively, our data suggest that Six1 controls Dusp6 expression and the duration of ERK1 signaling in SCs during the regeneration process.
Dusp6 controls SC return to quiescence
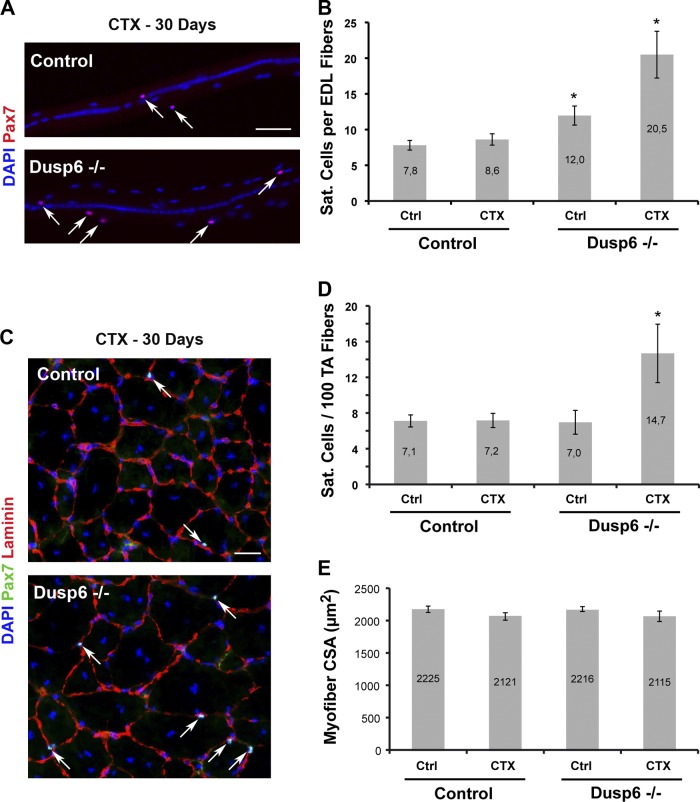
To assess the role of Dusp6 in SC self-renewal, we analyzed Dusp6−/− mice. 2-mo-old Dusp6−/− mice did not exhibit visible defects in skeletal muscle tissue. Evaluation of SC niche occupancy in Dusp6−/− muscles showed that Dusp6−/− EDL myofibers presented a moderate increase in SC content compared with control mice whereas TA muscles from Dusp6−/− and control mice contained the same number of quiescent sublaminar Pax7+ SCs (n = 4 mice, >400 cells scored; Fig. 9, B and D).
Figure 9.
Dusp6 is required for restoring the SC pool during regeneration. TA and EDL muscles of control and Dusp6−/− mice were injured by a single CTX injection and analyzed 30 d after the injury. (A) Single myofibers isolated from 30-d regenerated EDL muscles. Renewed Pax7+ SCs are located around host myofibers (arrows). (B) The SC pool is increased 2.4-fold on regenerated in Dusp6−/− myofibers compared with control myofibers. (C) Cryosections of 30-d regenerated TA muscles. Immunolocalization of Pax7 and Laminin proteins allows visualization of sublaminar renewed SCs (arrows). (D) The SC pool is increased twofold within regenerated Dusp6−/− muscles compared with control muscles. (E) Quantification of muscle fiber caliber in uninjured and regenerated TA muscles of Dusp6−/− and control animals. No muscle defects were observed in mutant mice. Error bars indicate standard deviations. *, P < 0.004. Bars, 50 µm.
We then analyzed regenerated skeletal muscle tissues from control and Dusp6−/− animals, sampled 1 mo after CTX injury. Strikingly, examination of SCs on EDL myofibers (Fig. 9 A) revealed that self-renewed SCs in Dusp6−/− muscles were 2.4-fold more frequent compared with control muscle (n = 4 animals, >350 cells scored per sample, P = 0.0004; Fig. 9 B). We then scored the number of quiescent sublaminar Pax7+ SCs on cryosections of regenerated TA muscles (Fig. 9 C) and observed a twofold increase in muscle stem cell niche occupancy in Dusp6−/− regenerated muscles compared with control regenerated muscles or Dusp6−/− contralateral undamaged muscles (n = 4 animals, >200 cells scored per sample, P = 0.005; Fig. 9 D).
Of note, Erk1−/− and Dusp6−/− mice are constitutive KO animals, meaning that during postnatal muscle growth, the SC dynamic has been challenged in these genetic backgrounds, before SC entry into quiescence in adulthood. The observations performed in these animals without injury reflects history of SCs, whereas experiments performed in Six1-cKO mice do not reflect SC history because these cells were “wild type” until conditional Six1 gene disruption at 8 wk of age. Moreover, we did not find any differences in myofiber CSA between contralateral and regenerated TA muscle of Dusp6−/− and control mice, which suggests that Dusp6 is dispensable for growth and regeneration of skeletal myofibers but has a unique function in regulating SC niche occupancy in vivo.
Discussion
Our data support a model in which Six1 controls the behavior of SCs during skeletal muscle regeneration (Fig. 10). Notably, Six1 regulates myogenic commitment and differentiation of SC-derived myoblasts through the timely induction of MyoD and Myogenin expressions. Six1 subsequently plays a critical role in regulating the size of the renewing SC population during regeneration, as compromised Six1 function results in increased SC numbers. Our data suggests that Six1 does not induce a specific amplification of the satellite stem cell population but acts as a rheostat for SC niche occupancy by dampening the level of ERK1 signaling through Dusp6.
Figure 10.
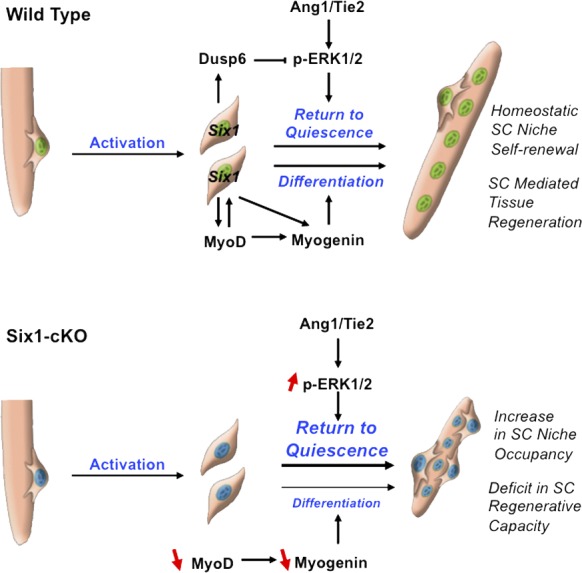
Proposed model for the role of Six1 in regulating muscle tissue repair and SC niche occupancy.
We demonstrate that Six1 function in muscle stem cells is necessary for proper myofiber repair after acute injury. Six1 disruption in SCs perturbs their myogenic potential, and results in reduced myogenic fusion and insufficient myonuclei accretion during the repair process. Unsuccessful repair after tissue damage leads to the derangement of normal structure, and an increased development of fibroblasts during the regeneration process was previously linked with defects in the myogenic process (Mann et al., 2011). Interestingly, ablation of SCs before muscle injury resulted in a complete loss of regenerated muscle, as well as misregulation of fibroblasts and a dramatic increase in connective tissue (Murphy et al., 2011). Our results confirm that impaired myofiber regeneration during the regeneration process leads to overproduction of interstitial cells and collagen deposition between the neomyofibers.
During embryonic myogenesis, Six1−/− embryos have impaired primary myogenesis, and activation of MRF gene expressions are reduced and delayed in migratory limb muscle precursors. Previous work from the laboratory demonstrated that Six1 lie genetically upstream of Myf5 and Myogenin, and thus control hypaxial myogenesis at multiple levels (Laclef et al., 2003; Giordani et al., 2007; Grifone et al., 2007). In contrast, our present study shows that in adult myogenic cells, Six1 appears to directly control MyoD and Myogenin activations. We did not detect differences in Myf5 expression levels nor in the proportion of Pax7+/Myf5+ cells in Six1-deficient SCs, and ChIP analysis did not reveal any binding of Six1 proteins on Myf5 limb enhancer regions. Six1 being upstream of MyoD but not Myf5 in adult SCs is in concordance with previous results demonstrating that MyoD and Myf5 possess defined specific roles in SC biology. MyoD has a role in myoblast differentiation potential, whereas Myf5 enables transient myoblast amplification (Megeney et al., 1996; Gayraud-Morel et al., 2007). Interestingly, Six1 has been shown to be a genetic target of MyoD, and Six1 transcription is down-regulated in MyoD−/− myoblasts (Berkes et al., 2004; Ishibashi et al., 2005). Collectively, these data suggest that a positive cross-regulatory loop between Six1 and MyoD exists during adult regenerative myogenesis. Our findings indicate that adult SCs and embryonic muscle progenitors have distinct requirements for Six1 functions. In contrast, fetal and perinatal myogenic progenitor expansion and survival depend on Pax7 (Seale et al., 2000; Kuang et al., 2006; Relaix et al., 2006), but when Pax7 is inactivated in adult mice, mutant SCs function normally (Lepper et al., 2009). Our data are consistent with the emerging view that prenatal and adult progenitors express or require different sets of myogenic transcription factors, or, alternatively, use these transcription factors to different extents through recruitment of stage-specific enhancers.
Targeted Six1 gene disruption resulted in a large increase in SC niche occupancy within the regenerated muscle tissue. We demonstrated that Six1 controls homeostasis of the SC pool during skeletal muscle regeneration by regulating the ERK1 pathway (Abou-Khalil et al., 2009). Our results indicate that Six1 may regulate autocrine signaling via the control of Ang1 secretion, and the integration of paracrine signals via the control of Dusp6 transcription, a negative regulator of intracellular ERK1/2 phosphorylation (Maillet et al., 2008). Our results are consistent with previous studies describing the crucial role of the tyrosine kinases coreceptor Syndecan4 (Syn4). Syn4 is expressed by quiescent SCs and their activated progeny (Cornelison et al., 2001), and Syn4−/− SCs have impaired niche occupancy and are defective in myogenic potential, failing to reconstitute damaged muscle in vivo (Cornelison et al., 2004). Furthermore, a recent study demonstrated that Sprouty1 (Spry1), a receptor tyrosine kinase signaling inhibitor, is required for the return to quiescence and homeostasis of the SC pool during repair (Shea et al., 2010). Interestingly, targeted disruption of Spry1 in the adult SCs leads to the loss of a subset of SCs after regeneration, whereas Dusp6−/− muscles have an increased SC population after regeneration. Moreover Spry1 is expressed by quiescent SC, and not by dividing cells, whereas quiescent SCs do not express Dusp6, whereas dividing cells strongly express Dusp6. Hence, known activators (Syn4) and inhibitors (Spry1 and Dusp6) of ERK signaling have distinct expression patterns and divergent impacts on the homeostasis of the SC pool during skeletal muscle regeneration. Future work will be dedicated to establishing a framework for study of the roles of different “flavors” of ERK signaling in SCs, as well as the timely requirement for inhibition/activation of the system for control of SC niche occupancy.
Characterization of transcription factors controlling SC niche occupancy is providing important insights into the molecular mechanisms regulating skeletal muscle regeneration. Our identification of a role for Six1 in regulating SC myogenic potential and self-renewal represents a significant advance in our understanding of muscle regeneration. Future experiments will investigate both the networks of genes controlled by Six1 in adult muscle stem cells, by whole-genome approaches, and the utility of modulating the ERK pathway in vivo to augment muscle regeneration toward ameliorating the loss of muscle function in neuromuscular disease.
Materials and methods
Mice and animal care
Animals were bred and handled as recommended by European Community guidelines. Experiments were performed in accordance with the guidelines of the French Veterinary Department and conducted on 8–12-wk-old mice. Myf5Cre/ROSA26-YFP mice were obtained by crossing the knockin Myf5Cre mice (this strain expresses Cre recombinase from the endogenous Myf5 locus; Tallquist et al., 2000) with the ROSA26-YFP reporter mice (these R26-stop-EYFP mutant mice have a loxP-flanked STOP sequence followed by the EYFP inserted into the Gt(ROSA)26Sor locus; when bred to mice expressing Cre recombinase, the STOP sequence was deleted and EYFP expression was observed in the cre-expressing tissues of the double mutant offspring; Srinivas et al., 2001). Six1KO mice were obtained by crossing the Tg:Pax7CreERT2 driver mice (in this strain, the mouse Pax7 promoter drives expression of a Cre recombinase fused with a mutated ligand-binding domain of the human estrogen receptor (ERT2). Upon the introduction of the drug TM, the Cre-ERT2 construct is able to penetrate the nucleus and induce targeted mutation in skeletal muscle SCs (Mourikis et al., 2012) with the Six1-LoxP line (these mice possess loxP sites on either side of exon 1 of the Six1 gene; when these mutant mice were bred to mice that express Cre recombinase, the resulting offspring will have exon 1 deleted in the Cre-expressing tissue; Fig. S1) backcrossed on C57BL/6 background. Dusp6-null mice (homozygous Dusp6 targeted mutant mice) were on a C57BL/6*SV129 mixed background (Maillet et al., 2008) and Erk1-null mice (homozygous Erk1 targeted mutant mice; provided by M. Gaudry, Institute Cochin, Paris, France) were on a C57BL/6 background (Pagès et al., 1999).
FACS
Flow cytometry analyses were performed at the Cochin Flow Cytometry Facility. Mononucleated muscle-derived cells were isolated from hind limb muscle of 6–8-wk-old mice. After removal of the skin and fascia from the ankle joint up toward the hip to expose underlying tissues, skeletal muscles of the hind limbs were dissected (hamstring muscle group, quadriceps, tibialis, EDL, gastrocnemius, soleus, and gluteus) with care to take off as much fat and connective tissue as possible. Muscles were transferred to a sterile 6-cm Petri dish on ice, mulched into a smooth pulp, and incubated in Collagenase B/DispaseII/CaCl2 solution (1.5 U/ml, 2.4 U/ml, and 2 M, respectively, in DME; Roche). After a 15-min incubation at 37°C in the culture incubator, the muscle pulp was triturated with heat-polished glass Pasteur pipettes, and this incubation/trituration step was repeated. The tissue digestion was stopped with the addition of FBS, and cells were filtered and washed twice with PBS. Erythrocytes were removed with the Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich). Antibody staining was performed as described previously (Le Grand et al., 2009). Cells were separated on a MoFlo cytometer (Dako) equipped with three lasers. Sorting gates were strictly defined based on single antibody-stained control cells as well as the forward and side-scatter (SSC) patterns of SCs. Dead cells and debris were excluded by Hoescht staining. FACS-purified SCs were CD34+, α7-Integrin+, CD31−, CD45−, and Sca1−, and were >95% pure. RNA was isolated from either freshly sorted SCs for quiescent SCs or from cells plated and maintained in culture for 3 d for proliferating myoblasts. Total RNA was prepared from cell populations by MicroRNA kit (QIAGEN).
Isolation of YFP± SC subsets
Lineage-tracing experiments identified a subpopulation of SCs that have never expressed Myf5 and function as a stem cell reservoir (Kuang et al., 2007). Satellite stem cells (Pax7+/Myf5−) represent ∼10% of the adult SC pool. To purify satellite stem cells and their committed counterparts, we generated Myf5-Cre*ROSA26-YFP mice. Hence SCs that have expressed Myf5 at any time during their developmental history also express the YFP protein. To purify YFP+ and YFP− SCs, we used our routine FACS strategy, and further separated [CD34+, α7-Integrin+, CD31−, CD45−, and Sca1−] SCs on the basis of Myf5-driven YFP expression.
Induction of Cre activity and muscle injury
I.p. injections of TM (150 µl, 10 mg/ml, diluted in maize oil; MP Biomedicals) were administrated to 2–3-mo-old mice daily for 4 d before injury. Muscle tissue injury was created by a single injection of 35 µl of CTX solution (12 µM; Latoxan) into TA muscle, and mice were allowed to recover for 4–30 d.
Isolation and culture of single EDL myofibers
EDL muscles were dissected from the legs by handling tendons only (cut the skin at the front of the leg from knee level to toes, cut the tendon across the top of the foot, cut the tendon at knee level, grab the foot end of the EDL tendon and pull to gently slide the muscle off). Muscles were then placed in Collagenase Type I solution (2 mg/ml in DME; Sigma-Aldrich) and incubated in shaking water bath at 35°C for 50 min. Muscles were transferred to DME-filled horse serum–coated Petri dishes (to prevent fiber attachment to the plastic) using heat-polished glass Pasteur pipettes (with bore sizes that are just big enough to let the muscle go through). One at a time, muscles were triturated until fibers were separated. The bulk of fibers were then transferred to a fresh Petri dish, and the dish containing the isolated fibers was placed in the culture incubator. The fibers should not be hypercontracted. Under the dissecting scope they should look long thin and shiny. Usually, 100 fibers can be freed from one EDL muscle. After 15 min, the damaged fibers shrink and the good fibers were transferred to a fresh DME-filled horse serum–coated Petri dish with the thinnest bore Pasteur pipette (under the dissecting scope), leaving behind any debris. The last step should be repeated twice until the fiber preparation only contains live undamaged fibers. Isolated myofibers were cultured in suspension for 3 d in horse serum–coated 6-well plates. Typically, SCs form 8–16 cell aggregates of clonal origin within 3 d, and the cells on myofibers committed to differentiation were Pax7−/MyoD+, whereas quiescent cells express Pax7 but not MyoD (Zammit et al., 2004). Alternatively, myofibers were allowed to adhere to Petri dishes coated with 20% Matrigel (BD) in DME (Invitrogen) and cultured for 6–9 d. Then SC-derived myogenic progenitors migrate off the myofibers and form myogenic colonies composed of proliferating myoblasts and differentiated myotubes. Fibers were incubated in plating medium consisting of 15% FBS (Hyclone) and 1% chick embryo extract (Accurate Chemicals) in DME. For in vivo activation of SCs, regeneration was induced by CTX injection into TA muscle, and 4 or 7 d later, individual myofibers were isolated from the neighboring EDL muscle.
Preparation of SC-derived myoblasts
The total cells from the muscle tissue, prepared similarly as per the FACS purification procedure, are resuspended in growth medium consisting of Ham’s F10 (Invitrogen) supplemented with 20% FBS and 2.5 ng/µl of basic FGF (R&D systems) and preplated onto a noncoated 10-cm plate (fibroblasts will adhere to the plate; myoblasts remain in suspension) for 2 h. At the end of the preplate procedure, the media is transferred onto collagen-coated Petri dishes. Cultures were maintained in growth medium until cells reached 80% confluence. The myoblast population was enriched by differential adhesion compared with fibroblasts (either by shaking off the myoblasts, by tapping the culture dishes, or by serial 20-min preplate procedures after trypsinization), and usually cultures are 95% pure after the fourth passage. Myoblasts were induced to differentiate into myotubes by shifting to low-mitogen medium consisting of 4% horse serum in DME.
Six1 silencing and overexpression
SC-derived myoblasts were re-fed 3 h before transfection performed in growth medium. Cells were re-fed with growth medium 6 h after transfection, and RNA was harvested after 24 h. Cells were transfected with siRNA duplexes (s201996; Ambion) at the final concentration of 10 nM each using Lipofectamine 2000 (Invitrogen) as a transfection reagent. Transfection efficiency was monitored using Cy3-labeled nonsilencing siRNA duplexes (AM4621; Ambion). Alternatively, cells were transfected with 2 µg of pCMV-Six1 plasmid encoding full-length mouse Six1 using FuGene (Roche) as a transfection reagent. Transfection efficiency was monitored using a pCMV-GFP plasmid. Modulation of gene expression was assessed by qRT-PCR.
Immunofluorescence
Single myofibers and cell cultures were fixed in 2% PFA in PBS, washed three times in PBS, and stored at 4°C in blocking solution (BS) consisting of 5% goat serum, 0.5% BSA, and 0.2% Triton X-100 in PBS (all from Sigma-Aldrich). Skeletal muscles were embedded in Tissue-Tek Compound (Gentaur), frozen on cold isopentane, and processed for cryostat sectioning. 10-µm sections were collected from the mid-belly of muscles. Cryosections were thawed at room temperature, fixed in 4% PFA, washed three times in PBS, and processed for antigen retrieval with the Antigen Unmasking Solution (Vector Laboratories) at 95°C for 15 min in a microwave with a thermostat. Cells and sections were washed twice with PBS and blocked for 1 h in BS at room temperature. This was followed by an overnight incubation at 4°C with primary antibodies diluted in BS. After three washes in PBS, the slides were incubated for 1 h at room temperature with secondary antibodies conjugated to a fluorescent dye (Alexa Fluor 488 or 568; Invitrogen and Molecular Probes) diluted in PBS. The staining was completed with three washes with PBS and incubation in DAPI solution to label cell nuclei. Primary antibodies used in this study were as follows: rat α7-Integrin (R&D Systems), rat CD34 (BD), goat Collagen Type I (SouthernBiotech), rabbit Desmin (Abcam), rabbit Dusp6 (Abcam), mouse Dystrophin (Novocastra), rabbit Ki67 (Abcam), rabbit Laminin (Sigma-Aldrich), rabbit Myf5 (Santa Cruz Biotechnology, Inc.), rabbit MyoD (Santa Cruz Biotechnology, Inc.), mouse Myogenin (Dako), mouse MyHC embryonic (Vector Laboratories), mouse MyHC total (Developmental Studies Hybridoma Bank), mouse Pax7 (Developmental Studies Hybridoma Bank), rabbit Phospho-ERK1/2 (Cell Signaling Technology), rabbit IgG (Santa Cruz Biotechnology, Inc.), Rabbit Six1 (Sigma-Aldrich), rabbit Six4 (Antibodies Online), and chicken Syndecan4 (a gift from B. Olwin, University of Colorado, Boulder, CO).
Image acquisition
Image acquisitions were performed in C. Desdouets’ Laboratory and in the Cochin Imaging Facility. Digital images were acquired using a microscope (Statif Eclipse E600; Nikon) with 40× magnification, a DXM1200 cooled charge-coupled device camera (Nikon), and ACT-1 (version 2.63; Universal Imaging). Alternatively, images were taken using a macroscope (AZ100; Nikon) with 5× magnification, a Digital sight DS-Ri1 camera (Nikon), and NIS-Element Br (Nikon). Images were taken at room temperature and the imaging medium was from Dako. Images were composed and edited in Photoshop (CS5; Adobe), in which background was reduced using brightness and contrast adjustments were applied to the whole image.
Histology and quantification
Transverse sections of experimental and contralateral muscles were cut with a cryostat (CM1850; Leica). The entire TA muscles were sectioned in order to compare experimental and contralateral muscles at the same level on serial sections (∼400 sections were obtained from each TA muscle). For Hemalun-Eosin and immunostaining, sections were fixed with 4% paraformaldehyde. For quantification of myofiber caliber and enumeration of myonuclei, pictures of Dystrophin-stained cryosections were assembled, individual fibers were outlined, and their CSA was determined with the public domain image analysis software ImageJ (National Institutes of Health). The SC enumeration was performed with Photoshop CS2 on pictures of Pax7 and Laminin co-immunostained cryosections, taken in regenerated areas where all the fibers had centrally located nuclei.
RNA isolation and real-time PCR
Total RNA was isolated from cells or muscle tissue using the RNEasy kits (QIAGEN) and subjected to on-column DNase digestion according to the manufacturer’s instructions. cDNA synthesis was performed using Superscript III reverse transcription and random hexamer primers (Invitrogen). PCR was carried on LightCycler 480 Real-Time PCR Systems (Roche) using a LightCycler 480 SYBR green I Master (Roche) with specific primers. The thermocycling conditions used were as follows: an initial step of 10 min at 95°C, 40 cycles of a 15-s denaturation at 94°C, 10 s annealing at 60°C, and a 15-s extension at 72°C. Transcript levels were normalized to 18 S transcript levels. Relative fold change in expression was calculated using the ΔΔCT method (CT values < 30). For relative transcript quantification, each cDNA sample was run on a 5-point standard curve as to assure a PCR efficiency of ≥95%.
Sequences of the primers used for real-time PCR were as follows: Ang1, 5′-GGGTACCTTGGTGAGCACAG-3′ and 5′-CCCTGAGGAGATGTGAAGGA-3′; Cyclophilin A, 5′-TTGCCATTCCTGGACCCAAA-3′ and 5′-ATGGCACTGGTGGCAAGTCC-3′; Dusp6, 5′-CAGCGACTGGAATGAGAACA-3′ and 5′-CTGCACGAGCCGTCTAGATT-3′; Myf5, 5′-TGAAGGATGGACATGACGGACG-3′ and 5′-TTGTGTGCTCCGAAGGCTGCTA-3′; MyoD, 5′-TACCCAAGGTGGAGATCCTG-3′ and 5′-CATCATGCCATCAGAGCAGT-3′; Myogenin, 5′-GAAAGTGAATGAGGCCTTCG-3′ and 5′-ACGATGGACGTAAGGGAGTG-3′; Pax7, 5′-CTGGATGAGGGCTCAGATGT-3′ and 5′-GGTTAGCTCCTGCCTGCTTA-3′; Etv4, 5′-GGGTACCTTGGTGAGCACAG-3′ and 5′-CCCTGAGGAGATGTGAAGGA-3′; Six1, 5′-TTAAGAACCGGAGGCAAAGA-3′ and 5′-GGGGGTGAGAACTCCTCTTC-3′; Six2, 5′-CTTCTCATCCTCGGAACTGC-3′ and 5′-CTTCTCATCCTCGGAACTGC-3′; Six3, 5′-CCTCACCCCCACACAAGTAG-3′ and 5′-GTCAGGCTGGACACACTGGT-3′; Six4, 5′-CAGGTCAGCAACTGGTTCAA-3′ and 5′-AGAGAGGCTGAGGTTGGTGA-3′; Six5, 5′-GCCAGGAAGATGGAACTCTG-3′ and 5′-GCCAGGAAGATGGAACTCTG-3′; Six6, 5′-TTCAGGACCCATATCCCAAC-3′ and 5′-ACAGAACCTGCTGCTGGAGT-3′; Tie2, 5′-GAACCTGACCTCGGTGCTAC-3′ and 5′-TTTCTGGTTGAGGAGGGAGA-3′.
ChIP analysis
Protein–DNA complexes were cross-linked with 1% formaldehyde and sheared by sonication. Processing of samples was performed according to the ChIP kit manufacturer’s instructions (EMD Millipore). 1,000–2,000 µg of protein–DNA complexes were immunoprecipitated with 4 µg of Six1 (HPA001893; Sigma-Aldrich) or MyoD (sc-304X; Santa Cruz Biotechnology, Inc.) antibodies, and control IgG overnight. DNA was recovered using PCR purification columns (QIAGEN). The immunoprecipitated DNA was subjected to real-time PCR assays, and results were normalized using a control locus representing DNA fragments that are immunoprecipitated nonspecifically to produce robust and reproducible results. Each DNA sample was analyzed in triplicate.
Sequences of the primers used for ChIP PCR were as follows: Dusp6_MEF3, 5′-AACCTCCCAACTCTGGTG -3′ and 5′-GGTTAGGAGGAGGAAGTG-3′; Dusp6_5kbUP, 5′-GGAAGCCTGTGGTXTGTCTC-3′ and 5′-CTAACTCGCTGTCGCAGTTG-3′; IL4_Intron, 5′-AGAATGAAAGGCCCCAAAGT-3′ and 5′-GGGAGGACAGATCTCTGGTG-3′; Myf5_Enhancer, 5′-AGGCATGACTAATTGCATGGTAACTGG-3′ and 5′-CTCATAATGATATGGTTTTAAGCCC-3′; MyoD_DRR, 5′-AGACTGGGTAGGGCAGAGGT-3′ and 5′-CATTTCAGCTCCCTTGGCTA-3′; and Myogenin_Promoter, 5′-GTTTCTGTGGCGTTGGCTAT-3′ and 5′-ATAGAAGTGGGGCTCCTGGT-3′.
Capillary isoelectric focusing (CIEF) immunoassay
Imaged CIEF was performed on a Nanopro1000 Analyzer (ProteinSimple) by P. Chafey at the Institut Cochin Proteomic Facility (Paris, France), as per the constructor protocol. The NanoPro is a multiplexed capillary-based isoelectric immunoassay with whole-column imaging detection. 3 × 105 primary myoblasts were used per sample, and the experiment was performed three times. We applied the NanoPro technology to rapidly measure relative charge distribution of ERK proteins with anti-ERK1/2 and anti-phospho-ERK1/2 antibodies.
Statistical analysis
A minimum of 3 and up to 5 replicates were used for the experiments presented. Data are presented with standard errors. Results were assessed for statistical significance using a Student’s t test (Excel; Microsoft) and differences were considered statistically significant at P < 0.05.
Online supplemental material
Fig. S1 shows schematic representation of the targeted insertion of LoxP sites in the Six1 gene, as well as the phenotype of mutant Six1Δ/Δ embryos obtained by crossing the conditional null Six1 allele with the ubiquitous EIIa-Cre mice. Fig. S2 shows α7-Integrin and CD34 immunostaining on WT and Six1KO single fibers as well as Six4 and Desmin immunostaining on WT and Six1KO primary myoblasts. Fig. S3 shows representative Hemalun-Eosin and collagen immunostaining of uninjured and regenerated WT and Six1KO TA muscles with quantifications of necrotic fibers and collagen area. Fig. S4 shows quantifications of Ki67-negative SCs, Six1-positive SCs, numbers SCs on single fibers, and relative increase in SC content in regenerated WT and Six1KO TA muscles, 1 mo after CTX injury. Also, the quantification of Pax7-positive cells during CTX-mediated regeneration of the TA muscle is shown. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201050/DC1.
Supplementary Material
Acknowledgments
We thank Bénédicte Chazaud, Rémi Mounier, and Athanassia Sotiropoulos for helpful critical reading of the manuscript. We thank M. Gaudry for the Erk1−/− mice and F. Relaix for the EIIa-Cre mice.
F. Le Grand was supported by a Young Researcher Fellowship by the Institut National pour la Santé et la Recherche Médicale (INSERM), and financial support was provided by INSERM, the Association Française contre les Myopathies (AFM), the Centre National de la Recherche Scientifique (CNRS), the Agence Nationale pour la Recherche (ANR no. RO5099KK and RPV09108KKA), and the FP6 MYORES European Network of Excellence. We also acknowledge the contribution of the Région Ile De France to the Institut Cochin Animal care facility. The authors declare that they have no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- CTX
- cardiotoxin
- CSA
- cross-sectional area
- DRR
- distal regulatory region
- EDL
- extensor digitorum longus
- ERK
- extracellular signal-regulated kinase
- MRF
- myogenic regulatory factor
- MyHC
- myosin heavy chain
- qRT-PCR
- quantitative RT-PCR
- SC
- satellite cell
- TA
- tibialis anterior
- TM
- tamoxifen
References
- Abou-Khalil R., Le Grand F., Pallafacchina G., Valable S., Authier F.J., Rudnicki M.A., Gherardi R.K., Germain S., Chretien F., Sotiropoulos A., et al. 2009. Autocrine and paracrine angiopoietin 1/Tie-2 signaling promotes muscle satellite cell self-renewal. Cell Stem Cell. 5:298–309 10.1016/j.stem.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 14:465–477 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Collins C.A., Olsen I., Zammit P.S., Heslop L., Petrie A., Partridge T.A., Morgan J.E. 2005. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 122:289–301 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Filla M.S., Stanley H.M., Rapraeger A.C., Olwin B.B. 2001. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev. Biol. 239:79–94 10.1006/dbio.2001.0416 [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Wilcox-Adelman S.A., Goetinck P.F., Rauvala H., Rapraeger A.C., Olwin B.B. 2004. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 18:2231–2236 10.1101/gad.1214204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., Tajbakhsh S. 2007. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 312:13–28 10.1016/j.ydbio.2007.08.059 [DOI] [PubMed] [Google Scholar]
- Giordani J., Bajard L., Demignon J., Daubas P., Buckingham M., Maire P. 2007. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc. Natl. Acad. Sci. USA. 104:11310–11315 10.1073/pnas.0611299104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M.J., Hamard G., Maire P. 2005. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 132:2235–2249 10.1242/dev.01773 [DOI] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P.X., Maire P. 2007. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 302:602–616 10.1016/j.ydbio.2006.08.059 [DOI] [PubMed] [Google Scholar]
- Gros J., Manceau M., Thomé V., Marcelle C. 2005. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 435:954–958 10.1038/nature03572 [DOI] [PubMed] [Google Scholar]
- Ishibashi J., Perry R.L., Asakura A., Rudnicki M.A. 2005. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 171:471–482 10.1083/jcb.200502101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Giacone E., Gayraud-Morel B., Jory A., Gomès D., Tajbakhsh S. 2005. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 19:1426–1431 10.1101/gad.345505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Chargé S.B., Seale P., Huh M., Rudnicki M.A. 2006. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 172:103–113 10.1083/jcb.200508001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 129:999–1010 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C., Hamard G., Demignon J., Souil E., Houbron C., Maire P. 2003. Altered myogenesis in Six1-deficient mice. Development. 130:2239–2252 10.1242/dev.00440 [DOI] [PubMed] [Google Scholar]
- Le Grand F., Rudnicki M.A. 2007. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 19:628–633 10.1016/j.ceb.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F., Jones A.E., Seale V., Scimè A., Rudnicki M.A. 2009. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 4:535–547 10.1016/j.stem.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Conway S.J., Fan C.M. 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 460:627–631 10.1038/nature08209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T.A., Fan C.M. 2011. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 138:3639–3646 10.1242/dev.067595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M., Purcell N.H., Sargent M.A., York A.J., Bueno O.F., Molkentin J.D. 2008. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J. Biol. Chem. 283:31246–31255 10.1074/jbc.M806085200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C.J., Perdiguero E., Kharraz Y., Aguilar S., Pessina P., Serrano A.L., Muñoz-Cánoves P. 2011. Aberrant repair and fibrosis development in skeletal muscle. Skelet Muscle. 1:21 10.1186/2044-5040-1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173–1183 10.1101/gad.10.10.1173 [DOI] [PubMed] [Google Scholar]
- Mourikis P., Sambasivan R., Castel D., Rocheteau P., Bizzarro V., Tajbakhsh S. 2012. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 30:243–252 10.1002/stem.775 [DOI] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. 2011. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 138:3625–3637 10.1242/dev.064162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès G., Guérin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., Pouysségur J. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 286:1374–1377 10.1126/science.286.5443.1374 [DOI] [PubMed] [Google Scholar]
- Pallafacchina G., François S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D., Buckingham M. 2010. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. (Amst.). 4:77–91 10.1016/j.scr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. 2005. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 435:948–953 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- Relaix F., Montarras D., Zaffran S., Gayraud-Morel B., Rocancourt D., Tajbakhsh S., Mansouri A., Cumano A., Buckingham M. 2006. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 172:91–102 10.1083/jcb.200508044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A.F., Demignon J., Sakakibara I., Pujol J., Favier M., Strochlic L., Le Grand F., Sgarioto N., Guernec A., Schmitt A., et al. 2011. Genesis of muscle fiber-type diversity during mouse embryogenesis relies on Six1 and Six4 gene expression. Dev. Biol. 359:303–320 10.1016/j.ydbio.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. 2008. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 456:502–506 10.1038/nature07384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. 2011. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 138:3647–3656 10.1242/dev.067587 [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Shea K.L., Xiang W., LaPorta V.S., Licht J.D., Keller C., Basson M.A., Brack A.S. 2010. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 6:117–129 10.1016/j.stem.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Demignon J., Porteu A., Kahn A., Concordet J.P., Daegelen D., Maire P. 1998. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. USA. 95:14220–14225 10.1073/pnas.95.24.14220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M.D., Weismann K.E., Hellström M., Soriano P. 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 127:5059–5070 [DOI] [PubMed] [Google Scholar]
- Tapscott S.J., Lassar A.B., Weintraub H. 1992. A novel myoblast enhancer element mediates MyoD transcription. Mol. Cell. Biol. 12:4994–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco F.S., Dellavalle A., Diaz-Manera J., Messina G., Cossu G. 2010. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 120:11–19 10.1172/JCI40373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H., Motohashi N., Ono Y., Sato S., Ikeda K., Masuda S., Yada E., Kanesaki H., Miyagoe-Suzuki Y., Takeda S., Kawakami K. 2010. Six family genes control the proliferation and differentiation of muscle satellite cells. Exp. Cell Res. 316:2932–2944 10.1016/j.yexcr.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Zammit P.S., Golding J.P., Nagata Y., Hudon V., Partridge T.A., Beauchamp J.R. 2004. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166:347–357 10.1083/jcb.200312007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.