Abstract
The increased demand on protein folding in the endoplasmic reticulum (ER) during bacterial infection activates the unfolded protein response (UPR). OCTR-1—a G protein-coupled catecholamine receptor expressed in neurons—suppresses innate immunity by downregulating a non-canonical UPR pathway and the p38 MAPK pathway. Here, we show that OCTR-1 also regulates the canonical UPR pathway, which is controlled by XBP-1, at the organismal level. Importantly, XBP-1 is not under OCTR-1 control during development, only at the adult stage. Our results indicate that the nervous system temporally controls the UPR pathway to maintain ER homeostasis during development and immune activation.
Keywords: ER stress, innate immunity, nervous system, OCTR-1, unfolded protein response
Introduction
In eukaryotic cells, the endoplasmic reticulum (ER) is an important intracellular organelle in which newly synthesized transmembrane and secretory proteins are folded, assembled and matured. When protein homeostasis is disrupted by environmental stresses or changes in physiological conditions, a series of unfolded protein response (UPR) pathways are activated. Three parallel branches of the UPR help to maintain ER homeostasis using a number of sophisticated mechanisms to control translation, protein degradation, and the levels of chaperones [1].
Proteostasis and the health of the ER are key for organismal homeostasis. When the increased demand for protein folding is not met with an appropriate UPR, the accumulation of damaged proteins can result in neurodegeneration, cancer, and metabolic diseases [2–4]. Increasing evidence also indicates that a well-balanced ER is critical for maintaining not only cell integrity, but also immune homeostasis. In response to pathogen infection, increased demand on protein folding causes stress in the ER, which must be alleviated by UPR pathways to restore cellular homeostasis [5–9].
The inositol-requiring protein (IRE) branch of the UPR, which is the most conserved branch of the UPR across species, has been very well characterized in the nematode Caenorhabditis elegans. Conformational changes induced by unfolded proteins trigger IRE’s endoribonuclease activity, which results in the removal of an intron in the mRNA that encodes the UPR transcription factor named X-box binding protein-1 (XBP-1) [10, 11]. The spliced xbp-1 mRNA encodes the active transcription factor that controls the expression of downstream UPR genes required for ER homeostasis [10–12]. While the cell autonomous mechanisms that sense ER stress and activate the XBP-1-mediated UPR pathway to prevent cellular damage and subsequent organismal failure have been elucidated, it is unknown whether XBP-1 is also controlled at the organismal level.
We have taken advantage of the conservation of the XBP-1-mediated UPR to investigate the organismal control of XBP-1 during normal development and during response to infection in C. elegans. OCTR-1, which is a G protein-coupled catecholamine receptor expressed in the nervous system that controls immune homeostasis by regulating the p38 mitogen-activated protein kinase pathway and an XBP-1-independent pathway [13], was found to inhibit XBP-1 at the young adult stage. OCTR-1 did not control XBP-1 activity during development, indicating that the nervous system gains control of the canonical UPR once the high demand for protein folding that takes place during development ends.
Results and Discussion
OCTR-1 does not control XBP-1 during development
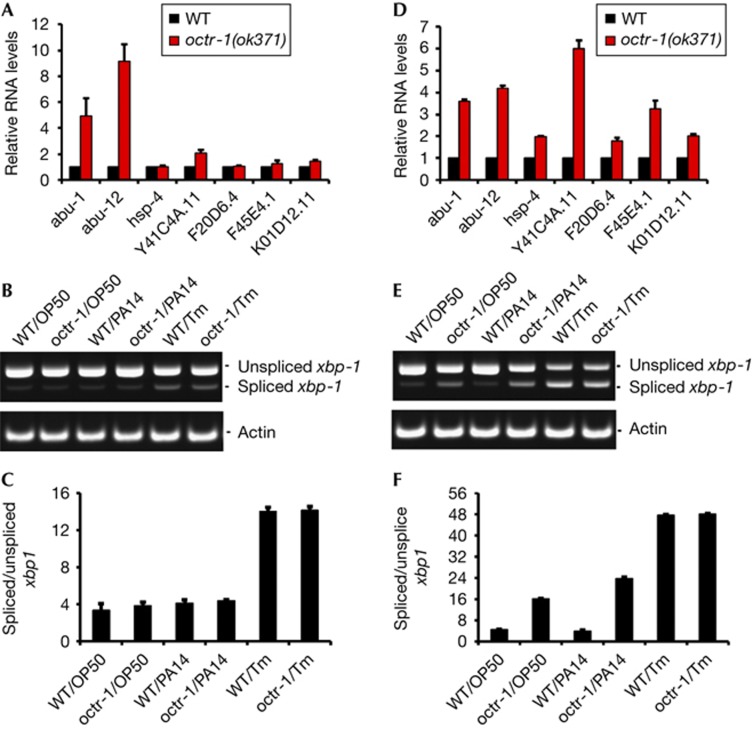
To investigate whether XBP-1 is controlled by the nervous system differently depending on the animal developmental stage, we measured the activity of XBP-1 by various means at the last larval stage (L4) and at the adult stage during infection. We first studied whether the transcriptional activity of XBP-1 was altered in octr-1(ok371) mutant animals infected at the L4 stage. octr-1(ok371) animals lack OCTR-1, a G protein-coupled receptor normally expressed in the cilia of neurons located in sensory openings of C. elegans [14]. In addition, we have recently reported that OCTR-1 functions in sensory neurons ASH and ASI to suppress a family of genes classified as abu (activated in blocked UPR) because they are activated in xbp-1 mutant animals when ER stress is induced by tunicamycin treatment [12, 13]. The abu genes encode UPR proteins that function in parallel with the canonical UPR pathway and that are required for immunity against infection by the human opportunistic pathogen Pseudomonas aeruginosa [12, 13]. As shown in Fig 1A, the expression levels of five XBP-1-dependent genes, hsp-4, Y41C4A.11, F20D6.4, F45E4.1, and K01D12.11 [12], were similar in octr-1(ok371) L4 animals and wild-type L4 animals exposed to P. aeruginosa. The levels of abu-1 and abu-12, two XBP-1-independent genes that are repressed by OCTR-1 [13], were higher in the octr-1(ok371) L4 animals than in wild-type L4 animals (Fig 1A).
Figure 1.
XBP-1-mediated UPR pathway is regulated by OCTR-1 at the adult stage. The expression of XBP-1-dependent and XBP-1-independent genes was studied in L4 (A,B,C) and 1-day-old animals (D,E,F). (A,D) Quantitative reverse transcription polymerase chain reaction analysis of abu-1, abu-12, hsp-4, Y41C4A.11, F45E4.1, K01D12.11, and F20D6.4 expression in octr-1(ok371) relative to wild-type (WT) animals exposed to Pseudomonas aeruginosa PA14. N=3 independent experiments; bar graphs correspond to mean±s.e.m. (B,E) Image of xbp-1 splicing in WT and octr-1(ok371) animals exposed to Escherichia coli OP50 or P. aeruginosa PA14 and/or treated with tunicamycin (Tm). Actin was used as a loading control. (C,F) Bar graph represents the mean ratio of spliced xbp-1/total xbp-1 transcript, multiplied by a factor of 100, and in three independent experiments. Error bars represent s.e.m. UPR, unfolded protein response; XBP-1, X-box binding protein 1.
Next, we investigated whether the activation of XBP-1 by removal of an intron from xbp-1 mRNA through splicing was controlled by OCTR-1 at the L4 stage in response to pathogen infection. Fig 1B,C show that the levels of spliced xbp-1 in octr-1(ok371) L4 animals were comparable to those in wild-type L4 animals. As expected, treatment with tunicamycin, an ER stress-inducing glycosylation inhibitor, increased the levels of spliced xbp-1. Taken together, these results indicate that while the nervous system has an important role in the control of XBP-1-independent UPR mechanisms during postembryonic development, the canonical XBP-1-mediated UPR pathway is not controlled by neuronal influences.
OCTR-1 gains control of XBP-1 after development
Because XBP-1 has a central role during development in both vertebrates and invertebrates [15–17], we hypothesized that OCTR-1 in the C. elegans nervous system might gain control of the XBP-1-mediated UPR pathway after development. Consistent with this hypothesis, C. elegans at the L4 stage exhibits a robust immune response against bacterial infections that might require constitutive activation of the XBP-1 pathway to cope with the higher demand of protein folding [18, 19]. In addition, recent studies indicate that basal immune activity during development causes ER stress that is exacerbated in the absence of intact UPR function [20]. To address the possibility that the nervous system, through OCTR-1, controls XBP-1 after larval development, we examined the expression levels of the five aforementioned XBP-1-dependent genes in octr-1(ok371) and wild-type adult animals in the presence of P. aeruginosa. As shown in Fig 1D, expression of the five genes was found to be significantly increased in octr-1(ok371) animals compared with wild-type animals. In addition, we found that the levels of spliced xbp-1 in the octr-1(ok371) animals grown on P. aeruginosa were significantly greater than those in the wild-type animals (Fig 1E,F). Similar results were obtained when the animals were grown on Escherichia coli (Fig 1E,F). As expected, tunicamycin treatment increased the levels of spliced xbp-1 (Fig 1E,F).
As adult animals contain eggs whose xbp-1 splicing might be differentially regulated by OCTR-1, we performed quantitative real time PCR (qRT–PCR) analysis on the five aforementioned XBP-1-dependent genes in embryos collected from wild-type and octr-1(ok371) adult animals that were infected with P. aeruginosa PA14 for 4 h at 25 °C. Supplementary Fig S1 online shows that the expression of XBP-1-dependent genes is not higher in octr-1(ok371) embryos than in those from wild-type animals, indicating that changes in gene expression at the embryonic stage cannot account for the differences observed in adult animals. These results indicate that OCTR-1 might inhibit the XBP-1-mediated UPR pathway during adulthood, but not during embryonic and postembryonic development.
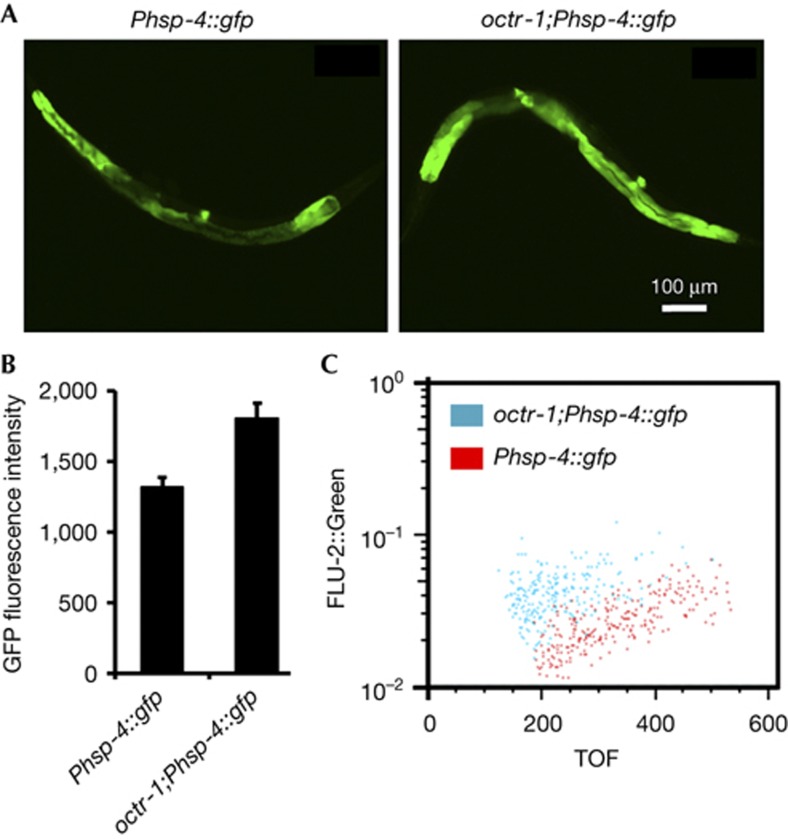
To study the in vivo role of OCTR-1 in the regulation of XBP-1 during the complete course of postembryonic development in the presence and absence of an infection, we use the transgenic transcriptional reporter Phsp-4::GFP(zcIs4). The C. elegans hsp-4 gene encodes a homologue of mammalian BiP/GRP-78 and the intestinal expression of the transgene Phsp-4::GFP(zcIs4) requires XBP-1 [10]. Consistent with the in vitro results shown in Fig 1, octr-1(ok371);Phsp-4::GFP(zcIs4) adult animals exhibited higher levels of Phsp-4::GFP expression than wild-type animals infected with P. aeruginosa (Fig 2A,B). In the absence of an infection, octr-1 mutation also slightly induced Phsp-4::GFP expression in adult animals (supplementary Fig S2 online). Large particle flow cytometry (COPAS Biosort instrument) confirmed that octr-1(ok371);Phsp-4::GFP(zcIs4) adult animals exhibit higher levels of Phsp-4::GFP expression than wild-type animals infected with P. aeruginosa (Fig 2C). Thus, we conclude that the nervous system controls, through OCTR-1, proteostasis in the intestinal cells of adult animals under normal physiological conditions and during the ER stress caused by bacterial infection.
Figure 2.
Loss of OCTR-1 signalling enhances the activation of XBP-1-mediated UPR pathway. (A) Images of Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) 1-day-old animals exposed to Pseudomonas aeruginosa PA14. Animals that best represent the fluorescence level of the population were shown. (B). GFP quantification from Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) 1-day-old animals exposed to P. aeruginosa PA14. Binary mean intensity of the region of interest (ROI) that corresponds to an entire animal was measured by NIS-Elements AR 3.2 software. N=10–20, error bars represent s.e.m. octr-1(ok371);Phsp-4::GFP(zcIs4) versus Phsp-4::GFP(zcIs4) on PA14: P<0.05. (C) GFP fluorescence intensity (FLU-2) of Phsp-4::GFP(zcIs4) and octr-1;Phsp-4::GFP(zcIs4) 1-day-old animals exposed to P. aeruginosa was plotted against animal size, measured as time of flight (TOF). Each dot represents an individual animal. GFP, green fluorescent protein; UPR, unfolded protein response; XBP-1, X-box binding protein-1.
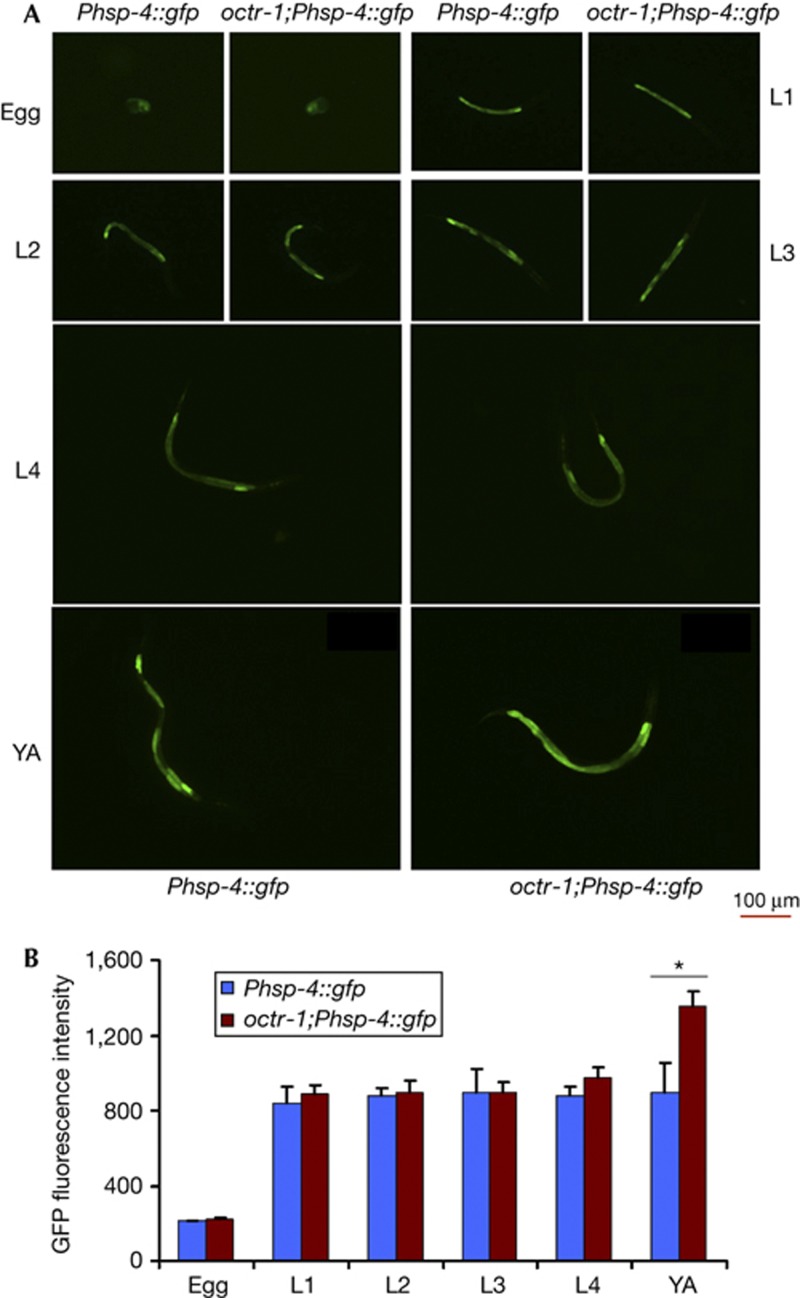
As shown in Fig 3 and supplementary Fig S3 online, the ER stress caused by P. aeruginosa infection is not controlled by OCTR-1 at the L1, L2, L3, or L4 stage. The expression of Phsp-4::GFP in eggs from infected octr-1(ok371);Phsp-4::GFP(zcIs4) animals was also comparable to that observed in eggs from infected wild-type animals (Fig 3). Similarly, the levels of Phsp-4::GFP expression during development were comparable in octr-1(ok371);Phsp-4::GFP(zcIs4) and wild-type animals grown on E. coli (supplementary Fig S4 online), indicating that the control of XBP-1 by OCTR-1 arises at the young adult stage under normal physiological conditions.
Figure 3.
OCTR-1 controls XBP-1-mediated UPR in infected young adult animals. (A) Images of Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) egg, L1, L2, L3, L4, and young adult (YA) animals grown on Pseudomonas aeruginosa PA14. Animals that best represent the fluorescence level of the population were shown. (B) GFP quantification from Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) egg, L1, L2, L3, L4, and YA animals exposed to P. aeruginosa PA14. Binary mean intensity of the region of interest that corresponds to an entire animal was measured by NIS-Elements AR 3.2 software. N=10–20, error bars represent s.e.m. Asterisk indicates significant difference. octr-1(ok371);Phsp-4::GFP(zcIs4) YA versus Phsp-4::GFP(zcIs4) YA on PA14: P<0.05. GFP, green fluorescent protein; UPR, unfolded protein response; XBP-1, X-box binding protein-1.
OCTR-1 blocks XBP-1-mediated resistance to pathogens
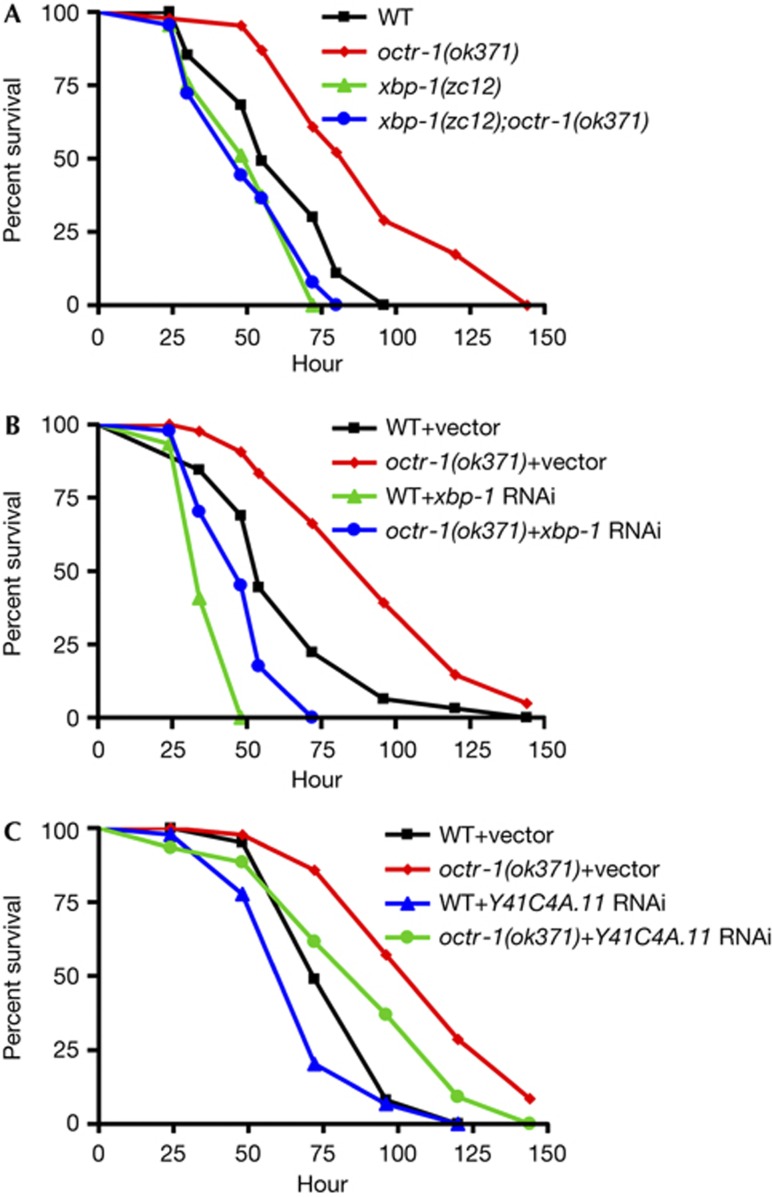
As OCTR-1 controls the XBP-1-mediated UPR pathway at the adult stage, we reasoned that xbp-1 mutation could suppress the enhanced resistance to P. aeruginosa-mediated killing phenotype exhibited by octr-1(ok371) animals [13]. To test this hypothesis, wild-type, octr-1(ok371), xbp-1(zc12), and xbp-1(zc12);octr-1(ok371) adult animals were exposed to P. aeruginosa and scored over time. As expected, octr-1(ok371) adult animals exhibited enhanced resistance to P. aeruginosa-mediated killing compared with wild-type animals (Fig 4A). In addition, this enhanced resistant phenotype of octr-1(ok371) animals was completely suppressed by xbp-1 mutation (Fig 4A). Likewise, xbp-1 RNA-mediated interference (RNAi) suppressed the enhanced resistance to P. aeruginosa of octr-1(ok371) animals (Fig 4B), suggesting that octr-1(ok371) animals are more resistant to pathogen infection than wild-type animals due to a higher protein folding capacity that is mediated by XBP-1.
Figure 4.
Inhibition of XBP-1-mediated UPR suppresses the enhanced resistance to Pseudomonas aeruginosa infection of octr-1(ok371) animals. (A) WT, octr-1(ok371), xbp-1(zc12), and xbp-1(zc12);octr-1(ok371) animals were exposed to P. aeruginosa PA14 and scored for survival over time. P-values are relative to octr-1(ok371) animals: WT (P<0.0001), xbp-1(zc12) (P<0.0001), xbp-1(zc12);octr-1(ok371) (P<0.0001). Shown is a representative assay of four independent experiments. (B) WT and octr-1(ok371) animals grown on double-stranded RNA (dsRNA) for vector control or dsRNA for xbp-1 were exposed to P. aeruginosa PA14 and scored for survival over time. P-values are relative to octr-1(ok371)+vector: WT+vector (P<0.0001), WT+xbp-1 RNAi (P<0.0001), octr-1(ok371)+xbp-1 RNAi (P<0.0001). Shown is a representative assay of three independent experiments. (C) WT and octr-1(ok371) animals grown on double-stranded RNA (dsRNA) for vector control or dsRNA for Y41C4A.11 were exposed to P. aeruginosa PA14 and scored for survival over time. P-values are relative to octr-1(ok371)+vector: WT+vector (P<0.0001), WT+Y41C4A.11 RNAi (P<0.0001), octr-1(ok371)+Y41C4A.11 RNAi (P<0.01). Shown is a representative assay of two independent experiments. RNAi, RNA-mediated interference; UPR, unfolded protein response; WT, wild-type; XBP-1, X-box binding protein-1.
We have previously reported that suppression of XBP-1-independent genes diminishes the enhanced resistance to P. aeruginosa of octr-1(ok371) animals [13]. To address whether XBP-1-dependent genes are required for resistance to P. aeruginosa of octr-1(ok371) animals, we studied hsp-4 and Y41C4A.11. As shown in Fig 1D, these two XBP-1-dependent genes are inhibited by OCTR-1. We found that while RNAi inhibition of hsp-4 had no effect on the enhanced resistance to P. aeruginosa-mediated killing of octr-1(ok371) animals, inhibition of Y41C4A.11 by RNAi partially suppressed it (Fig 4C; supplementary Fig S5 online). hsp-4 RNAi might not suppress the octr-1(ok371) phenotype due to chaperon redundancy or because hsp-4 is not highly upregulated in the absence of OCTR-1. RNAi inhibition of Y41C4A.11, which is the most highly upregulated gene in the absence of OCTR-1 (Fig 1D), suppressed the enhanced resistance to P. aeruginosa of adult octr-1(ok371) animals. Using proteomics, it has been shown that Y41C4A.11 is increased 4.6-fold in C. elegans exposed to Cry5B, a pore-forming toxin produced by Bacillus thuringiensis [7]. Taken together, these results indicate that the homeostasis of the ER by the activation of XBP-1-mediated UPR during immune response against bacterial infections is controlled at the organismal level by OCTR-1.
Concluding remarks
We have shown a temporal regulation of the canonical XBP-1 pathway involved in the UPR by the neurally expressed receptor OCTR-1. The results indicate that XBP-1 is not under neural control during embryonic and postembryonic development. It is possible that the high demand for protein synthesis and folding that takes place during development necessitates a constitutively active XBP-1 pathway. Indeed, deficiencies in the folding capacity of the ER have been linked to developmental defects not only in invertebrates but also in vertebrates [15–17].
Animals have evolved sophisticated mechanisms to modify specific properties in response to internal changes such as those that take place during development, or in response to changes in the external environment such as those that occur in response to bacterial infections. The nervous system, which can rapidly respond to many types of environmental stimuli, might integrate inflammatory signals to maintain immune homeostasis by regulating antimicrobial pathways and the UPR. Previously, we reported that OCTR-1 functions in ASH and ASI neurons to suppress XBP-1-independent genes and the p38 mitogen-activated protein kinase pathway [13]. Further experimentation will be required to understand the mechanisms that allow OCTR-1 to control the XBP-1-independent UPR during development, while gaining control of the XBP-1-mediated UPR only in adulthood.
Methods
Nematode and bacterial strains. The following C. elegans strains were cultured under standard conditions and fed E. coli OP50 [21]. Wild-type animals were C. elegans Bristol N2. octr-1(ok371) and phsp-4::gfp were obtained from the Caenorhabditis elegans Genetics Center (University of Minnesota, Minneapolis, MN). xbp-1(zc12) was obtained from Raffi Aroian’s laboratory (University of California, San Diego). The mutants octr-1(ok371);Phsp-4::GFP(zcIs4) and octr-1(ok371);xbp-1(zc12) were constructed using standard genetic techniques
E. coli strain OP50 and P. aeruginosa strain PA14 [18] were grown in Luria-Bertani broth at 37 °C.
C. elegans killing assay. C. elegans wild-type animals and mutants were maintained as hermaphrodites at 20 °C, grown on modified nematode growth medium (NGM) agar plates (0.35% instead of 0.25% peptone) and fed with E. coli OP50 as described [21]. The bacterial lawn used for C. elegans killing assays were prepared by placing a 50 μl of an overnight culture of the bacterial strains on modified NGM agar on plates 3.5 cm in diameter. Plates were incubated at 37 °C for 12–16 h. Plates were cooled down at room temperature for at least 1 h before seeding with synchronized animals. The killing assays were performed at 25 °C and live animals were transferred daily to fresh plates. Animals were scored at the times indicated and were considered dead when they failed to respond to touch.
RNA interference. RNA interference was used to generate loss-of-function RNAi phenotypes by feeding nematodes with E. coli strain HT115(DE3) expressing double-stranded RNA that is homologous to a target gene [22, 23]. Briefly, E. coli with the appropriate vectors were grown in Luria-Bertani broth containing ampicillin (100μg/ml) at 37 °C overnight, and plated onto NGM plates containing 100μg/ml ampicillin and 3 mM isopropyl β-D-thiogalactoside. RNAi-expressing bacteria were allowed to grow overnight at 37 °C. L2 or L3 larval animals were placed on RNAi or vector control plates for 2 days at 20 °C until nematodes became gravid. Gravid adults were then transferred to fresh RNAi-expressing bacterial lawns and allowed to lay eggs for 2 h to synchronize a second-generation RNAi population. The gravid adults were removed and eggs were allowed to develop at 20 °C to reach L4 stage or young adult stage for subsequent assays. Clone identity was confirmed by sequencing. unc22 RNAi was included as a positive control in all experiments to account for RNAi efficiency.
Tunicamycin treatment. Tunicamycin plates were prepared by making NGM plates containing 5 μg/ml of tunicamycin dissolved in dimethylsulphoxide. Control plates were prepared using dimethylsulphoxide alone. One-day-old plates that were seeded with E. coli OP50 or P. aeruginosa PA14 were used. Wild-type and octr-1(ok371) L4 or 1-day-old adult animals were transferred to tunicamycin plates or to control plates for 4 h at 25 °C. The animals were collected from the plates at the end of the treatment and used for RNA extraction.
RNA isolation. Gravid adult wild-type and octr-1(ok371) animals were lysed using a solution of sodium hydroxide and bleach (ratio of 5:2), the eggs were washed, and synchronized for 22 h in S basal liquid medium at room temperature. Synchronized L1 larval animals were placed onto NGM plates seeded with E. coli OP50 and grown at 20 °C until the animals had reached L4 or 1-day-old adult stage. Animals were collected and washed with M9 buffer before being transferred to NGM plates containing P. aeruginosa PA14 with or without tunicamycin for 4 h at 25 °C. After 4 h, animals were collected and washed with M9 buffer, and RNA was extracted using QIAzol lysis reagent (Qiagen Company) and purified following RNeasy Plus, Universal Handbook. Residual genomic DNA was removed by DNase treatment (Ambion Austin, TX).
Quantitative real-time PCR. Total RNA was obtained as described above and subjected to reverse transcription as suggested by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was conducted using the Applied Biosystems Two-Step Real-time PCR protocol using SYBR Green fluorescence (Applied Biosystems) on an Applied Biosystems 7900HT real-time PCR machine in 96-well plate format. Fifty nanograms of cDNA were used for real-time PCR. Twenty-five microlitre reactions were set-up and performed as outlined by the manufacturer (Applied Biosystems). Relative fold-changes for transcripts were calculated using the comparative CT(2−ΔΔCT) method [24] and normalized to pan-actin (act-1, -3, -4). Cycle thresholds of amplification were determined by StepOnePlus software (Applied Biosystems). All samples were run in triplicate. Primer sequences are available on request.
xbp-1 splicing experiments. xbp-1 RT–PCR analysis was carried out as previously described [25]. A set of published primers encompassing the non-canonical intron of the xbp-1 transcript was used [25]. The primers generate two PCR products: an approximately 200-bp PCR product of amplified unspliced xbp-1 transcript and an approximately 180-bp PCR product of amplified spliced xbp-1 transcript. PCR products were analysed on 6% native polyacrylamide gels. Quantification analysis was conducted using ImageJ and Excel.
Fluorescence imaging. Gravid adults Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) were transferred to NGM plates seeded with E. coli OP50 or P. aeruginosa PA14 for 1 h at 25 °C to lay eggs. Gravid adults were removed from plates and the eggs were allowed to hatch at 20 °C. Eggs, L1, L2, L3, L4, and young adult animals were observed under Nikon Eclipse 80i microscope and fluorescence images were taken using NIS-Elements AR 3.2 software. In addition, gravid adult Phsp-4::GFP(zcIs4) and octr-1(ok371);Phsp-4::GFP(zcIs4) were transferred to NGM plates seeded with E. coli OP50 for 1 h at 25 °C to lay eggs. Gravid adults were removed from NGM plates and the eggs were allowed to hatch at 20 °C. Young adults were then exposed to P. aeruginosa PA14 or E. coli OP50 at 20 °C. Animals were observed under Nikon Eclipse 80i microscope and fluorescence images were taken using NIS-Elements AR 3.2 software. Binary mean intensity was measured by NIS-Elements AR 3.2 software.
COPAS Biosorter GFP analysis. Expression levels of the transgenic transcriptional reporter Phsp-4::GFP(zcIs4) and octr-1;Phsp-4::GFP(zcIs4) were analysed using the COPAS Biosort instrument (Union Biometrica). Synchronized eggs, L1, L2, L3, L4, young adult, and 1-day-old animals were exposed to P. aeruginosa PA14 and washed in M9 buffer before analysis. Plots were constructed using FlowJo flow cytometry analysis software (Tree Star).
Statistical analysis. Animal survival was plotted as a nonlinear regression curve using the PRISM (version 4.00) computer program. Survival curves were considered different than the appropriate control indicated in the main text when P-values were <0.05. Prism uses the product limit or Kaplan–Meier method to calculate survival fractions and the log-rank test, which is equivalent to the Mantel–Heanszel test, to compare survival curves. A two-sample t-test for independent samples was used to analyse results of qRT–PCR, xbp-1 splicing quantification and binary mean fluorescence intensity; P-values <0.05 are considered significant. All the experiments were repeated at least three times, unless otherwise indicated. The analysis of the flow cytometry data was performed using FlowJo flow cytometry analysis software (Tree Star). FlowJo uses two algorithms (Kolmogorov–Smirnov and Cox χ2 related to Probability Binning) to determine the statistical difference between samples.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center (University of Minnesota) for strains used in this study. We thank R. Aroian (University of California, San Diego) for providing xbp-1(zc12). We thank J. Tenor, X. (Candy) Yang, and Y. Chen for technical assistance on the COPAS Biosorter. We thank Y. Zou and R. Beerman for technical assistance on the Nikon Eclipse 80i Microscope. AA is funded by the Dana Foundation and the National Institutes of Health (grant GM070977).
Author contributions: A.A. designed experiments, analysed data, and wrote the paper; J.S. designed and performed experiments, analysed data and wrote the paper; Y.L. helped with experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS (2008) Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 3: 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM (2004) The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer 4: 966–977 [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D (2006) Endoplasmic reticulum stress signaling in disease. Physiol Rev 86: 1133–1149 [DOI] [PubMed] [Google Scholar]
- Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A (2008) Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell 15: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH (2010) An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463: 1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof LJ, Kao CY, Los FC, Gonzalez MR, Shen Z, Briggs SP, van der Goot FG, Aroian RV (2008) Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathogens 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH (2010) TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA, Ron D, Tabas I (2009) Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol 11: 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Shen X et al. (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D (2002) A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol 158: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, Aballay A (2011) Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332: 729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wragg RT, Hapiak V, Miller SB, Harris GP, Gray J, Komuniecki PR, Komuniecki RW (2007) Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. J Neurosci 27: 13402–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souid S, Lepesant JA, Yanicostas C (2007) The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol 217: 159–167 [DOI] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC (2007) Atg5 and Bcl-2 provide novel insights into the interplay between apoptosis and autophagy. Cell Death Differentiation 14: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Reimold AM et al. (2000) An essential role in liver development for transcription factor XBP-1. Genes Dev 14: 152–157 [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA 96: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aballay A, Yorgey P, Ausubel FM (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol 10: 1539–1542 [DOI] [PubMed] [Google Scholar]
- Richardson CE, Kinkel S, Kim DH (2011) Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet 7: e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(–ΔΔC(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA 107: 9730–9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.