Abstract
During the stringent response, Escherichia coli enzyme RelA produces the ppGpp alarmone, which in turn regulates transcription, translation and replication. We show that ppGpp dramatically increases the turnover rate of its own ribosome-dependent synthesis by RelA, resulting in direct positive regulation of an enzyme by its product. Positive allosteric regulation therefore constitutes a new mechanism of enzyme activation. By integrating the output of individual RelA molecules and ppGpp degradation pathways, this regulatory circuit contributes to a fast and coordinated transition to stringency.
Keywords: ribosome, L11, ppGpp, alarmone, signalling
Introduction
Cells adapt to environmental changes predominantly by changing proteome composition through transcription and translation [1, 2]. Post-translational control systems, such as allosteric regulation of enzyme activity, are considerably faster than expression-level control and therefore are often used to regulate transcriptional networks [3]. In bacteria, the stringent response system is a regulatory hub that integrates several sensory inputs, such as amino acid, carbon source and iron limitation through modulation of the intracellular concentration of the alarmone nucleotide ppGpp [4]. Various enzymes are modulated by ppGpp, with RNA polymerase being a key target [4]. In Escherichia coli, the stringent response enzyme RelA senses amino-acid starvation by directly monitoring the aminoacylation status of the ribosomal A-site tRNA, which when in a deacylated state strongly induces RelA-mediated production of ppGpp from GDP and ATP and pppGpp from GTP and ATP [5]. Rapid conversion of pppGpp to ppGpp by guanosine pentaphosphate phosphohydrolase (gpp) results in the latter acting as the primary regulatory nucleotide in E. coli [6].
Close examination of time courses of ppGpp production by RelA in in vitro systems reveals deviations from linearity in early time points, displaying a more or less pronounced lag effect [5, 7]. This prompted us to suggest that its product, ppGpp, directly modulates the activity of RelA.
Results
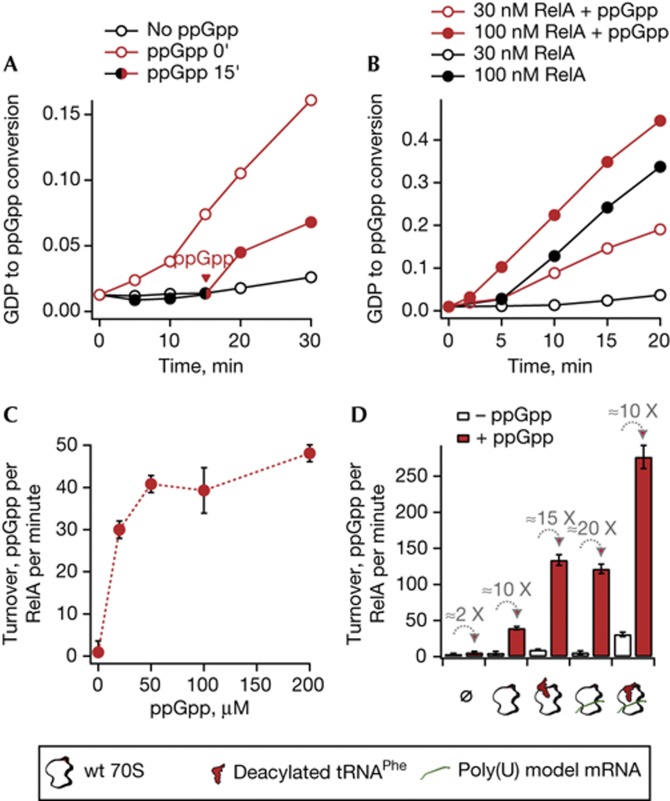
We prove this hypothesis by in vitro stringent response experiments: in the presence of 70S ribosomes, addition of ppGpp to final concentration of 100 μM results in pronounced activation of the enzymatic activity of RelA (Fig 1A). The same effect is observed when ppGpp is added during the reaction time course (Fig 1A, solid circles, ppGpp added at the 15-min time point), demonstrating that activation is not reliant on RelA:70S preincubation at 37 °C. In situ-formed ppGpp also activates RelA, as is evident from the kink in the time course of ppGpp production (Fig 1B, black solid circles). This auto-activating effect is more pronounced at higher RelA concentrations, where ppGpp is produced more rapidly (Fig 1B, solid circles versus hollow circles). As in situ production masks the effects of added ppGpp, all the following experiments were performed at a low RelA concentration (30 nM), which is in the range of in vivo estimates [8], and the range of time points was chosen to avoid ppGpp buildup during the course of the experiment. To determine the ppGpp concentration necessary to activate RelA, we titrated the reaction mixture with ppGpp, monitoring the effects on ppGpp production (Fig 1C). A concentration of 50 μM is enough to fully activate RelA, which is well within the range of cellular ppGpp concentrations [9].
Figure 1.
ppGpp activates the 70S-dependent synthetic activity of RelA. (A) Time course of 70S-dependent ppGpp synthesis in the absence of 100 μM ppGpp added (hollow black circles) and in the presence of ppGpp added at 0 (hollow red circles) or at 15 min (solid circles, black and red). Reaction mixture contains 30 nM RelA, 0.5 μM 70S ribosomes, 300 μM 3H-GDP and 0.5 mM ATP. (B) Time course of 70S-dependent ppGpp synthesis by RelA in the absence (black circles) and presence (red circles) of 100 μM ppGpp using 30 nM (hollow circles) and 100 nM (solid circles) RelA. (C) 70S-dependent RelA synthetic activity as a function of ppGpp concentration. (D) Effects of 70S ribosomes (0.5 μM), poly(U) (0.06 μg/μl) and deacylated tRNAPhe (1 μM) on ppGpp production in the presence (solid red bars) and absence (hollow bars) of 100 μM ppGpp. In C and D, error bars represent standard deviations of the turnover estimates by linear regression. Each experiment was performed at least three times. wt, wild-type.
By using the classical model for studying RelA enzymology in vitro, the poly(U)-programmed translational system [5, 10, 11], we addressed the question of whether ppGpp-mediated activation is complimentary or mutually exclusive with the effects of previously known RelA activators, that is, A-site-bound deacylated tRNAPhe and mRNA. We see a strong enhancement of RelA activity by ppGpp when the reaction is performed in the presence of poly(U)-programmed 70S ribosomes in complex with deacylated tRNAPhe (Fig 1D). While ppGpp does slightly elevate ppGpp production by RelA alone, this effect is amplified by 70S ribosomes and deacylated tRNAPhe, demonstrating that ppGpp is required for the full activation of RelA even in the presence of other activators.
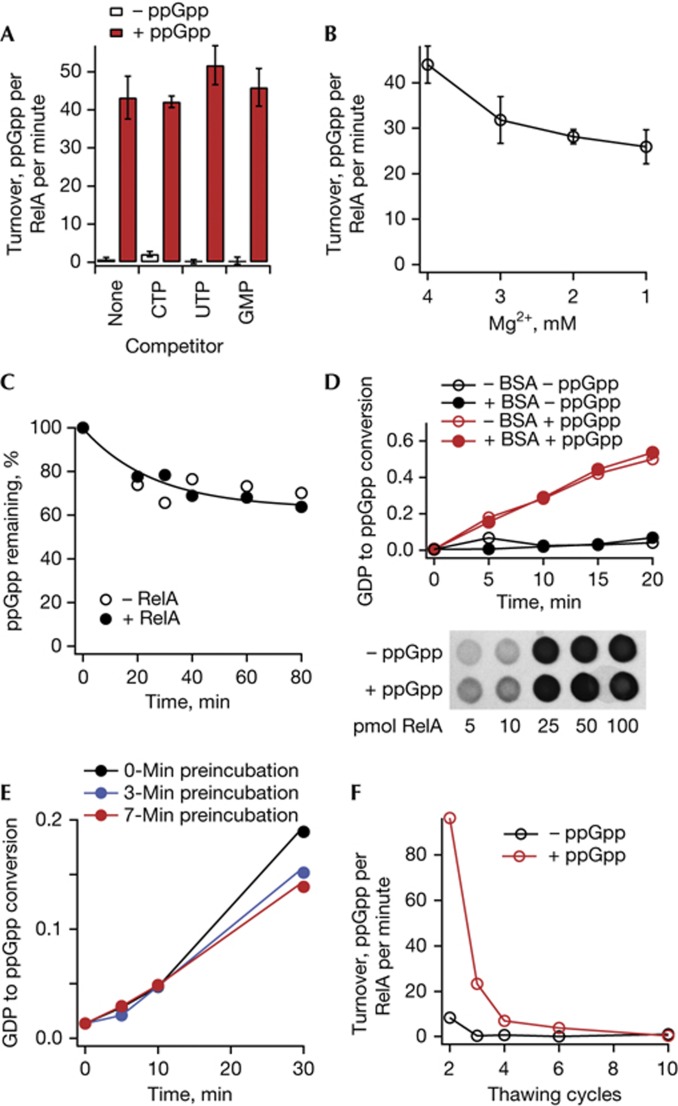
As E. coli RelA is an exceedingly challenging protein for in vitro work—it exhibits high propensity for aggregation, it loses activity during storage and it nonspecifically absorbs to charged surfaces such as glass [8]—we have taken extra precautions and performed exhaustive control experiments. First, we show that activation is specific for ppGpp and is not inhibited by excess amounts of CTP, UTP and GMP (Fig 2A). Second, reduction of the free magnesium ion concentration through potential Mg2+ chelation by the phosphate groups of ppGpp does not stimulate RelA activity. The magnesium ion is known to be an important cofactor for RSH proteins, having different effects on E. coli RelA [12] and bifunctional RSH Rel from Mycobacterium tuberculosis [13]. In agreement with Cochran and Byrne [12], less Mg2+ decreases RelA activity, and thus cannot be accountable for the strong activating effect we observe (Fig 2B). Third, we confirm that our RelA preparations have no detectable hydrolytic activity (Fig 2C), consistent with an inactive hydrolytic domain described previously [14]. This rules out a mass action effect, in which extra ppGpp saturates the hydrolytic activity and shifts the system towards production of ppGpp. Fourth, as RelA is susceptible to nonspecific aggregation [8] we tested the effects of addition of bovine serum albumin solution on RelA-mediated ppGpp production in our system (Fig 2D, top) and whether the concentration of the soluble RelA is different in the presence and absence of ppGpp (Fig 2D, bottom); we observed no significant effects. Finally, we kept the preincubation time at 37 °C (before initiating the reaction by addition of ATP) to a constant 5 min in all experiments, even though variation in this time did not affect the results (Fig 2E), as opposed to the protein Rel from Mycobacterium tuberculosis where it did affect results [15]. RelA activity is lost after just a few thawing cycles (Fig 2F), and the instability of RelA preparations is a well-known phenomenon [8]. Therefore, all experiments are conducted with fresh (up to 3 weeks at −80 °C, aliquoted and never passed through thaw/freeze cycles) preparations, and we regularly tested activity and response to ppGpp of our RelA preparations. All estimates of turnover rates in the same panels of all our figures are directly comparable as they depict experiments that were always performed in parallel.
Figure 2.
Activation of RelA enzymatic activity by ppGpp is specific. (A) Effects of CTP, UTP and GMP on RelA ppGpp synthetic activity in the presence (solid red bars) and absence (hollow bars) of 100 μM ppGpp. The reaction mixture contains 30 nM RelA, 0.5 μM 70S, 100 μM ppGpp, 300 μM 3H-GDP, 0.5 mM ATP and 500 μM competing nucleotides. (B) RelA ppGpp synthetic activity in the presence of ppGpp as function of Mg2+ concentration. ppGpp production was followed in the presence of 1 μM 70S, 1 mM ATP, 300 μM GDP, 100 μM ppGpp and 100 nM RelA. (C) ppGpp degradation in the presence (solid circles) and absence (hollow cycles) of RelA. The concentration of RelA (1 μM) was 30 times higher than in the other experiments, and ppGpp concentration was 400 μM. (D) Top: ppGpp synthesis in the presence (red circles) and absence (black circles) of added ppGpp in the presence (solid circles) and absence (hollow circles) of 0.5 mg/ml bovine serum albumin (BSA). The reaction mixture contains 30 nM RelA, 0.5 μM 70S, 100 μM ppGpp, 300 μM 3H-GDP and 0.5 mM ATP. Bottom: anti-6His western blot analysis of the RelA concentration in solution. Increasing amounts of the 0.1-μM RelA solution (5, 10, 25, 50 and 100 pmol) was spotted onto a nitrocellulose membrane in the presence and absence of 100 μM ppGpp. (E) Effects of RelA preincubation in the reaction mixture (RelA, GDP, ppGpp, 70S) at 37 °C for 0 min (black trace), 3 min (blue trace) or 7 min (red trace) before addition of ATP on the ppGpp synthesis kinetics. The reaction mixture contains 30 nM RelA, 0.5 μM 70S, 100 μM ppGpp, 300 μM 3H-GDP and 0.5 mM ATP. (F) Effect of thawing/freezing cycles on RelA activity in the presence (solid red circles) and absence (hollow black circles) of ppGpp. Experiments were performed with 30 nM RelA, 100 μM ppGpp and 0.5 μM 70S. In A and B, error bars represent standard deviations of the turnover estimates by linear regression. Each experiment was performed at least three times.
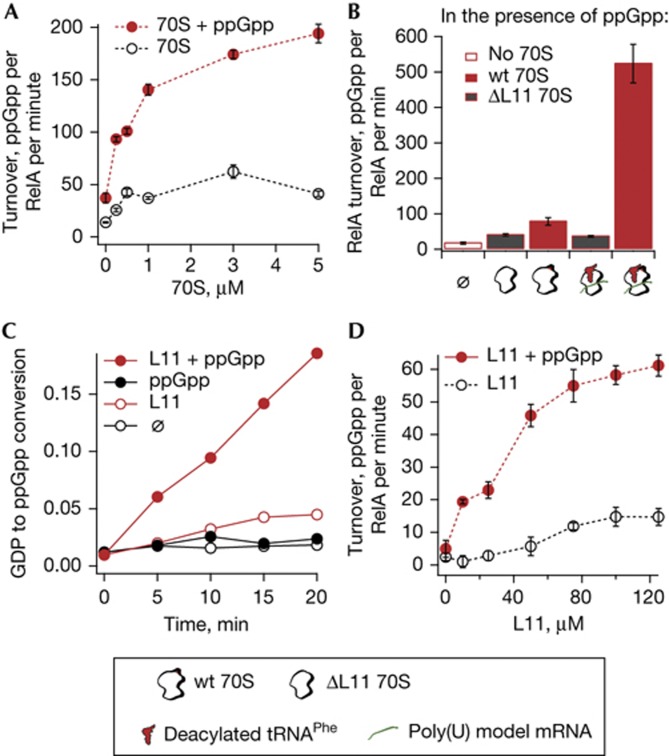
After establishing the specificity of the ppGpp-mediated RelA activation, we investigated the nature of this effect: does ppGpp merely increase susceptibility of RelA to other activators, that is, does it decrease its Michaelis constant Km to 70S ribosomes, or does it regulate the intrinsic catalytic efficiency of RelA by increasing its catalytic rate constant kcat? To address this question, we measured RelA turnover rate as a function of 70S ribosomes concentration, both in the presence and absence of externally added 100 μM ppGpp (Fig 3A). The two titration curves never converge at high 70S concentrations and maximum ppGpp production rates plateau at much higher turnover values in the presence of ppGpp (Fig 3A). At the same time, plateaus are achieved at similar 70S ribosomes concentrations; that is, the addition of ppGpp does not markedly affect RelA sensitivity to 70S ribosomes, which is consistent with similar affinities of RelA to the 70S ribosomes in the presence and absence of ppGpp (supplementary Fig S1 online). Taken together, these observations show that RelA indeed becomes a much more efficient enzyme in the presence of ppGpp.
Figure 3.
ppGpp-mediated activation of RelA is not masked by effects of 70S ribosomes, deacylated tRNAPhe and poly(U) mRNA, and is strictly L11-dependent. (A) RelA synthetic activity as a function of the 70S ribosomes concentration in the presence (solid red cycles) and absence (hollow black circles) of 100 μM ppGpp. (B) Effects of L11 on RelA synthetic activity in the presence of ppGpp, 70S ribosomes and 70S ribosomes programmed with poly(U) and tRNAPhe. Experiments performed in the presence of 0.5 μM wild-type (wt) 70S (red fill), 0.5 μM ΔL11 ribosomes (grey fill), and in absence of 70S ribosomes (hollow bars). (C) Time course of ppGpp synthesis in the absence (black stroke) and in the presence of 100 μM ppGpp (red stroke), and in the absence (hollow circles) and presence (solid circles) of 100 μM L11. (D) RelA synthetic activity as a function of L11 concentration in the presence (solid red cycles) and absence (hollow black circles) of 100 μM ppGpp. In A,B and D, error bars represent standard deviations of the turnover estimates by linear regression. Each experiment was performed at least three times.
We investigated which components of the ribosome are essential in ppGpp-mediated activation of RelA. It is well documented that RelA is dependent on the ribosomal protein L11 [11, 16]. Both naked 70S ribosomes lacking L11 (ΔL11 70S ribosomes) and ribosomes lacking L11 programmed with poly(U) mRNA and deacylated tRNAPhe induce very weak and tRNAPhe-insensitive RelA activation as compared with RelA:ppGpp alone (Fig 3B, compare grey bars versus hollow bar). This underscores the role of L11 as an essential element of the system, which cannot be substituted by positive feedback activation of ppGpp alone in the absence of L11. It has been shown that the N-terminal region of L11 alone activates RelA [17], suggesting that the minimal system comprising of just L11 and RelA can reproduce at least some of the characteristics of the RelA-mediated stringent response. Indeed, the ppGpp activation effect can be faithfully reconstructed in a simplified system consisting of L11 and RelA (Fig 3C). Titration by L11 shows the difference in plateau levels in RelA turnover in the presence or absence of ppGpp, similarly to the 70S ribosome titrations (compare Fig 3A,D). However, the concentrations of L11 and 70S ribosomes necessary to achieve the maximum catalytic efficiency differ by almost two orders of magnitude. Taken together with the weak and almost tRNAPhe-insensitive activation effect of the ΔL11 ribosomes, this suggests that the body of the ribosome acts mainly as a scaffold for positioning RelA for efficient activation by L11 and tRNA, as well as by ppGpp.
Discussion
Enzyme regulation by its product is commonly mediated through negative feedback auto-inhibition, with examples of enzyme activation by its product being exceedingly rare [18, 19]. There is, however, a critical difference between RelA activation by ppGpp and the two enzymes described in references [18, 19]. Both of the enzymes are activated by NAD+ by means of oxidation, not through allosteric regulation. RelA’s product, ppGpp, is highly unlikely to act by means of oxidation and is known to be an allosteric regulator of enzymes other than RelA [20, 21], suggesting allosteric nature of the ppGpp-mediated activation of RelA.
A positive feedback loop acting on the enzymatic level is inherently faster than positive feedback on the transcriptional level, as de novo production of mRNA and proteins is not required. This mechanism enables rapid amplification of a small input response and fast synchronization between the entire cellular population of RelA enzymes. This synchronizing mechanism explains why in our recent single-molecule in vivo tracking experiments we did not detect simultaneously both fast (that is, enzymatically active, freely diffusing) and slow-moving (that is, enzymatically inactive, ribosome-bound) RelA populations at the same time in the same cell [22]. If enough ppGpp is produced to trigger the positive feedback response, the cell commits to stringency, rapidly traversing intermediate ppGpp levels, and thus ensuring a fast coordinated response on the cellular level leading to the binary nature of starved/non-starved states [23]. This positive feedback regulatory system, similar to all bistable switches, requires a threshold filter so that small fluctuations in the signal do not induce an all-out response. ppGpp degradation by the RelA homologue SpoT establishes this filter, and not surprisingly, the deletion of the SpoT gene is lethal [24].
Once stringency is established and ppGpp reaches the concentration at which it efficiently binds to the RNA polymerase, the cell’s transcription programme is switched onto the stringency escape route. ppGpp is not a master regulator of RelA activity. Instead, the nature of the ribosomal complex (that is, presence of the A-site deacylated tRNA) is the ultimate regulator (Fig 1D). Production of amino acids and the consequent increase in tRNA charging levels results in resumption of translation, which inhibits RelA-mediated ppGpp production [5], providing a slow negative feedback loop [25]. This combination of a fast positive feedback loop controlled by a slow negative loop provides a very sensitive toggle system: such coupled feedback circuits rapidly turn on response to the stimulus, robustly maintain its status, and turn off the reaction when the stimulus disappears [26].
It is highly likely that direct positive feedback loops similar to that described here are involved in other fast cellular responses, but are not yet discovered. Indeed, identification of such a system requires rather counterintuitive experimentation: the product concentration should be kept low to avoid masking the effects of auto-activation (Fig 1B). Bacteria use numerous regulatory enzymes generating low-molecular-weight secondary messengers, which are controlling stress responses, virulence, biofilm formation and much more [27]. To our knowledge, RelA represents the first example of such an enzyme regulated through direct positive feedback control by its product. Thus, the present discovery adds yet another mechanism to the systems biology toolbox of control mechanisms with profound consequences for the stringent response and growth control by the signal-molecule ppGpp in growing bacterial cells.
Methods
In vitro system for RelA-mediated ppGpp production. Wild-type and ΔL11 E. coli 70S ribosomes (strain courtesy of Dr Scott Blanchard [28]) were prepared according to Antoun et al [29]. Absence of L11 in ΔL11 70S was validated by mass spectrometry, and functionality was restored on addition of the L11 protein (supplementary Fig S2 online), confirming specificity of the ΔL11 effect. C-terminally 6His-tagged RelA was overexpressed and purified according to Knutsson Jenvert and Holmberg Schiavon [10], aliquoted, shock-frozen with liquid nitrogen and stored at −80 °C in buffer containing 20 mM HEPES-KOH, pH 7.5, 600 mM KCl, 5 mM MgCl2 and 2 mM dithiothreitol. Deacylated tRNAPhe was purchased from Chemical Block. ppGpp was prepared using the RelSeq enzyme from Streptococcus equisimilis as described in Mechold et al. [30]. The Poly(U) model mRNA was purchased from Amersham Biosciences.
Thin layer chromatography (TLC) analysis of ppGpp synthesis. TLC analysis of ppGpp synthesis by RelA was performed according to Mechold et al [30] using 3H-labelled GDP (GE Healthcare). Progress of the reaction was quantified as 3H-GDP to 3H-ppGpp conversion, [3H-ppGpp]/([3H-ppGpp]+[3H-GDP]), ranging from 0 (no 3H-ppGpp is produced) to 1.0 (all the 3H-GDP is converted to 3H-ppGpp). Experiments were performed in Polymix buffer [29, 31] using low-bind eppendorf tubes (Eppendorf). In the experiments using the poly(U) system, the Mg2+ concentration was increased to 15 mM to promote complex formation. If not stated otherwise, experiments were performed with 30 nM RelA, 0.5 μM 70S, 0.3 mM 3H-labelled GDP (GE Healthcare), 0.5 mM ATP, 100 μM ppGpp, 100 μM L11, 1 μM tRNAPhe and 0.06 μg/μl (∼1 μM) poly(U), and reaction mixture was preincubated at 37 °C for 5 min before the reaction was started by addition of ATP. All experiments were performed from three to five times with at least two different RelA preparations.
Separation of 70S:RelA from unbound RelA by analytical gel-filtration. The S-300 gel-filtration method [11] was adapted to study the effects of ppGpp on RelA binding to 70S ribosomes. Binding experiments were performed in a total volume of 100 μl, containing 0.5 μM ribosomes, 50 nM RelA, 1 mM ATP, 300 μM GDP and 100 μM ppGpp in Polymix buffer [31]. The reaction mixture was preincubated for 5 min at 37 °C before loading on pre-equilibrated Illustra MicroSpin S-300 sephacryl columns (GE Healthcare). The first 100 μl fraction containing ribosomes was eluted by step-wise centrifugation, and 70S-associated RelA was detected by western blot analysis using the Dot Blot Convertible Filtration Manifold System (Life Technologies; for details see supplementary Methods online).
Supplementary Material
Acknowledgments
We are grateful to L. Holmberg for sharing the overexpression construct for RelA–6His, to A. Draycheva for her efforts in setting up the in vitro system for RelA-mediated ppGpp production, to J. Beljantseva and A. Kuzmenko for experimental help, T. Tammsalu for help with mass spectrometry, G. Atkinson for discussions and comments, to S. Blanchard for sharing the E. coli strain for purification of ΔL11 70S ribosomes and to M. Cashel for sharing the overexpression construct for the RelSeq enzyme and detailed protocols for ppGpp purification. This work was supported by the Estonian Science Foundation grants (6768 to T.T. and 7616, 9012 to V.H., and DoRa 6 travel grants to V.S., S.T. and A.E.) and European Regional Development Fund through the Center of Excellence in Chemical Biology (V.H. and T.T.), Swedish Institute (V.S.), European Research Council (J.E.), the Foundation for Strategic Research (J.E.), the Swedish Research Council (J.E.), the Knut and Alice Wallenberg Foundation (J.E.) and a Human Frontier Science Program cross-disciplinary fellowship (B.P.E.).
Author contributions: V.H., J.E. and M.E. designed research; V.H., B.P.E. and J.E. conceived the study; V.S., S.T., A.E., B.P.E., P.K. and V.H. performed research; V.H., J.E., T.T. and M.E. coordinated research; T.T., J.E. and M.E. contributed reagents; V.H. and B.P.E. wrote the manuscript with contributions from J.E., T.T. and M.E.
Footnotes
The authors declare that they have no conflict of interest.
References
- Guell M, Yus E, Lluch-Senar M, Serrano L (2011) Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat Rev Microbiol 9: 658–669 [DOI] [PubMed] [Google Scholar]
- Weiss RB, Atkins JF (2011) Molecular biology. Translation goes global. Science 334: 1509–1510 [DOI] [PubMed] [Google Scholar]
- Ray JC, Tabor JJ, Igoshin OA (2011) Non-transcriptional regulatory processes shape transcriptional network dynamics. Nat Rev Microbiol 9: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62: 35–51 [DOI] [PubMed] [Google Scholar]
- Haseltine WA, Block R (1973) Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA 70: 1564–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ahmed A (1979) Mutants of Escherichia coli defective in the degradation of guanosine 5′-triphosphate, 3′-diphosphate (pppGpp). Mol Gen Genet 169: 315–323 [DOI] [PubMed] [Google Scholar]
- Payoe R, Fahlman RP (2011) Dependence of RelA-mediated (p)ppGpp formation on tRNA identity. Biochemistry 50: 3075–3083 [DOI] [PubMed] [Google Scholar]
- Pedersen FS, Kjeldgaard NO (1977) Analysis of the relA gene product of Escherichia coli. Eur J Biochem 76: 91–97 [DOI] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H (2008) Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol 190: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsson Jenvert RM, Holmberg Schiavone L (2005) Characterization of the tRNA and ribosome-dependent pppGpp-synthesis by recombinant stringent factor from Escherichia coli. FEBS J 272: 685–695 [DOI] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH (2002) Dissection of the mechanism for the stringent factor RelA. Mol Cell 10: 779–788 [DOI] [PubMed] [Google Scholar]
- Cochran JW, Byrne RW (1974) Isolation and properties of a ribosome-bound factor required for ppGpp and ppGpp synthesis in Escherichia coli. J Biol Chem 249: 353–360 [PubMed] [Google Scholar]
- Sajish M, Tiwari D, Rananaware D, Nandicoori VK, Prakash B (2007) A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins. J Biol Chem 282: 34977–34983 [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6: e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H (2005) Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44: 9913–9923 [DOI] [PubMed] [Google Scholar]
- Parker J, Watson RJ, Friesen JD (1976) A relaxed mutant with an altered ribosomal protein L11. Mol Gen Genet 144: 111–114 [DOI] [PubMed] [Google Scholar]
- Jenvert RM, Schiavone LH (2007) The flexible N-terminal domain of ribosomal protein L11 from Escherichia coli is necessary for the activation of stringent factor. J Mol Biol 365: 764–772 [DOI] [PubMed] [Google Scholar]
- Coleman KJ, Cornish-Bowden A, Cole JA (1978) Activation of nitrite reductase from Escherichia coli K12 by oxidized nicotinamide-adenine dinucleotide. Biochem J 175: 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V, Veeger C (1961) Studies on the reaction mechanism of lipoyl dehydrogenase. Biochim Biophys Acta 48: 33–47 [DOI] [PubMed] [Google Scholar]
- Kanjee U et al. (2011) Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J 30: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PS, Raghavan A, Chatterji D (1995) Evidence for a ppGpp-binding site on Escherichia coli RNA polymerase: proximity relationship with the rifampicin-binding domain. Mol Microbiol 15: 255–265 [DOI] [PubMed] [Google Scholar]
- English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J (2011) Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci USA 108: E365–E373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T (2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68: 1128–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M (1991) Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266: 5980–5990 [PubMed] [Google Scholar]
- Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, Conway T (2011) Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol 79: 830–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kwon YK, Cho KH (2007) Coupled positive and negative feedback circuits form an essential building block of cellular signaling pathways. Bioessays 29: 85–90 [DOI] [PubMed] [Google Scholar]
- Pesavento C, Hengge R (2009) Bacterial nucleotide-based second messengers. Curr Opin Microbiol 12: 170–176 [DOI] [PubMed] [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC (2010) Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol 399: 576–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Tenson T, Ehrenberg MM (2004) Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol Proced Online 6: 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Murphy H, Brown L, Cashel M (2002) Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol 184: 2878–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc PC, Kurland CG (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA 76: 3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.