Abstract
LeuT serves as the model protein for understanding the relationships between structure, mechanism and pharmacology in neurotransmitter sodium symporters (NSSs). At the present time, however, there is a vigorous debate over whether there is a single high-affinity substrate site (S1) located at the original, crystallographically determined substrate site or whether there are two high-affinity substrates sites, one at the primary or S1 site and the other at a second site (S2) located at the base of the extracellular vestibule. In an effort to address the controversy over the number of high-affinity substrate sites in LeuT, one group studied the F253A mutant of LeuT and asserted that in this mutant substrate binds exclusively to the S2 site and that 1 mM clomipramine entirely ablates substrate binding to the S2 site. Here we study the binding of substrate to the F253A mutant of LeuT using ligand binding and X-ray crystallographic methods. Both experimental methods unambiguously show that substrate binds to the S1 site of the F253A mutant and that binding is retained in the presence of 1 mM clomipramine. These studies, in combination with previous work, are consistent with a mechanism for LeuT that involves a single high-affinity substrate binding site.
Keywords: neurotransmitter uptake, sodium-coupled transporter, x-ray crystallography
Introduction
Neurotransmitter sodium symporters (NSSs) have a critical role in the central nervous system, and are involved in many disorders of the nervous system that include depression [1], Parkinson’s disease [1] and epilepsy [2]. They are also the primary drug target of clinical antidepressants such as Prozac, tricyclic antidepressants, sertraline and fluoxetine [3, 4] and illegal drugs that include cocaine and amphetamines [5]. As the only NSS homologue amenable to high-resolution structural studies, LeuT [6] is a paradigm for both sodium-dependent and -independent transporters [7, 8], and is most importantly a molecular blueprint for structural and functional studies of eukaryotic NSSs [9–12].
At the present time, however, there is a controversy over the number of high-affinity substrate binding sites in LeuT [13]. Crystal structures of LeuT in detergent micelles [6, 14, 15] and in lipid bicelles [16], together with ligand binding studies [17], reveal a single high-affinity substrate binding site, termed the S1 site. By contrast, molecular dynamics, substrate binding and single-molecule experiments have been interpreted in terms of a model in which there is a second, or S2, high-affinity substrate binding site [18–22]. In an effort to disrupt substrate binding to the S1 site, Javitch and colleagues [18, 21] studied the F253A mutant, a residue that lines a portion of the S1 binding site [6, 14]. On the basis of their experiments on the F253A mutant, the authors make several assertions. First, they state that F253A mutation ablates substrate binding to the S1 site and has ‘all-or-nothing’ effect, and that substrate only binds to the S2 site [18, 21]. Second, they argue that the tricyclic antidepressant clomipramine (CMI), which has been shown to bind within the extracellular vestibule from analyses of high-resolution cocrystal structures from multiple laboratories [15, 23], entirely competes with substrate binding to the S2 site of the F253A mutant [18]. Finally, they argue that substrate binding to the S2 site of LeuT is lost if the protein is subjected to a concentration of the detergent n-dodecyl-β-D-maltoside (C12M) that is greater than 0.15% [18].
In this study, we performed substrate binding experiments using protein prepared according to the methods of Javitch and colleagues in C12M or in the recently developed lauryl maltose neopentyl glycol (MNG-3) detergent [24]. We carried out the binding experiments in C12M in an attempt to replicate the experiments of Javitch and colleagues, and we explored the use of the MNG-3 detergent because recent studies have shown that MNG-3 preserves the solubility and ligand binding capacity of LeuT to a greater extent than C12M [24]. In the context of both detergents, we find that CMI does not ablate the binding of leucine to the F253A mutant, yet we do show that preincubation of LeuT with CMI diminishes the extent of leucine binding. To better understand this phenomenon, we examined the time course of leucine binding and the solubility of LeuT in the presence of CMI. Furthermore, we solved crystal structures of the LeuT F253A mutant in complex with leucine and the substrate analogue selenomethionine (SeMet) in the context of lipid bicelles, finding that substrate binds unambiguously to the S1 site. Taken together, our studies clearly demonstrate that substrate binds to the S1 site of the LeuT F253A mutant and also provide plausible explanations for the discrepancies in experimental results between the Javitch and Gouaux research groups.
Results
Saturation binding assay
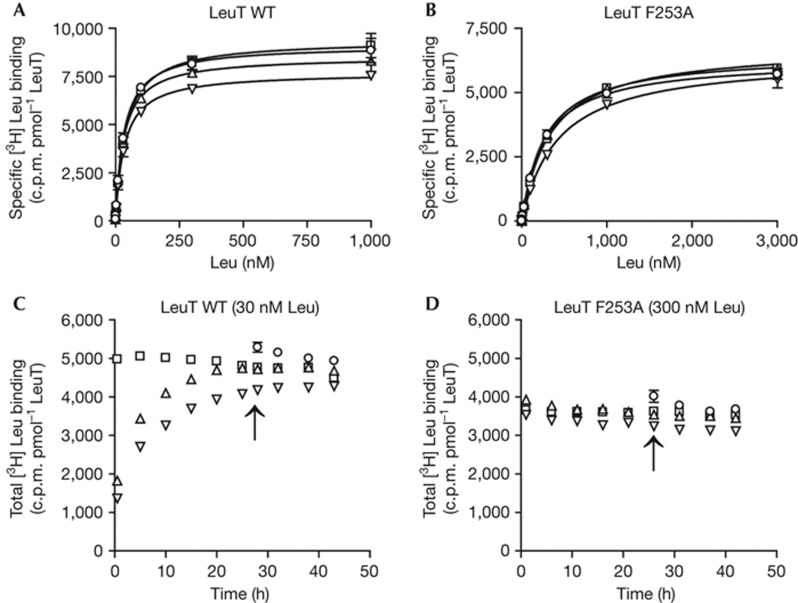
To probe the binding of substrate we began by examining saturation binding of 3H leucine using the scintillation proximity assay [25] and unconcentrated protein solubilized in 0.05% C12M. Under these conditions, the Kd for leucine binding to wild-type LeuT (LeuT WT) is 31.7±0.9 nM and to the F253A mutant is 99.4±5.1 nM (supplementary Fig S1 online, supplementary Table I online), values similar to those previously reported by the Javitch and Gouaux groups [14, 17, 18]. Because the Javitch group reports that CMI has a pronounced effect on the binding of leucine to the WT and F253A forms of LeuT, we next carried out saturation binding experiments under conditions where 1 mM CMI was added before or at the same time as leucine, or after equilibrium is achieved (supplementary Fig S1 online). For LeuT WT, inclusion of CMI yields similar Kd values, yet when CMI is added before leucine we see a decrease in Bmax by ∼20% (supplementary Fig S2 online). For the mutant, the effects of CMI are more profound, causing both an increase in Kd and a decrease in Bmax, particularly when CMI is added before leucine (supplementary Figs S1 and S2 online). However, we find no substantial differences in the Bmax for both LeuT WT and F253A mutant if CMI is added simultaneously with leucine or after equilibrium is achieved (supplementary Fig S2 online). More importantly, we find that under all conditions, inclusion of CMI in the binding experiment does not ablate binding of leucine to the F253A mutant, a finding in direct contradiction to that reported by the Javitch group [18].
Because we observed a decrease in Bmax on preincubation with CMI for both the WT and mutant transporters, we hypothesized that apo LeuT might be prone to unfolding and/or aggregation in C12M, and thus we asked whether a different detergent would better stabilize the protein. We therefore carried out the same leucine saturation binding experiments as described above using the new detergent MNG-3, which has previously been shown to better stabilize LeuT in comparison to C12M [24]. For both the WT and the F253A mutant, we measured Kd values in the absence and presence of CMI (Fig 1). The Kd values for the WT transporter are similar to those measured with the protein in C12M, whereas we observed an increase in Kd to 406.7±33.5 nM for the F253A mutant in MNG-3 detergent (supplementary Table I online). Most importantly, we found that in MNG-3 the Bmax values did not decrease as much as in C12M on preincubation of the WT and mutant proteins with CMI (supplementary Fig S2 online), consistent with the notion that MNG-3 better stabilizes both the WT and F253A mutant of apo LeuT [24].
Figure 1.
Substrate binds to LeuT wild-type (WT) and the F253A mutant in the absence and presence of CMI in lauryl maltose neopentyl glycol (MNG-3) detergent. (A) Measurement of 3H-leucine saturation binding to LeuT WT and (B) to the F253A mutant in the absence (squares) and presence of 1 mM CMI by using the scintillation proximity assay. CMI was added before leucine (inverted triangle), simultaneously with leucine (triangles) or after equilibrium (23 h; circles). (C) Time course for binding of 30 nM 3H leucine to LeuT WT and (D) 300 nM 3H leucine to the F253A mutant in the absence and presence of CMI. Symbols are as in A and B. The time points at which CMI was added are labelled by arrows. Error bars, s.e.m., n=6–9.
To probe if we are allowing sufficient time for the scintillation proximity assay (SPA) binding experiments to reach equilibrium, and to also determine whether preincubation of LeuT with CMI alters the rate at which the binding reactions reach equilibrium, we carried out time-course assays using a concentration of 3H leucine near the Kd values for the WT and mutant transporters in both MNG-3 and C12M detergents (Fig 1; supplementary Fig S1 online). In both detergents we find that for LeuT WT, CMI slows the rate at which the WT transporter reaches equilibrium, requiring ∼20 h for complete equilibration. By contrast, the F253A mutant shows no similar lag to reaching equilibrium in the presence of CMI. Taken together, these experiments define appropriate times at which to carry out saturation ligand binding experiments, values that we then used in the aforementioned equilibrium binding assays.
Nevertheless, because we observed a modest yet reproducible decrease in Bmax on preincubation of LeuT with CMI, we probed the solubility of LeuT in the presence of CMI. Owing to the fact that the absorbance spectrum of CMI overlaps with that of LeuT we cannot simply measure A280 to estimate protein concentration. Therefore, we incubated WT and the F253A mutant of apo LeuT in the absence and presence of 1 mM CMI and estimated the amount of LeuT protein remaining in the solution by integration of protein elution profiles following size-exclusion chromatography and monitoring protein presence by tryptophan fluorescence [26]. We found that after preincubating with CMI, the peak areas of LeuT WT in MNG-3 and C12M decreased by 12% and 15%, respectively, whereas for LeuT F253A, the peak areas decreased by 20% and 30% in MNG-3 and C12M, respectively (supplementary Fig S3 online). Thus our results provide one possible explanation for why preincubation with CMI leads to a decrease in Bmax for 3H-leucine binding.
Structure of LeuT F253A with leucine and SeMet
We next set out to unambiguously visualize the location of substrate bound to the F253A mutant of LeuT by crystallizing the substrate-bound LeuT in bilayer-like bicelles. To do this, we purified the F253A mutant in the presence of the substrate leucine or in the presence of the anomalously scattering substrate analogue SeMet (supplementary Fig S4 online). We obtained crystals of both complexes and solved the resulting structures of the leucine and SeMet complexes to resolutions of 2.6 and 3.0 Å, respectively (Table 1). Both structures were refined to satisfactory crystallographic statistics, and analysis of omit maps at the position of residue 253 clearly showed the presence of an alanine residue (supplementary Fig S5 online). Moreover, both structures are very similar to each other, to the original LeuT structure (PDB code 2A65) determined in detergent micelles, and to the recently determined structure of WT LeuT in bicelles (PDB code 3USG, Table 2).
Table 1. Data collection and refinement statistics*.
| LeuT F253A–Leu | LeuT F253A–SeMet † | |
|---|---|---|
|
Data collection
| ||
| Wavelength (Å) | 0.970 | 0.979 |
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 122.1, 89.9, 81.3 | 122.8, 90.50, 82.0 |
| α, β, γ (°) | 90.0, 103.0, 90.0 | 90.0, 103.2, 90.0 |
| Resolution (Å) | 50.0–2.60 (2.69–2.60) | 30.0–3.00 (3.11–3.00) |
| Rmerge | 0.09 (0.53) | 0.11 (0.51) |
| I/σI | 13.0 (2.0) | 23.3 (3.4) |
| Completeness (%) | 97.9 (98.6) | 98.7 (99.8) |
| Redundancy | 5.0 (4.9) | 5.0 (5.2) |
| No. of reflections | 25,839 | 17,292 |
|
Refinement
| ||
| Resolution (Å) | 50.0–2.60 | 30.0–3.00 |
| Rwork/Rfree | 0.21/0.22 | 0.20/0.22 |
| No. of atoms | ||
| Protein | 3,968 | 3,962 |
| Ligand/ion | 1 Leu, 2 Na | 1 SeMet, 2 Na |
| Water | 64 | — |
| B-factors (Å2) | ||
| Protein | 51.4 | 75.6 |
| Ligand/ion | 44.3 | 74.7 |
| Water | 51.7 | — |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | 0.010 |
| Bond angles (°) | 0.542 | 0.578 |
| *Values in parentheses correspond to the highest resolution shells. | ||
| †SeMet: the substrate analogue selenomethionine. | ||
Table 2. Comparison of LeuT F253A and LeuT wild-type (WT) crystal structures*.
| LeuT β-OG † | LeuT–Leu ‡ | LeuT F253A–Leu | LeuT F253A–SeMet | |
|---|---|---|---|---|
| LeuT β-OG | 0 | |||
| LeuT–Leu | 0.72 | 0 | ||
| LeuT F253A–Leu | 0.74 | 0.10 | 0 | |
| LeuT F253A–SeMet | 0.75 | 0.12 | 0.11 | 0 |
| *Numbers in the table are r.m.s. deviation values for main chain atoms following superposition (Å). | ||||
| †LeuT β-OG: LeuT WT crystallized in n-octyl-β-D-glucopyranoside (PDB code 2A65). | ||||
| ‡LeuT–Leu: LeuT WT crystallized in DMPC-CHAPSO bicelles (PDB code 3USG). | ||||
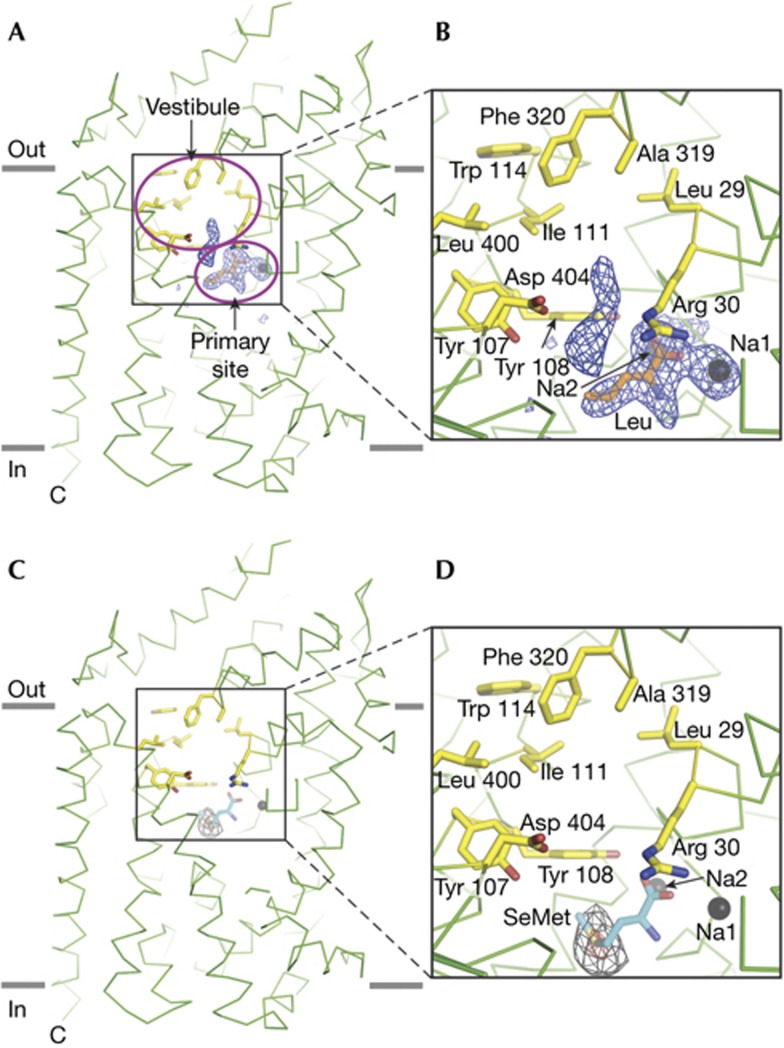
Inspection of difference electron density maps derived from the F253A mutant in complex with leucine clearly revealed density for a leucine molecule in the S1 binding site (Fig 2A,B). A single leucine molecule was readily fit into the S1 site electron density and subsequent crystallographic refinement proceeded smoothly. Immediately adjacent to the S1 site density for leucine and near the site of the F253A mutation, we observed sausage-shaped electron density (supplementary Fig S6A,B online). We attempted to model a leucine molecule to this electron density feature, but we obtained neither a satisfactory fit of the leucine structure to the density nor reasonable crystallographic refinement results (supplementary Fig S7 online). After refinement of a second leucine molecule, we consistently observed prominent difference electron density peaks and B-factors approximately twofold higher than those of the surrounding atoms. We observed no other prominent electron density features in or near the putative S2 substrate site.
Figure 2.
Substrate binds at the S1 site of the LeuT F253A mutant. (A) ‘Omit’ electron density difference map for the primary binding site and the base of the extracellular vestibule in the LeuT F253A complex with leucine. The substrate leucine in the S1 site is in stick representation, and two sodium ions are shown as grey spheres. Key residues in the vestibule are in stick representation. The Fo-Fc difference electron density map is displayed at 3.5σ, depicted in blue mesh and calculated with the substrate leucine and the two sodium ions omitted from the structure factor calculation. (B) Close-up of primary binding site and the base of the extracellular vestibule. (C) Anomalous difference Fourier electron density map for the primary substrate site and extracellular vestibule derived from diffraction data measured from LeuT F253A–SeMet cocrystals. The map is contoured at 4σ and 12σ and depicted in black and red mesh, respectively. SeMet (cyan) was positioned in the primary site and is shown in stick representation. Sodium ions Na1 and Na2 are illustrated as grey spheres. (D) Close-up of primary binding site and the base of the extracellular vestibule.
To more sensitively visualize bound substrate to the F253A mutant, we analysed anomalous difference electron density maps derived from the complex with SeMet, a substrate analogue [14, 16]. Binding experiments show that 1 mM SeMet can suppress the specific binding of leucine to the protein to the same extent as 5 mM Ala (supplementary Fig S4 online), suggesting SeMet is a faithful analogue of substrate as previously proposed [16]. Inspection of these maps showed a 17σ peak in the S1 binding site that could readily be fit to the Se atom of a SeMet molecule in the S1 site (Fig 2C,D), thus providing further evidence that substrate binds to the S1 site of the F253A mutant. Further scrutiny of the anomalous difference maps, especially in the region of the putative S2 site and the extracellular vestibule, revealed no additional peaks >3.5σ.
Discussion
We have carried out simple and straightforward experiments to examine the substrate binding properties of the WT and F253A mutant of LeuT. In both MNG-3 and C12M detergents, we performed direct substrate binding experiments in the presence and absence of CMI. Consistent with previous results [15], we demonstrate that the non-competitive inhibitor CMI affects only the kinetics of substrate binding, and that Bmax is not substantially changed in the presence of CMI for LeuT WT. To explain the discrepancy between the Gouaux and Javitch groups on the effect of 1 mM CMI on substrate binding stoichiometry for LeuT WT, we show in our time-course experiment that if time points are taken early, before equilibration, the data erroneously indicate that CMI competes for 3H-leucine binding. The unchanged leucine Kd for LeuT WT in the presence of CMI suggests that ‘on’ and ‘off’ rates were affected to the same extent. The CMI effect on the leucine binding time course might also explain the apparently non-competitive binding of other tricyclic compounds to LeuT reported previously [23].
More importantly, we demonstrate that inclusion of CMI in the binding experiment does not ablate binding of leucine to the F253A mutant, in contradiction to the results of Javitch and colleagues [18, 21]. Interestingly, the apparent kinetics of leucine binding to the F253A mutant are not altered by the presence of CMI. We speculate that the off-rate of substrate is faster for the F253A mutant in comparison to LeuT WT and thus the binding reaction comes to equilibrium faster than that observed for the WT transporter. We further observed different substrate binding properties in MNG-3 and C12M with and without CMI. In MNG-3, the Kd for leucine binding to F253A is ∼400 nM, and both the Kd and Bmax are not changed by inclusion with CMI. By contrast, when the F253A mutant is prepared in C12M, the Kd is ∼100 nM and, on inclusion of CMI, the Kd increases by approximately fivefold. In addition, when the protein is preincubated with CMI, the Bmax decreases by ∼20%. This last result suggests that consequences of incubating LeuT with CMI are more pronounced in C12M than in MNG-3, and that MNG-3 is a superior detergent for LeuT ligand binding studies. Our fluorescence-detection size-exclusion chromatography experiment (supplementary Fig S3 online) further demonstrates that 1 mM CMI can diminish the protein concentrations to a greater extent in C12M than in MNG-3, an observation that is also consistent with the notion MNG-3 better stabilizes apo LeuT [24]. The mechanism by which CMI affects the solubility of LeuT is presently unclear. Nevertheless, we note that earlier cocrystal structures of LeuT with CMI revealed several binding sites [15], thus suggesting the possibility that CMI could bind to several sites on LeuT and perturb its solubility. Thus, not only does CMI reduce the solubility of LeuT, but it also decreases the binding affinity for leucine—effects most pronounced for apo LeuT in C12M—therefore providing possible explanations why the Javitch laboratory does not see binding of 3H leucine to the F253A mutant in the presence of CMI.
The undeniable electron density for substrate in the S1 site of LeuT F253A mutant provides new and reinforcing evidence for a single high-affinity substrate binding site in LeuT. While the position of unmodeled density in the extracellular vestibule is somewhat larger in the mutant structure than in the WT structures (supplementary Fig S6A,B online), we note that the density feature is not located near the putative S2 binding site defined by Ile 111 and Leu 400 (supplementary Fig S7A online) [19]. Moreover, the mutation of phenylalanine to alanine creates space for the nonspecific binding of small non-substrate molecules, such as water, precipitant molecules or the ‘tails’ of lipids. Most importantly, in the SeMet cocrystal structure, we see no evidence for SeMet in the S2 site. We therefore suggest that the non-protein electron density feature near the S1 site is either a portion of a lipid acyl chain or a solvent molecule, as proposed before [16].
Taken together, our findings provide more evidence against the supposition that there is a second or S2 high-affinity substrate binding site in LeuT [18–22]. The results of the Javitch group related to the S2 site, the properties of the F253A mutant and the role of the putative S2 site in the mechanism of LeuT and NSSs are, at present, without satisfactory explanation. In contrast, the results reported here are in agreement with previous results from this group and with the conclusion that there is a single high-affinity substrate site in LeuT [6, 14, 16, 17]. Our results cast further doubt on the relevance of the S2 site to the mechanism of LeuT and NSSs.
Methods
Protein expression and purification. The LeuT WT and LeuT F253A mutant were expressed as described previously for LeuT WT [6, 17]. Cell membranes were solubilized using a buffer composed of 25 mM Tris–HCl (pH 8.0), 250 mM NaCl and 1% n-dodecyl-β-D-maltoside (C12M), and the protein was then purified by immobilized-metal affinity chromatography in the presence of 0.05% C12M and in the absence of substrate. For binding studies, unconcentrated protein from immobilized-metal affinity chromatography fractions was directly desalted by Zeba Desalt Spin Columns in buffer I (150 mM TrisMes, pH 7.5, 50 mM NaCl, 20% (vol/vol) glycerol, 1 mM TCEP (tris(2-carboxyethyl)phosphine) and 0.05% C12M) [19, 20].
Crystallization. For crystallization, 1 mM L-leucine or 1 mM L-SeMet was included in the size-exclusion chromatography buffer, and C12M was the only detergent utilized throughout the purification. Protein crystallization in bicelles was performed in a similar way as before [16]. LeuT F253A–Leu (8–10 mg ml−1) in the presence of 7% (wt/vol) DMPC-CHAPSO bicelles [27] was crystallized by vapour diffusion at 20 °C with the crystallization reservoir solution containing 100 mM sodium acetate (pH 4.7), 29% (vol/vol) MPD (2-methyl-2,4 pentanediol) and 10% (vol/vol) PEG400 and 50 mM MgCl2. LeuT F253A–SeMet crystals in the presence of 7% (wt/vol) DMPC-DMPE-CHAPSO bicelles [16] were grown in crystallization buffer with 100 mM sodium acetate (pH 4.7), 36% (vol/vol) MPD, 10% (vol/vol) PEG400 and 50 mM MgCl2. Crystals were directly flash frozen in liquid nitrogen before collection of X-ray diffraction data.
Data collection and structure elucidation. Diffraction data sets were collected at the Advanced Photon Source (Argonne National Laboratory, beamlines 24-ID-C and 24-ID-E) and Stanford Synchrotron Radiation Lightsource (beamline 12-2). Data sets were indexed, integrated and scaled using HKL2000 software [28]. The LeuT F253A–Leu and LeuT F253A–SeMet structures were determined by molecular replacement using the LeuT–Leu (PDB code 3USG) as a search probe using Phaser [29] in CCP4 suite [30]. The refinements were carried out using Phenix [31], and manual adjustments were made using Coot [32]. The structure quality analysis was carried out using Molprobity [33]. For both structures, Ramachandran geometry is excellent, with at least 97% of the residues in the most favoured regions and none in disallowed regions. Omit map was calculated by Phenix [31], with the substrate leucine and the two sodium ions omitted from the structure factor calculation. For the LeuT F253A–SeMet crystallographic analysis, the anomalous difference maps were calculated using the fast Fourier transform in the CCP4 suite [30]. All structure figures were generated with PyMOL (DeLano Scientific).
Binding assays. Scintillation proximity assays were performed as before [17] using 10 nM unconcentrated protein with 2 mg ml−1 Cu-Ysi SPA beads in both buffer I and buffer II (150 mM TrisMes, pH 7.5, 50 mM NaCl, 20% (vol/vol) glycerol, 1 mM TCEP and 0.02% lauryl MNG-3) in the presence of 0.3–3,000 nM [3H] Leu (21.7 Ci mmol−1). 1 mM CMI was added before ligand (1 h incubation), simultaneously with ligand or after the equilibrium was achieved (after 23 h) in the binding assays. Nonspecific binding was measured in the presence of 5 mM Ala. Plate readings were taken at every 4 h using a Wallac Microbeta plate counter. Data after 25–30 h incubation was analysed by GraphPad Prism4 and fit into a single-site binding function. Experiments were performed three times, each in triplicate.
Fluorescence-detection size-exclusion chromatography. Fluorescence-detection size-exclusion chromatography was performed as previously described [26]. After incubating with 1 mM CMI for 1 h and spinning down at 40 k r.p.m., 20 pmol LeuT WT or LeuT F253A in buffer I and buffer II was loaded onto a Superose-6 column preequilibrated with 20 mM Tris–HCl, pH 8.0, 150 mM NaCl and 1 mM C12M. The elution was monitored by a fluorometer, which was set as excitation at 295 nm and emission at 335 nm. The protein in the absence of CMI was used as control.
Supplementary Material
Acknowledgments
We thank M. Kavanaugh, D. Claxton, A. Penmatsa and K. Wang for comments, L. Vaskalis for assistance with illustrations and H. Owen for proofreading the manuscript. We also thank the beamline staff at the Advanced Photon Source (Argonne National Laboratory, beamlines 24-ID-C and 24-ID-E) and Stanford Synchrotron Radiation Lightsource (beamline 12-2). This work was supported by the National Institutes of Health. E.G. is an Investigator with the Howard Hughes Medical Institute.
Author contributions: H.W. and E.G. contributed to all aspects of the project.
Footnotes
The authors declare that they have no conflict of interest.
Accession codes. The atomic coordinates and merged structure factors for LeuT F253A–Leu and LeuT F253A–SeMet have been deposited in the Protein Data Bank with the accession codes 4FXZ and 4FY0.
References
- Hahn MK, Blakely RD (2002) Monoamine transporter gene structure and polymorphisms in relation to psychiatric and other complex disorders. Pharmacogenomics J 2: 217–235 [DOI] [PubMed] [Google Scholar]
- Richerson G, Wu Y (2004) Role of the GABA transporter in epilepsy. Adv Exp Med Biol 548: 76–91 [DOI] [PubMed] [Google Scholar]
- White K, Walline C, Barker E (2005) Serotonin transporters: implications for antidepressant drug development. AAPS J 7: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen L (2006) Neurotransmitter transporters and their impact on the development of psychopharmacology. Br J Pharmacol 147: 82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara SG, Sonders MS (1998) Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend 51: 87–96 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437: 215–223 [DOI] [PubMed] [Google Scholar]
- Boudker O, Verdon G (2010) Structural perspectives on secondary active transporters. Trends Pharmacol Sci 31: 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Kramer R, Ziegler C (2011) The structural basis of secondary active transport mechanisms. Biochim Biophys. Acta (BBA)—Bioenergetics 1807: 167–188 [DOI] [PubMed] [Google Scholar]
- Kanner BI, Zomot E (2008) Sodium-coupled neurotransmitter transporters. Chem Rev 108: 1654–1668 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy H, Piscitelli CL, Gouaux E (2009) Unlocking the molecular secrets of sodium-coupled transporters. Nature 459: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63: 585–640 [DOI] [PubMed] [Google Scholar]
- Rudnick G (2011) Cytoplasmic permeation pathway of neurotransmitter transporters. Biochemistry 50: 7462–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes N, Tavoulari S (2011) To be, or not to be two sites: that is the question about LeuT substrate binding. J Gen Physiol 138: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Piscitelli CL, Yamashita A, Gouaux E (2008) A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322: 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E (2007) Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448: 952–956 [DOI] [PubMed] [Google Scholar]
- Wang H, Elferich J, Gouaux E (2012) Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol 19: 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscitelli CL, Krishnamurthy H, Gouaux E (2010) Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature 468: 1129–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Shi L, Zehnpfennig B, Weinstein H, Javitch JA (2012) Experimental conditions can obscure the second high-affinity site in LeuT. Nat Struct Mol Biol 19: 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Winther A-ML, Shi L, Nissen P, Weinstein H, Javitch JA (2009) Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci 106: 5563–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA (2008) The Mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell 30: 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, Javitch JA (2011) Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature 474: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA (2010) Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465: 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith MEA, Wang DN (2007) LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317: 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae PS et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. (2010) Nat Meth 7: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Javitch JA (2007) Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci 104: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Gouaux E (2006) Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14: 673–681 [DOI] [PubMed] [Google Scholar]
- Faham S, Bowie JU (2002) Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol 316: 1–6 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected inoscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Adams PD et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.