Abstract
Current research on the cytokine-mediated signalling towards the polarization and differentiation of a T-helper cell lineage lacks mechanistic insights on the transcriptional regulation of cytokine receptor genes. Here, we propose a new mechanism for the transcriptional regulation of the interferon gamma receptor 1 gene via long-range intrachromosomal interactions with the Ifnγ locus mediated by the protein CTCF. These interactions sustain the monoallelic expression of the differentially methylated IfnγR1 gene and are persistent on blockade of active transcription. Our findings suggest that regulatory elements for a cytokine gene locus can also positively regulate the transcription of its receptor.
Keywords: CD4+ cells, ChIP-loop, IFN-γ signalling, RNA–DNA FISH, 3C
Introduction
Naïve CD4+ T cells can differentiate into different cell lineages such as the T-helper type 1 (TH1), TH2 and TH17 cells, depending on the antigenic stimulus and the cytokine milieu they encounter. These discrete differentiation programmes are initiated and maintained based on the master transcriptional regulators expressed by the different cell subtypes and the signature cytokines secreted [1].
T-cell receptor-mediated activation of naïve CD4+ T cells leads to transcriptional activation of GATA3, signal transducer and activator of transcription (STAT)6 activation and interleukin-2 (IL-2) production. Their cumulative effect is the production of IL-4 during the initiation of the TH2 polarization process [2]. On the other hand, the TH1-specific differentiation programmes are initiated by STAT1, which is activated in response to interferon-γ (IFN-γ) and IL-27. STAT1 activates T-bet, the master transcriptional activator of the TH1 cell lineage, which upregulates the production of IFN-γ and the expression of the IL-12 receptor-β2 [3]. Crosslinking of the IL-12 cytokine to its receptor leads to the activation of STAT4, which together with T-bet bind to regulatory elements of the Ifnγ gene locus and activates Ifnγ expression [4, 5]. Thus, this positive feedback loop regulates the generation of TH1 cells.
Importantly, in TH1 cells, STAT1 activation on IFN-γ engagement requires a functional IFN-γ receptor consisted of two subunits, namely the IFN-γR1 (ligand-binding chain) and IFN-γR2 [6, 7]. The IFN-γ–STAT1 signalling pathway leads to antiproliferative effects on a wide range of cell types preventing tumour progression and killing of pathogen-infected cells [6]. In contrary, TH1 cells that are among the main producers of IFN-γ are shielded from its antiproliferative effects, owing to downregulation of the IFN-γR2 chain.
So far, research concerning TH1 cell differentiation programmes has focused mainly on specific epigenetic changes regulating the expression of the Ifnγ gene locus and the master transcriptional regulator T-bet [8], but there is limited information about the transcriptional regulation of the Ifnγ receptor genes, which are of fundamental importance, as their expression leads to proper IFN-γ–STAT1 signalling and thus TH1 cells polarization.
In this report, we present a long-range monoallelic intrachromosomal interaction between the Ifnγ and IfnγR1 gene loci in non-differentiated CD4+ cells and cells of the TH1 cell lineage but not in TH2 cells. Chromosome conformation capture (3C) experiments revealed that the IfnγR1 gene promoter comes in close proximity with the Ifnγ gene as well as previously characterized downstream regulatory elements of the latter. Remarkably, this monoallelic DNA interaction supports the monoallelic expression of the IfnγR1 gene. Such an intrachromosomal interaction is not dependent on active transcription as it is not affected on RNA Polymerase II blockade in IfnγR1-expressing cells. CTCF protein, whose IfnγR1 binding might be facilitated by T-bet, mediates the Ifnγ–IfnγR1 interaction. In addition, the monoallelic IfnγR1 interaction and expression could be owing to the differential methylation of its promoter region. The mechanism we present here, where regulatory elements for a cytokine gene locus might directly regulate the expression of the cytokine receptor gene, provides insights as of how a positive feedback regulatory loop, responsible for TH1 cell differentiation, is regulated at the transcription level.
Results and Discussion
Cell-specific colocalization of Ifnγ and IfnγR1 loci
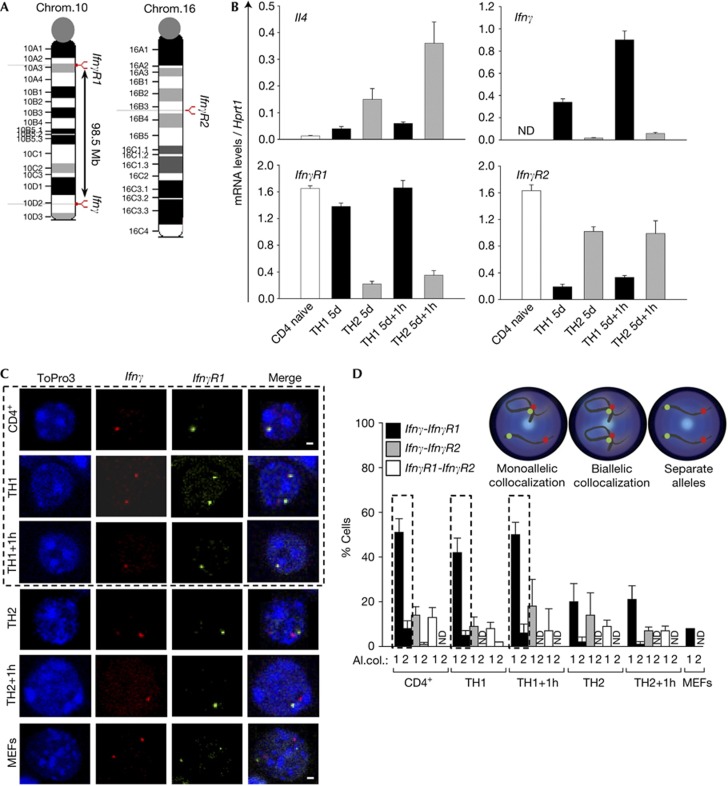
The aim of our study was to decipher regulatory mechanisms that govern the transcriptional activation of the IfnγR1 and IfnγR2 genes located on mouse chromosomes 10 and 16, respectively (Fig 1A). In the present study, we have used the well-established in vitro differentiation system of CD4+ T cells into the TH1 and TH2 cell lineages. Initially, we analysed the expression profile of the signature cytokine genes Il4 and Ifnγ that were found to be specifically expressed in TH2 and TH1 cells, respectively, as expected (Fig 1B). We then analysed the messenger RNA expression levels of the two IfnγR genes. IfnγR1 was highly expressed in naive CD4+ cells and TH1 cells while IfnγR2 was highly expressed in naive CD4+ and TH2 cells (Fig 1B).
Figure 1.
Monoallelic intrachromosomal colocalization of the Ifnγ-IfnγR1 gene loci. (A) Schematic representation of the Ifnγ and IfnγR1 gene loci. (B) Quantitative RT–PCR analysis for the mouse Il4, Ifnγ, IfnγR1 and IfnγR2 genes. Results are the mean±s.e.m. from triplicate samples of four independent experiments. (C) Single optical sections from confocal analysis of DNA-FISH. (D) Quantitative analysis of the DNA-FISH experiments presented in (C) and supplementary Fig S3A,B online. Al.col., allelic colocalization (1=a cell with one allele of each locus colocalized, 2=a cell with both alleles of one locus colocalized with both alleles of the other locus). DNA-FISH, DNA fluorescence in situ hybridization; MEFs, mouse embryonic fibroblasts; mRNA, messenger RNA; ND, not detected; RT–PCR, reverse transcriptase PCR.
To study the subnuclear localization pattern of the Ifnγ cytokine gene and its receptor gene loci IfnγR1 and IfnγR2, we performed DNA fluorescence in situ hybridization (DNA-FISH). We performed DNA-FISH for the Ifnγ and IfnγR1 gene loci (Fig 1C). We counted the percentage of cells with either mono- or biallelic colocalization of the two loci. The most CD4+ cells and cells of the TH1 cell lineage showed monoallelic versus biallelic colocalization. On the contrary, decreased levels of colocalization were detected in the TH2 cell lineage and mouse embryonic fibroblasts (MEFs) (Fig 1C,D). On the basis of the quantitative reverse transcriptase PCR (RT–PCR) results (Fig 1B), we observed that the IfnγR1-expressing cell types (CD4+, TH1 cells) showed considerable levels of colocalization between the Ifnγ and IfnγR1 gene loci. Therefore, we speculated that there might be an underlying mechanism for the transcriptional regulation of the IfnγR1 gene from the Ifnγ locus.
The cell volume of the different cell types under study was similar (supplementary Fig S1A online). We also measured the distance between the two proximal signals of the Ifnγ-IfnγR1 alleles as well as for the more distant ones, and we found that the median for the distance of the proximal alleles was much lower in CD4+ cells and TH1 cells compared with the TH2 cells (supplementary Fig S1B online). The Kolmogorov–Smirnov test for the distribution of allele distances in the different T-cell types confirmed these results (supplementary Fig S2 online). Therefore, we concluded that the differences in the percentage of cells with colocalized Ifnγ-IfnγR1 alleles were not due to differences in the cell volume of the different cell types but colocalization rather represented a cell-specific effect.
To extend our hypothesis about long-range chromosomal interactions regulating gene expression, we performed DNA-FISH experiments for the Ifnγ and IfnγR2 gene loci and found that only a low percentage of cells showed colocalization of the two loci (Fig 1D and supplementary Fig S3 online). In line with the previous experiments, we performed DNA-FISH experiments for the IfnγR1 and IfnγR2 gene loci, and the percentage of cells with colocalized alleles was also low (Fig 1D; supplementary Fig S3B online).
Our next question was whether the colocalization between the Ifnγ and IfnγR1 gene loci, which both lie on the same chromosome, was intra- or interchromosomal. To answer this question, we performed DNA-FISH for the Ifnγ and IfnγR1 gene loci in combination with chromosome painting for mouse chromosome 10 (supplementary Fig S3C online). We found the interaction to occur between the two loci within the same chromosome in 100% of CD4+ cells and in 99% of the TH1 cells examined. Therefore, the colocalization observed between the Ifnγ and IfnγR1 gene loci is intrachromosomal.
Physical proximity of the Ifnγ and IfnγR1 gene loci
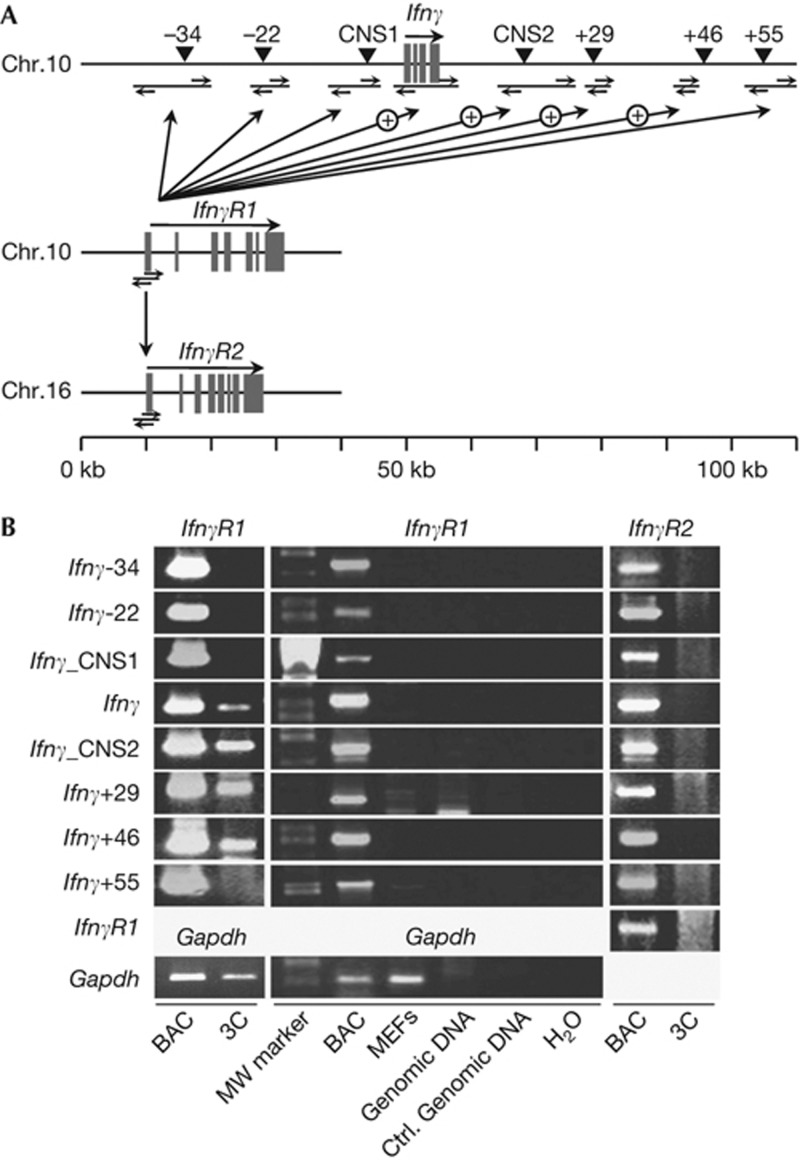
Complementary to our DNA-FISH results, to identify the interacting DNA elements between the Ifnγ and IfnγR1 gene loci, we performed the 3C assay. We digested the chromatin of non-differentiated CD4+ T cells to generate genomic fragments that contained known regulatory elements [8–10]. We detected specific interactions between the IfnγR1 promoter-containing fragment and various fragments mapping on the Ifnγ locus containing the Ifnγ gene, the conserved non-coding sequence 2, and two regions with DNAse I hypersensitive sites +29 and +46 kb downstream of the Ifnγ transcription start site (Fig 2).
Figure 2.
Long-range intrachromosomal physical proximity of the Ifnγ-IfnγR1 gene loci revealed by 3C. (A) Schematic representation of the loci used for the 3C analysis. (B) 3C analysis in non-differentiated CD4+ cells for the Ifnγ-IfnγR1, Ifnγ-IfnγR2, IfnγR1-IfnγR2 and Gapdh gene loci. Representative PCR reactions (one out of four) for each pair of fragments analysed.BAC, positive control using digested and ligated BAC DNA; Ctrl. Genomic DNA, naked mouse genomic DNA digested and ligated; Gapdh, 3C analysis of two closely located (<1 kb) fragments of the mouse Gapdh gene locus. MEF, mouse embryonic fibroblasts (3C template prepared from mouse embryonic fibroblasts); 3C, chromosome conformation capture (CD4+ cells used as template).
Ifnγ-receptor genes are monoallelically expressed
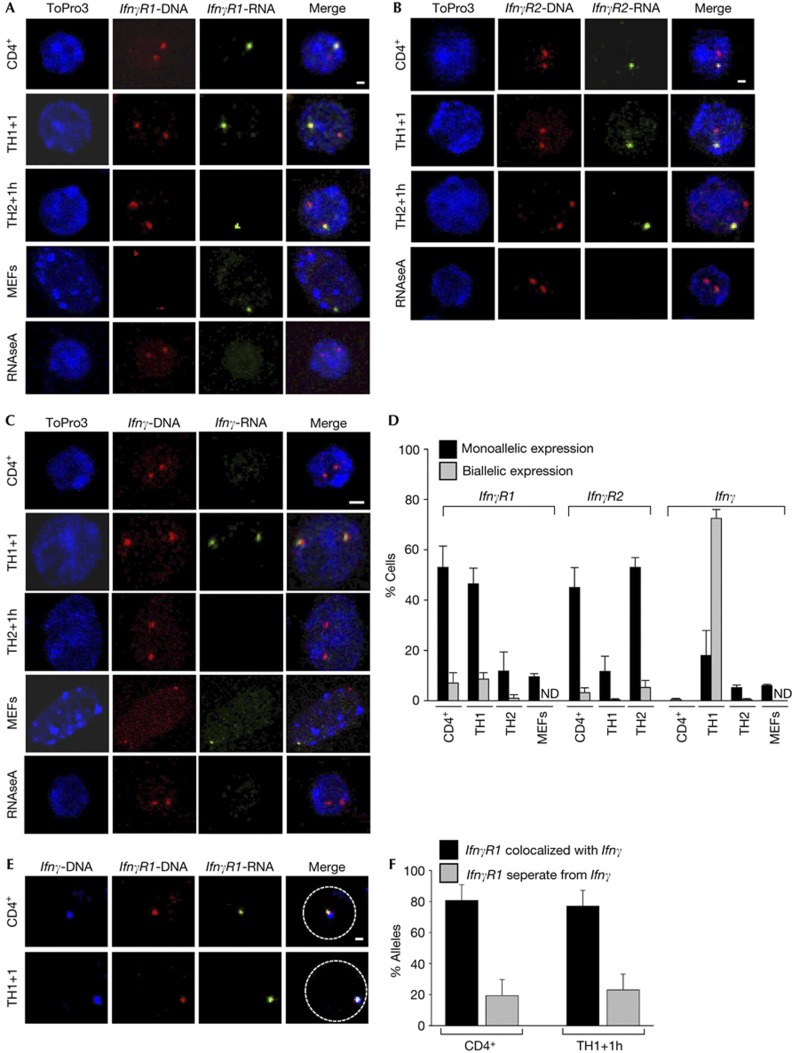
The 3C technology is based on formaldehyde crosslinking and is able to detect close physical interactions mediated by protein factors in the range of 2.4Å distance [11]. Therefore, we speculated that the IfnγR1 gene might use the interacting regulatory elements of the Ifnγ locus for its transcriptional regulation. To test this hypothesis, we studied the allelic expression profile of the IfnγR1 gene using RNA–DNA FISH experiments and found that IfnγR1 was expressed mainly by non-differentiated CD4+ cells and TH1 cells in a monoallelic manner (Fig 3A,D). Importantly, the total mRNA levels of the IfnγR1 gene as quantitated with the RT–PCR experiments (Fig 1B) were in accordance with its allelic expression profile as deduced by the RNA–DNA FISH experiments.
Figure 3.
Monoallelic IfnγR1 gene expression from the allele colocalized with the Ifnγ locus. (A) RNA–DNA FISH for the IfnγR1 gene. Scale bar, 1 μm for T-cell types and 1.5 μm for MEFs. (B) RNA–DNA FISH for the IfnγR2 gene. Scale bar, 1 μm. (C) RNA–DNA FISH for the Ifnγ gene. Scale bar, 2 μm for T-cell types and 2.5 μm for MEFs. (D) Quantitative analysis for the experiments depicted in (A–C). (E) DNA-FISH for the Ifnγ and IfnγR1 gene loci combined with RNA-FISH for the IfnγR1 gene. Scale bar, 1 μm. (F) Quantitative analysis for the IfnγR1-expressing alleles from (E). DNA was counterstained with ToPro3 and pseudocolored blue. DNA-FISH, DNA fluorescence in situ hybridization; MEFs, mouse embryonic fibroblasts; RNA-FISH, RNA fluorescence in situ hybridization.
Next, we studied the allelic expression profile of the IfnγR2 gene by RNA–DNA FISH experiments (Fig 3B) and found that, in accordance with the RT–PCR results, non-differentiated CD4+ cells and TH2 cells monoallelically expressed IfnγR2 (Fig 3D). We concluded that the IfnγR1 and IfnγR2 genes are alternatively expressed in TH1 and TH2 cells in a monoallelic manner while they are both expressed in non-differentiated CD4+ cells. As far as IFN-γ expression pattern is concerned, it was found to be expressed mainly in a biallelic manner in TH1 cells (Fig 3C,D).
To gain insight into the functional significance of the intrachromosomal interaction between the Ifnγ-IfnγR1 gene loci and the regulation of the monoallelic IfnγR1 gene transcription, we performed DNA-FISH for the two loci combined with RNA fluorescence in situ hybridization for the IfnγR1 gene (Fig 3E). We found that a high percentage of the IfnγR1-expressing alleles colocalized with the Ifnγ locus in non-differentiated CD4+ cells and in TH1 cells (Fig 3F). Taken together, our data suggest that the monoallelic interaction between the Ifnγ and IfnγR1 gene loci is highly correlated with the monoallelic expression of the latter.
Ifnγ-IfnγR1 interactions are independent of transcription
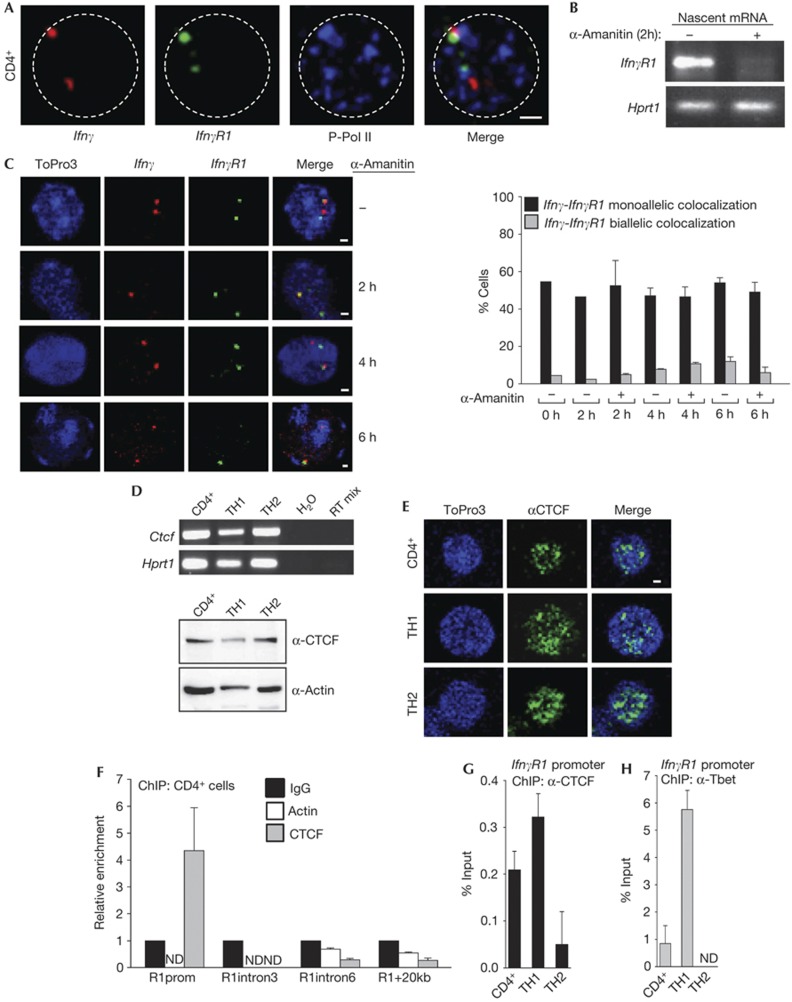
A common mechanism of transcriptional regulation is that coregulated gene loci colocalize with RNA Polymerase II (RNA-PolII) factories [12] by looping out of their chromosome territories. To test this hypothesis, we performed immunohistochemistry experiments with specific antibodies for the phosphorylated and active form of RNA-PolII, in combination with DNA-FISH for the Ifnγ and IfnγR1 gene loci in non-differentiated CD4+ T cells. We found that 75% of the Ifnγ alleles that monoallelically colocalized with the IfnγR1 locus interacted with RNA PolII factories. Moreover, 88% of the IfnγR1 alleles monoallelically colocalizing with the Ifnγ locus interacted with RNA PolII factories (Fig 4A). Statistical analysis of the experiment revealed that there is a statistically significant tendency (P-value=0.01) for the colocalizing Ifnγ-IfnγR1 alleles to interact with RNA PolII factories. As the IfnγR1 gene is expressed in these cells, it is possible that regulatory elements of the Ifnγ locus are utilized by the IfnγR1 gene to positively regulate its transcription.
Figure 4.
IfnγR1 promoter binds CTCF and colocalizes with Ifnγ in RNA-PolII factories. (A) DNA-FISH for the Ifnγ and IfnγR1 gene loci in combination with immunofluorescence for the phosphorylated form of RNA Polymerase II in CD4+ cells. (B) Semiquantitative RT–PCR for the nascent IfnγR1 transcript in CD4+ cells treated with 50 μg/ml α-Amanitin. (C) DNA-FISH analysis for Ifnγ-IfnγR1 loci in CD4+ cells treated with 50 μg/ml α-amanitin. Scale bar, 1 μm. (D) Semiquantitative RT–PCR for the Ctcf gene. Lower panel: western blot analysis for the CTCF protein. (E) Immunofluorescence analysis for CTCF. (F) Quantitative PCR analysis for CD4+ cells on chromatin immunoprecipitation. Primer sets are specific for the IfnγR1 promoter/intron 3/intron 6 and a region 20-kb downstream the IfnγR1 gene. (G) Quantitative PCR analysis on chromatin immunoprecipitation using a CTCF antibody and primers specific for the promoter region of the IfnγR1 gene. (H) Quantitative PCR analysis as in (G) using an antibody against T-bet. ChIP, chromatin immunoprecipitation; DNA-FISH, DNA fluorescence in situ hybridization; mRNA, messenger RNA; RT–PCR, reverse transcriptase PCR.
Moreover, we wanted to test if the Ifnγ-IfnγR1 loci were found colocalized owing to the transcription process, or whether colocalization was an active process that takes place to facilitate gene regulation. Towards this direction, we treated non-differentiated CD4+ cells with the RNA polymerase inhibitor α-amanitin. We performed semiquantitative RT–PCR experiments for the IfnγR1 nascent RNA and found that 2-h treatment of the cells with α-Amanitin was enough to block its transcription (Fig 4B). We then performed DNA-FISH for the Ifnγ and IfnγR1 gene loci and found that the colocalization was also not affected by the addition of α-Amanitin (Fig 4C). Therefore, blocking of transcription by RNA PolII inhibition did not disrupt Ifnγ–IfnγR1 intrachromosomal interactions, but negatively affects IfnγR1 expression. A possible explanation is that once the long-range interactions are formed on transcription factories they are stable and other protein factors exist that are responsible for their maintenance. Taking into account these results, we conclude that the Ifnγ–IfnγR1 interactions take place in RNA PolII factories but are independent of active transcription of the two loci.
CTCF-mediated intrachromosomal interactions
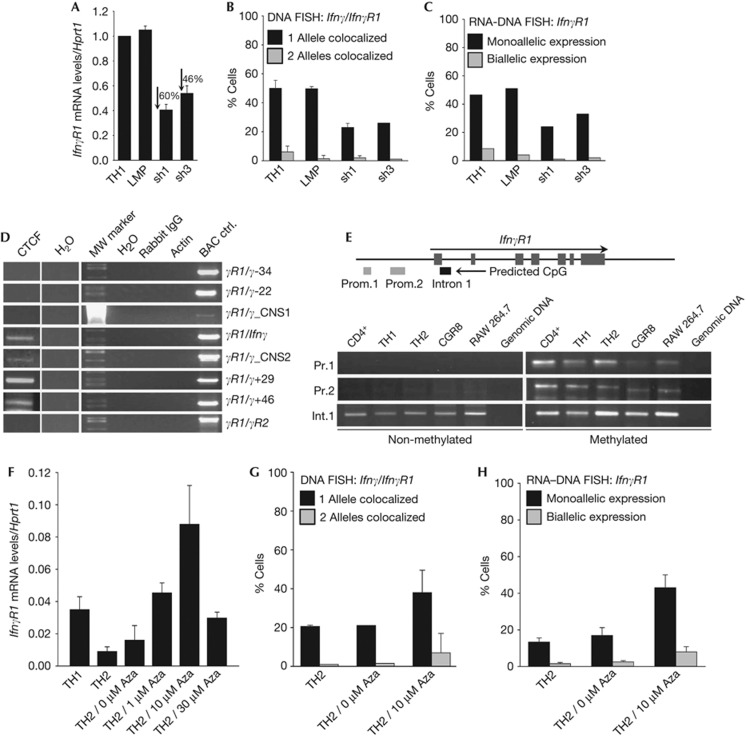
Long-range chromosomal interactions are mediated by transcription factor complexes and proteins with a general chromatin organizing activity [13], such as CTCF [14], so we tested whether CTCF is involved in the regulation of the Ifnγ–IfnγR1 interactions. CTCF was expressed in all T-cell lineages under study (Fig 4D,E). Therefore, although CTCF was expressed in all T-cell types examined, the Ifnγ–IfnγR1 interaction was detected only in non-differentiated CD4+ cells and TH1 cells. To test for differences in the TH2 cells, we performed chromatin immunoprecipitation (ChIP) experiments for CTCF and found that it was recruited on the IfnγR1 promoter in non-differentiated CD4+ cells (Fig 4F), TH1 cells and in much lower levels in TH2 cells (Fig 4G). We also performed ChIP against T-bet and found that it was also recruited on the IfnγR1 promoter in the same cell types as CTCF (Fig 4H). More importantly, targeting of CTCF mRNA using retrovirally expressed short-hairpin RNAs in TH1 cells (supplementary Fig S4 online) resulted in the reduction of the IfnγR1 mRNA levels (Fig 5A), the reduction in the colocalization of the Ifnγ-IfnγR1 gene loci (Fig 5B) and also the reduction of the IfnγR1-expressing TH1 cells (Fig 5C).
Figure 5.
CTCF mediates the Ifnγ–IfnγR1 intrachromosomal interactions in CD4+ T cells. (A) Quantitative RT–PCR for the IfnγR1 mRNA levels in TH1 cells before and after retroviral transduction of short-hairpin RNAs targeting CTCF. Results are the mean±s.e.m. from triplicate samples from one representative of three independent experiments. (B) DNA-FISH for the Ifnγ-IfnγR1 loci in cells as in (A). Bars are mean values for the percentage of cells with s.d. from six independent experiments for TH1 cells, three experiments for TH1/LMP cells and three experiments for TH1/sh1-CTCF cells. One representative of two conducted is shown for TH1/sh3-CTCF cells. A total of 1178 cells have been scored in total. (C) RNA–DNA FISH for the IfnγR1 gene in cells as in (B). Bars indicate the percentage of cells from one representative experiment of two conducted for each treatment. A total of 568 cells have been scored in total. (D) ChIP-loop analysis using a CTCF and control antibodies and primers designed to detect physical interactions between fragments of the Ifnγ locus and the promoter region of the IfnγR1 gene. (E) PCR analysis of bisulphate-treated genomic DNA [19]. (F) Quantitative RT–PCR for the IfnγR1 mRNA levels in TH2 cells treated with increased concentrations of the DNA demethylating agent 5-Aza-2′deoxycytidine. Results are the mean±s.e.m. from triplicate samples from one representative of three independent experiments. (G) DNA-FISH for the Ifnγ-IfnγR1 loci in TH2 cells, before and after treatment with 5-Aza-2′deoxycytidine. Bars are mean values for the percentage of cells with s.d. from seven independent experiments for TH2 cells and three experiments for TH2 10 μM Aza. The results for TH2 0 μM Aza-treated cells are from one representative experiment of two conducted for this treatment. A total of 1248 cells have been scored in total. (H) RNA–DNA FISH for the IfnγR1 gene in TH2 cells as (G). ChIP, chromatin immunoprecipitation; DNA-FISH, DNA fluorescence in situ hybridization; LMP, empty retroviral vector control; mRNA, messenger RNA; RT–PCR, reverse transcriptase PCR.
To test the hypothesis that CTCF protein is recruited on these loci and allows the Ifnγ–IfnγR1 interaction, we performed a ChIP-loop assay [15]. The assay was based on CTCF immunoprecipitation of chromatin from non-differentiated CD4+ cells and subsequent 3C analysis. We could detect the CTCF-bound chromatin fragments between the IfnγR1 promoter fragment and the same chromatin regions of the Ifnγ gene locus (Fig 5D) as we have previously detected with the 3C approach (Fig 2). Taken together, the above experiments clearly demonstrate the participation of CTCF in the IfnγR1 expression and in the intrachromosomal Ifnγ–IfnγR1 interactions.
As these results support the hypothesis that CTCF is involved in mediating the Ifnγ–IfnγR1 interactions, we wondered if this could be explained by the differential recruitment of CTCF onto one of the two IfnγR1 alleles owing to differential DNA methylation. Therefore, we employed bisulphite modification of DNA and we found that a CpG-predicted IfnγR1 promoter region was both non-methylated and methylated (Fig 5E). These results correlate well with the monoallelic expression of the IfnγR1 gene and suggest that the non-methylated IfnγR1 allele might allow CTCF and T-bet binding that in turn mediate a monoallelic intrachromosomal interaction with the Ifnγ locus. To further support the latter findings, we studied the effect of 5-Aza-2′-deoxycytidine (5-Aza), a drug known to cause DNA demethylation, on IfnγR1 transcriptional regulation in TH2 cells, which lack considerable IfnγR1 mRNA expression and Ifnγ-IfnγR1 colocalization. Indeed, we found that 5-Aza treatment of TH2 cells resulted in increased IfnγR1 mRNA levels (Fig 5F), increased Ifnγ-IfnγR1 intrachromosomal colocalization (Fig 5G) and increased percentage of cells with mono- and biallelic expression of the IfnγR1 gene as shown by RNA–DNA FISH experiments (Fig 5H).
On the basis of our data, we propose a model as of how a cytokine receptor gene is transcriptionally regulated in the three-dimensional nucleus by regulatory elements necessary for the proper expression of the cytokine gene itself. We suggest that in non-differentiated CD4+ T cells and TH1 cells there is a long-range intrachromosomal interaction where, upon looping out the intervening sequences, the IfnγR1 gene comes in close proximity to the Ifnγ gene and its downstream regulatory elements in order to be expressed. Differential methylation in the promoter region of the IfnγR1 gene determines the T-bet- and CTCF-specific binding, and sets a pattern of monoallelic colocalization and positive transcriptional regulation (supplementary Fig S5 online).
Methods
DNA fluorescence in situ hybridization. Cells were attached onto poly-L-lysine-coated coverslips, fixed with 4% paraformaldehyde/1 × phosphate-buffered saline (PBS) for 12 min, washed three times with 1 × PBS and permeabilized with 0.5% Triton X-100/1 × PBS for 10 min. Cells were then equilibrated with 20% glycerol/1 × PBS for 30 min and were subsequently freeze–thawed for three cycles in liquid nitrogen. Histones were removed by 5-min incubation in 0.1 N HCl, rinsed in 2 × SSC and stored at 70% ethanol overnight. Hybridization was carried out in 50% formamide, 2 × SSC, 10% dextran sulphate, 1 μg mouse Cot-1 (Invitrogen) and 100 ng fluorescently labelled BAC probe, overnight at 37°C. Slides were washed three times for 5 min each in 2 × SSC and mounted in Prolong Gold antifade reagent with DAPI (Invitrogen). For the generation of fluorescently labelled BAC DNA probes ((Ifnγ, RP24-352N22), (IfnγR1, RP23-238E1), (IfnγR2, RP23-276H8)), 2μg of DNA were labelled using a Nick translation kit (Vysis) and either 0.025 mM Spectrum Orange dUTP, 0.025 mM Spectrum Green dUTP (Vysis) or 0.025 mM OBEA dCTP-647 (Invitrogen).
RNA–DNA FISH. Cells attached onto poly-L-lysine-coated coverslips were incubated for 3 min in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes, 0.5% TritonX-100, 1 mM EGTA and 2 mM vanadyl-ribonucleoside complex), fixed with 4% paraformaldehyde/1 × PBS for 10 min, washed three times with 70% ethanol and stored in 70% ethanol at −20°C overnight. Hybridization was done overnight at 37°C after a 5 min denaturation at 73°C. The coverslips were washed at 37°C sequentially with 2 × SSC/50% formamide, 2 × SSC, 1 × SCC and 4 × SSC. RNA signals were amplified using the Renaissance TSATM Biotin System (PerkinElmer). Nick-translated biotin-labelled cDNA for the IfnγR1, IfnγR2 and Ifnγ genes was used as a probe for the detection of the RNA signals.
3C and ChIP-loop assay. 3C assay was performed as previously described [16, 17]. The 3C and ChIP experiments were combined for ChIP-loop as previously described [18]. In brief, on immunoprecipitation of digested chromatin, the complexes were eluted from the beads twice with 25 μl 10mM DTT, for 20 min at 37°C. Supernatants were diluted to obtain a DNA concentration of less than 3 ng/μl and ligation was performed overnight at 16°C. The samples were further treated with RNAse A at 37°C for 30 min and 250 μg/ml Proteinase K at 65°C for 6 h, followed by phenol/chloroform extraction and ethanol precipitation. PCR amplification was performed using the same primer sets as in 3C
A description of extra reagents and detailed protocols are available in the supplementary information online.
Supplementary Material
Acknowledgments
We would like to thank J. Moschandreas for instructions about the SPSS software, M. Gialitakis, J. Papamatheakis, A. Kretsovali and the members of our lab for critically reading the manuscript, and G. Vrentzos for cell culture assistance. C.G.S. is supported by a Cancer Research Institute Investigator Award, by a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme (FP7/2007-2013) under grant agreement no 239339 and by IMBB intramural funds.
Author contributions: C.D. and C.G.S. performed the experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Zhu J, Yamane H, Paul WE (2010) Differentiation of effector CD4 T cell populations. Annu Rev Immunol 28: 445–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24: 607–656 [DOI] [PubMed] [Google Scholar]
- Intlekofer AM et al. (2005) Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6: 1236–1244 [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman GC, Glimcher LH (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100: 655–669 [DOI] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W (2003) GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity 18: 415–428 [DOI] [PubMed] [Google Scholar]
- Bach EA, Aguet M, Schreiber RD (1997) The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol 15: 563–591 [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75: 163–189 [DOI] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB (2009) CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity 31: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M (2009) Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 9: 91–105 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P (2010) The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Nunez E, Fu XD, Rosenfeld MG (2009) Nuclear organization in the 3D space of the nucleus—cause or consequence? Curr Opin Genet Dev 19: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky Y, Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S (2009) Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol Biol 567: 171–188 [DOI] [PubMed] [Google Scholar]
- Spilianakis C, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Spilianakis C, Lalioti M, Town T, Lee GR, Flavell RA (2005) Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.