Abstract
Oligodendroglial Myelin Basic Protein (MBP) synthesis is essential for myelin formation in the central nervous system. During oligodendrocyte differentiation, MBP mRNA is kept in a translationally silenced state while intracellularly transported, until neuron-derived signals initiate localized MBP translation. Here we identify the small non-coding RNA 715 (sncRNA715) as an inhibitor of MBP translation. SncRNA715 localizes to cytoplasmic granular structures and associates with MBP mRNA transport granule components. We also detect increased levels of sncRNA715 in demyelinated chronic human multiple sclerosis lesions, which contain MBP mRNA but lack MBP protein.
Keywords: myelin basic protein, oligodendrocyte, multiple sclerosis, microRNA, translational control
Introduction
The myelination of central nervous system (CNS) axons by oligodendrocytes enables fast and energy-efficient propagation of action potentials as well as axonal support [1, 2]. The absence of the second most abundant CNS myelin protein MBP results in severe hypomyelination, shivering symptoms and premature death in rodents [3]. MBP compacts myelin membranes, binds to cytoskeletal proteins and is involved in oligodendroglial calcium signalling [4, 5]. Moreover, it regulates the molecular composition of the glial plasma membrane, facilitating the formation of lipid-rich myelin domains required for effective axon insulation [6, 7]. Therefore, appropriate MBP levels are important to maintain oligodendroglial homeostasis as well as functional myelin membrane formation. During oligodendrocyte maturation, MBP transcription precedes translation by about 1 day [8, 9], but the mechanisms underlying this translational repression of MBP remain poorly understood. Axon–glial signalling mechanisms have been described that induce MBP translation. These involve axonal L1-CAM, laminins and action potentials signalling through F3/contactin, β1-integrin and glutamate receptors, respectively [9–11]. The non-receptor tyrosine kinase Fyn has a pivotal role in the translation of axonal signals into MBP synthesis and myelination [12]. At the subcellular level, heterogeneous nuclear ribonucleoprotein (hnRNP) A2 binds to a defined sequence in the 3′ untranslated region (UTR) of MBP termed the A2 response element [13] resulting in cytoplasmic transport in RNA granules to the axon–glial contact site [14]. Several proteins associate with MBP mRNA granules during transport, including tumour overexpressed gene (TOG) and the hnRNPs E1, K and F [9, 15–17]. These may contribute to the translational inhibition during transport, but it is still only poorly understood how repression of MBP synthesis is generally mediated. The role of small non-coding RNAs (sncRNAs) such as microRNAs (miRNAs) or small interfering RNAs (siRNAs) in translational regulation has been intensively studied in recent years [18]. The absence of mature miRNAs in oligodendrocytes results in abnormal oligodendrocyte differentiation and impaired myelination [19–22] and a number of direct miRNA targets have been identified in oligodendrocytes [23].
Here we identify sncRNA715 as a new essential regulator of MBP translation. The levels of sncRNA715 and MBP protein inversely correlate in vitro and in vivo and MBP translation responds to experimentally altered sncRNA715 levels. We found endogenous sncRNA715 to be present in cytoplasmic granular structures and to associate with hnRNP A2 and MBP mRNA biochemically, implying sncRNA715-mediated translational repression during intracellular MBP mRNA transport. We demonstrate the functional relevance of sncRNA715 for oligodendroglial homeostasis as increased sncRNA715 levels affect cell morphology and reduce cell numbers by apoptosis. Importantly, we show that sncRNA715 is expressed in human brain and that its levels are increased in chronic inactive multiple sclerosis (MS) white matter lesions, which contain MBP mRNA but lack MBP protein.
Results and discussion
SncRNA715 and MBP protein levels inversely correlate
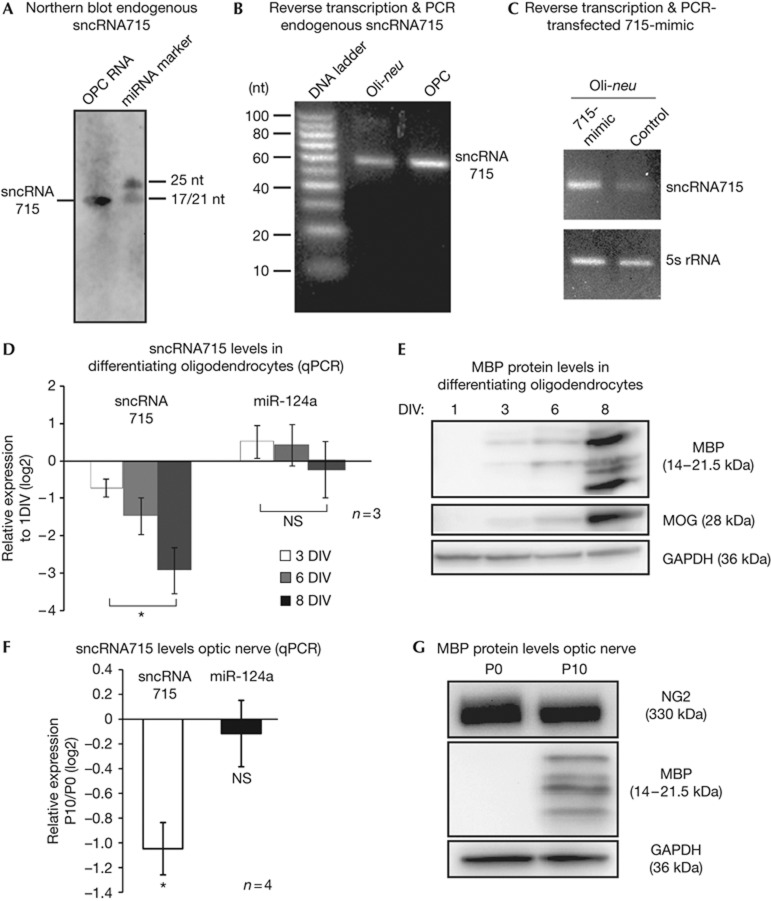
We used the MicroCosm target tool [24] to search for miRNAs that potentially regulate MBP mRNA and found that miR-715 has a predicted binding site in the 3′UTR of MBP mRNA that is conserved in mouse, rat and human. During the course of our studies it was revealed in miRBase [25], that the sequencing reads, which initially classified this RNA as a miRNA, are not consistent with the processing patterns of other miRNAs. At present, a clear distinction of sncRNAs including miRNAs, piRNAs, endogenous siRNAs and other non-canonical small RNAs is difficult [26]. We hence refer to this molecule as sncRNA715 rather than miR-715 and characterized its expression thoroughly. SncRNA715-specific northern blot analysis with a locked nucleic acid probe recognizing the 21-nt long sequence of the initially annotated mature miR715 (CUCCGUGCACACCCCCGCGUG) revealed the presence of this small RNA in cultured primary oligodendrocyte precursor cells (OPCs; Fig 1A). Reverse transcription and subsequent PCR with specific primers for sncRNA715 confirmed the presence of sncRNA715 in the OPC line Oli-neu [27] as well as in primary mouse OPCs (Fig 1B). We transfected Oli-neu cells with a synthetic sncRNA715 (715-mimic) or with a control siRNA. RNA extraction, polyadenylation and reverse transcription with oligo dT primers followed by PCR resulted in a single band of the same size for endogenous sncRNA715 (control transfected) and 715-mimic–transfected cells (Fig 1C). This suggests that the sequence of sncRNA715 is identical to the synthetic 715-mimic and did not reveal potential 3′-extensions as seen in precursor molecules (for example, pre-miRNA). Transfection of 715-mimic into primary OPCs also leads to strongly increased sncRNA715 levels, which we quantified by quantitative PCR (qPCR; supplementary Figs S1A,B online).
Figure 1.
SncRNA715 expression in oligodendrocytes. (A) Northern blot (band at ∼21 nucleotides, nt) and (B) specific PCR and EtBr-stained agarose gel show sncRNA715 in primary mouse OPCs (1 DIV) and Oli-neu cells. PCR products are ∼60-nt long due to the use of hairpin primers in the RT reaction. (C) EtBr-stained agarose gel of 715-mimic and control small interfering RNA-transfected Oli-neu cells showing PCR products as indicated on the right. (D–G) Total RNA or protein from cultured oligodendrocytes and mouse optic nerves was analysed for RNA/protein levels by qPCR and western blotting, respectively. qPCR results depict relative quantification (ΔΔCt) of indicated time points. In D,F and the following figures, number of experiments (n) are indicated and bar graphs represent mean values ±s.e.m. (D) Unpaired t-test; (F) one-sample t-test. *P (P-value)<0.05. DIV, day in vitro; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MBP, myelin basic protein; MOG, myelin oligodendrocyte glycoprotein; NS, not significant; OPC, oligodendrocyte precursor cell; qPCR, quantitative PCR; RT, reverse transcription; sncRNA715, small non-coding RNA 715.
We next analysed the relative levels of sncRNA715 during oligodendrocyte development in culture and in the optic nerve in vivo by qPCR. We found that sncRNA715 is significantly downregulated in mature oligodendrocytes and the optic nerve, while endogenous MBP protein levels increase (Fig 1D–G). As a control we analysed the levels of miR-124a, which are unaltered (Fig 1D,F). Cell-type–specific qPCR analysis revealed strikingly higher sncRNA715 levels in oligodendrocytes in comparison to neurons, astrocytes and microglial cells (supplementary Fig S1C online), leading to the conclusion that the observed changes in the optic nerve originate from oligodendrocytes. The inverse correlation between sncRNA715 and MBP expression further encouraged us to investigate a regulatory function of sncRNA715 on MBP translation.
SncRNA715 regulates MBP translation
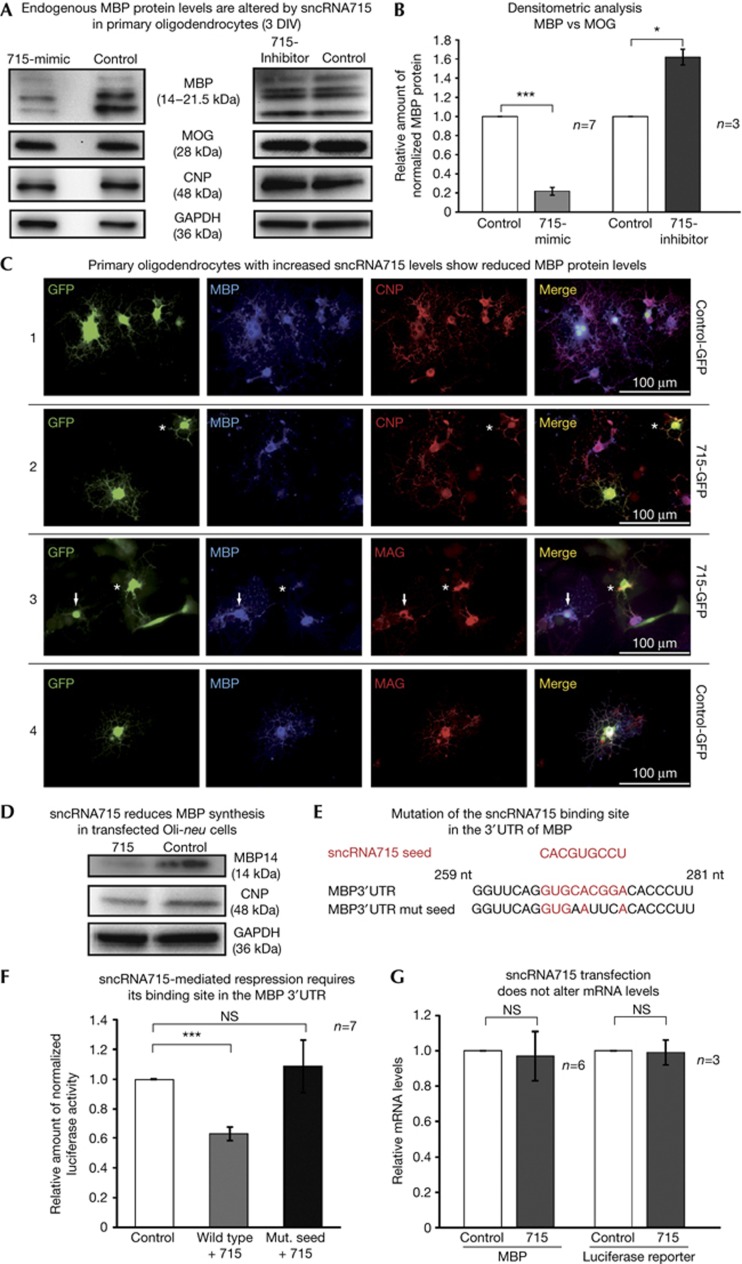
We increased the levels of sncRNA715 by transfection of synthetic 715-mimic into cultured primary mouse OPCs on the first day in vitro (DIV). A striking reduction of all MBP protein isoforms was detected 48 h later in western blots using equal amounts of total protein lysate (Fig 2A, left panel). Preventing the endogenous interaction of sncRNA715 with its targets by transfection of the complementary sequence (715-inhibitor) leads to an increase of MBP protein levels (Fig 2A, right panel). The levels of the myelin proteins CNP (2',3'-cyclic-nucleotide 3'-phosphodiesterase) and MOG (myelin oligodendrocyte glycoprotein) and the loading control GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were not affected in these experiments (Fig 2A). To distinguish between a specific effect of sncRNA715 on MBP protein synthesis or an influence on oligodendrocyte differentiation, we performed densitometric analyses from several western blots. We normalized the MBP to the MOG levels and related these normalized values in 715-mimic–transfected cells to control-transfected cells, as MOG is expressed simultaneously or later than MBP in cultured oligodendrocytes [28] (Fig 1E). A striking reduction of MBP protein levels in the 715-mimic–transfected cells can be observed, while 715 inhibition results in an increase of MBP levels (Fig 2B).
Figure 2.
SncRNA715 regulates MBP translation. (A) Western blots of 715-mimic, 715-inhibitor and control-transfected mouse OPCs with the indicated antibodies. (B) Densitometric analysis showing normalized MBP/MOG values related to control transfections; one-sample t-test, *P<0.05; ***P<0.001. (C) Rat OPCs were transfected with plasmids coexpressing control hairpin RNA and eGFP (C1,C4) or 715-hairpin RNA and eGFP (C2,C3) at 3 DIV (adherent Lonza 4D nucleofection) and immunostained 48 h later with the indicated antibodies. See text for details. (D) Western blots of Oli-neu cells co-transfected with MBP14 and a 715- or control-hairpin plasmid. (E) Predicted sncRNA715 seed region and its binding site in MBP mRNA, which was mutated as indicated. (F) Luciferase assays after co-transfection of either wild-type or mutated MBP 3′UTR reporter constructs with 715- or control-hairpin plasmid; Wilcoxon signed-rank test, ***P<0.001; (G) qPCR and relative quantification (ΔΔCt) of MBP or firefly luciferase mRNA in 715- versus control-transfected cells; one-sample t-test, NS, not significant (P>0.05). CNP, 2′,3′-cyclic-nucleotide 3′-phosphodiesterase; DIV, day in vitro; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; MBP, myelin basic protein; MOG, myelin oligodendrocyte glycoprotein; OPC, oligodendrocyte precursor cell; qPCR, quantitative PCR; sncRNA715, small non-coding RNA 715; UTR, untranslated region.
To analyse sncRNA715-mediated translational inhibition of MBP in single cells, we transfected primary rat oligodendrocytes (3 DIV) with 715-hairpin or control-hairpin plasmids, which coexpress enhanced GFP (eGFP) and immunostained these cells for the myelin proteins CNP, MAG (myelin-associated glycoprotein) and MBP (Fig 2C) 48 h later. Untransfected and control-hairpin-eGFP positive cells show normal levels for MBP, CNP and MAG. In contrast, 715-hairpin-eGFP positive cells show strongly reduced levels of MBP while they express MAG and CNP protein. Cells with lower levels of 715-eGFP still contain low levels of MBP (arrows in Fig 2C3), while cells with higher 715-eGFP levels lack MBP protein (asterisks in Fig 2C3). We conclude that sncRNA715 levels correlate with the degree of MBP reduction in single cells.
An additional approach confirmed the repressive effect of sncRNA715 on MBP synthesis by co-transfection of a plasmid coding for MBP14, including its 3′UTR with either a 715-hairpin or a control-hairpin plasmid into Oli-neu cells (Fig 2D).
Next we analysed if this translational inhibition is mediated through the interaction of sncRNA715 with its target site in the 3′UTR of MBP mRNA. The RNAhybrid tool [29] predicts a typical minimum free hybridization energy of −29 kcal/mol between sncRNA715 and mouse MBP mRNA (supplementary Fig S2A online). Co-transfection of a firefly luciferase reporter construct containing parts of MBP mRNA’s 3′UTR, including the binding site for sncRNA715 (Fig 2E) [11] with a 715-hairpin plasmid, showed a significant reduction in the amount of normalized luciferase activity when related to co-transfection with a control-hairpin plasmid (Fig 2F). Mutation of the sncRNA715 binding site (MBP 3′UTR mut. seed in Fig 2E) abolished the sncRNA715-mediated effect.
To assess if the sncRNA715-mediated effects are caused by mRNA degradation, we quantified MBP or luciferase reporter mRNA levels by qPCR and observed no changes (Fig 2G). We conclude that sncRNA715 regulates MBP translation through its binding site in the 3′UTR of MBP mRNA.
SncRNA715 affects morphology and cell numbers
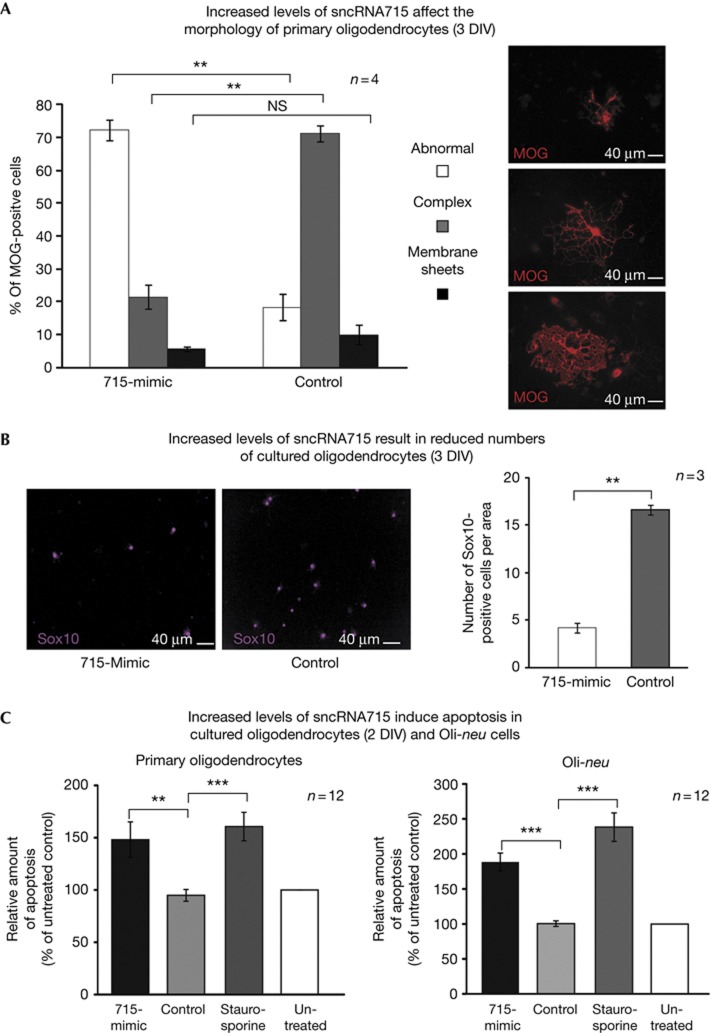
In the transfection experiments we noted a large number of morphologically altered oligodendrocytes with shorter or abnormally shaped processes (asterisks in Figs 2C 2) and quantified this by classifying oligodendrocytes into three different categories. We distinguished MOG-positive cells with a complex branching of cellular processes (complex), more mature cells that form membrane sheets (membrane sheets) and abnormally shaped cells that are normally found only very rarely in oligodendrocyte cultures (abnormal, Fig 3A). We found a significant increase in the number of abnormal cells and a significant reduction of complex cells after 715-mimic transfection compared with control-transfected cells (Fig 3A). The number of cells with membrane sheets was relatively low at this differentiation stage in culture and did not change significantly.
Figure 3.
SncRNA715 affects cell morphology and cell number. Primary mouse OPCs (1 DIV) were transfected with control RNA or 715-mimic and analysed 48 h later. (A) Cells were immunostained for MOG and classified by morphology as indicated (see text). The percentage of MOG-positive cells per category is plotted. (B) Cells were immunostained for Sox10 and counted per visible area. See alternative quantification approach in supplementary Fig S2C online. (C) Control cells were treated with staurosporine. Fluorimetric TUNEL assays were performed. The relative amount of signal from apoptotic cells (TUNEL positive) per total amount of cells (nuclear staining) was determined and the values were plotted as percentage of untreated control cells. A,B, paired t-test; C, unpaired t-test; NS, not significant; **P<0.01; ***P<0.001. DIV, day in vitro; MOG, myelin oligodendrocyte glycoprotein; OPC, oligodendrocyte precursor cell; sncRNA715, small non-coding RNA 715; TUNEL, TdT-mediated dUTP nick end labelling.
Furthermore, after 715-mimic transfection we observed a reduction in the counted number of Sox10-positive oligodendrocytes [30] (Fig 3B) and also quantified this fluorimetrically in primary OPCs and Oli-neu cells (supplementary Fig S2B online). As measured by fluorimetric TdT-mediated dUTP nick end labelling (TUNEL) assays, we found that 715-mimic transfection results in a significant increase of apoptotic oligodendrocytes (2 DIV) and Oli-neu cells comparable with the effects of the apoptosis-inducing reagent staurosporine (Fig 3C). Possibly, the observed morphological changes in cells with increased sncRNA715 levels represent cells undergoing apoptosis. Alternatively, they may result from reduced MBP levels in response to increased sncRNA715 levels. The importance of MBP for the elaboration of the distinct molecular composition of the oligodendroglial membrane (and myelin) has been demonstrated in detail previously [6] and MBP is associated with the actin and tubulin cytoskeleton [4]. It remains to be shown if sncRNA715 targets the synthesis pathway of other proteins in addition to MBP, which may also lead to the observed effects on cellular morphology and induction of apoptosis.
SncRNA715 localizes to cytoplasmic granular structures
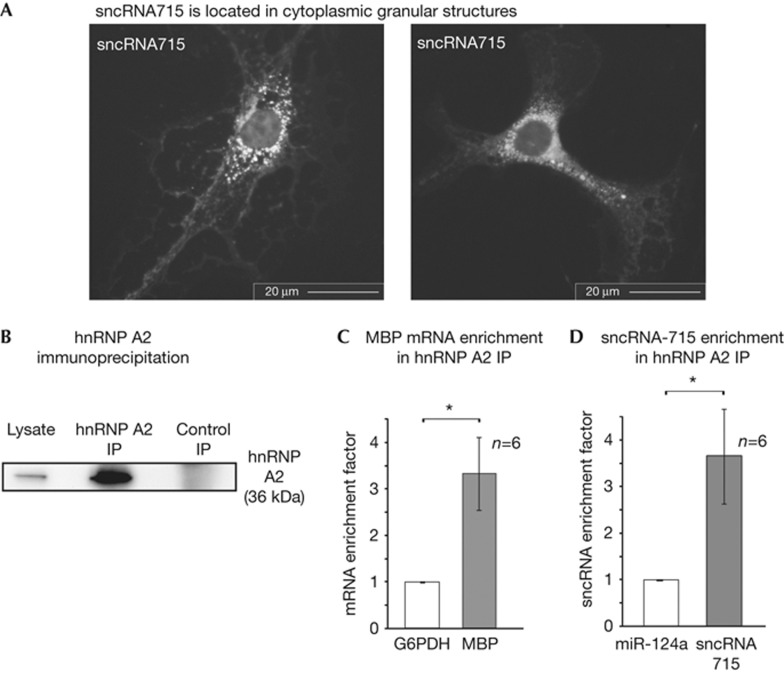
The RNA binding protein hnRNP A2 recruits MBP mRNA to RNA transport granules, which are transported from the nucleus to the periphery of the cell where the initiation of MBP translation is mediated by Fyn kinase [10, 11, 14]. Having shown the repressive effect of sncRNA715 on MBP translation, we next investigated if it is part of the MBP mRNA localization complex. Visualization of endogenous sncRNA715 in primary OPCs by fluorescence in situ hybridization shows its presence in granular cytoplasmic structures (Fig 4A). We analysed if sncRNA715 associates with hnRNP A2-dependent MBP mRNA granules. We purified hnRNP A2 by immunoprecipitation (IP; Fig 4B) and calculated the enrichment factor for copurifying RNAs after reverse transcription and qPCR as described previously [17, 31]. This factor relates the amount of RNA in the hnRNP A2 IP to the input and a control IP. MBP mRNA and sncRNA715 are similarly enriched in the hnRNP A2 IP compared to control RNAs glucose-6-phosphate dehydrogenase and miR-124a, respectively (Fig 4C,D). As additional specificity controls we analysed CNP and β-actin mRNAs, which are not enriched (supplementary Fig S3 online). Our results indicate that sncRNA715 is associated with hnRNP A2 and MBP mRNA and is present in cytoplasmic granular structures suggesting an involvement of sncRNA715 in translational repression during MBP mRNA transport.
Figure 4.
SncRNA715 associates with cytoplasmic MBP mRNA transport components. (A) Detection of sncRNA715 by fluorescence in situ hybridization in cytoplasmic granular structures in mouse oligodendrocytes (3 DIV). (B) Western blot showing hnRNP A2 levels before (lysate) and after hnRNP A2 or control immunoglobulin G immunoprecipitation (IP). (C,D) qPCR analysis and enrichment of MBP mRNA and sncRNA715 coimmunoprecipitating with hnRNP A2 (as in B). (C,D) Wilcoxon signed-rank test; *P<0.05. DIV, day in vitro; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; G6PDH, glucose-6-phosphate dehydrogenase; hnRNP, heterogeneous nuclear ribonucleoprotein; MBP, myelin basic protein; OPC, oligodendrocyte precursor cell; sncRNA715, small non-coding RNA 715.
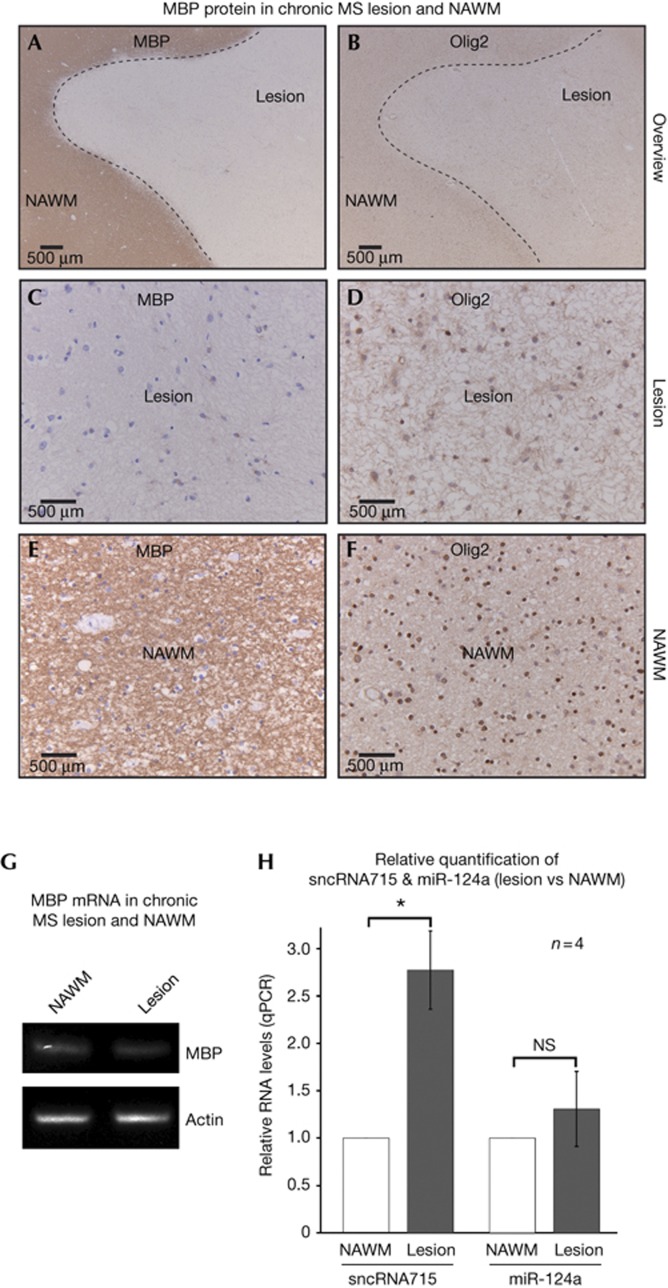
SncRNA715 is upregulated in chronic MS lesions
In the course of MS, focal demyelination in the CNS can be functionally compensated to a certain extent by remyelination [32]. This regenerative potential is ultimately lost and leads to neuronal degeneration correlating with clinical decline. In demyelinated lesions, activated CNS-resident OPCs increase the expression of transcription factors like Olig2 that are required for oligodendrocyte maturation [33]. It is not understood why these cells lose their remyelination potential with the progression of disease. In the light of our observed regulatory function of sncRNA715 on MBP translation and the importance of MBP for the myelination process, we addressed a potential correlation in MS. We analysed post-mortem brain tissue from four MS patients (supplementary Fig S4A online). As depicted in Fig 5A–F, chronic inactive MS lesions contain Olig2-positive cells, while MBP immunoreactivity is lacking in contrast to surrounding normal-appearing white matter (NAWM). This is in agreement with previous observations [34]. We analysed total RNA from these chronic lesions and surrounding NAWM. As expected, MBP mRNA is more abundant in the NAWM compared with the lesion (supplementary Fig S4B online), but we still detected relatively high amounts of MBP mRNA in the lesion samples (Fig 5G). Importantly, the levels of sncRNA715 are increased in the lesion compared with NAWM, while control miR124a levels are similar (Fig 5H). Our data indicate that high levels of sncRNA715 inhibit the translation of MBP mRNA in these chronic MS lesions. Previous studies have revealed the differential expression of miRNAs in MS lesions [35] indicating the importance of sncRNAs in MS pathology [36]. Our results show the role of an oligodendroglial sncRNA in the direct regulation of a key myelin protein essential for the myelination process.
Figure 5.
SncRNA715 levels are increased in chronic MS lesions. (A–F) MS lesion and surrounding normal-appearing white matter (NAWM) of a human brain section were immunostained for MBP and Olig2. (G) End point PCR from lesion and NAWM RNA with MBP and β-actin–specific primers. (H) qPCR from lesion and NAWM RNA and relative quantification (ΔΔCt) from four patients. One-sample t-test; *P<0.05; NS, not significant. MBP, myelin basic protein; MS, multiple sclerosis; qPCR, quantitative PCR; sncRNA715, small non-coding RNA 715.
In summary, we identified sncRNA715 as a new regulator of MBP translation, which is important for oligodendroglial homeostasis and myelination. Our study may contribute to a better understanding of remyelination failures in chronic MS lesions and the development of new therapeutical strategies for the treatment of MS.
Methods
Cell culture. Oli-neu cells (provided by J Trotter, Mainz) and primary rat or mouse oligodendrocytes were cultured as described in the supplementary information online.
qPCR. SncRNAs were reverse transcribed by the TaqMan MicroRNA Reverse Transcription Kit with stem-loop RT primers specific for sncRNA715, miR-124a or snoRNA-135 sequence (Applied Biosystems) and amplified with the Taqman Universal Master Mix (Roche) with specific primers and probes for the indicated sncRNAs (Applied Biosystems). The qPCR crossing points were used for relative quantification based on the ΔΔCt method using REST software [37]. SnoRNA-135, 5S, Renilla luciferase, β-actin and glucose-6-phosphate dehydrogenase were used as reference genes.
RNA analysis. RNA extraction, northern blots and locked nucleic acid–enzyme-labelled fluorescence–fluorescence in situ hybridization are described in supplementary information online.
Luciferase reporter assay. Luciferase assays were performed by using the Dual-Glo Luciferase Assay System (Promega), as described previously [12], with modifications that are described in supplementary information online.
Autopsy material. We collected chronic inactive lesions from four patients with clinically diagnosed and neuropathologically confirmed MS. The Netherlands Brain Bank received permission to perform autopsies for the use of tissue and for access to medical records for research purposes from the ethics committee of the VU Medical Center (Amsterdam, The Netherlands). See supplementary information online for details.
Supplementary Material
Acknowledgments
We thank J. Bruttger for initial experiments, N. Knauer for technical assistance. We thank C. Linington, W. Rigby, M. Simons, J. Trotter and M. Wegner for reagents. Funding was supported by the University of Mainz (level 1 support & MAIFOR to R.W.) and scholarships by Stipendienstiftung RLP to N.M.B. and F.T.N. Mainz to C.M.
Author contributions: N.M.B. performed most of the experiments and C.M., M.W., B.A. assisted with the experiments. N.M.B., J.v.H., P.v.d.V., H.J.L., R.W. analysed data and R.W. designed the study and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aggarwal S, Yurlova L, Simons M (2011) Central nervous system myelin: structure, synthesis and assembly. Trends Cell Biol 21: 585–593 [DOI] [PubMed] [Google Scholar]
- Nave KA (2010) Myelination and support of axonal integrity by glia. Nature 468: 244–252 [DOI] [PubMed] [Google Scholar]
- Readhead C, Hood L (1990) The dysmyelinating mouse mutations shiverer (shi) and myelin deficient (shimld). Behav Genet 20: 213–234 [DOI] [PubMed] [Google Scholar]
- Harauz G, Ladizhansky V, Boggs JM (2009) Structural polymorphism and multifunctionality of myelin basic protein. Biochemistry 48: 8094–8104 [DOI] [PubMed] [Google Scholar]
- Smith GS, Paez PM, Spreuer V, Campagnoni CW, Boggs JM, Campagnoni AT, Harauz G (2011) Classical 18.5-and 21.5-kDa isoforms of myelin basic protein inhibit calcium influx into oligodendroglial cells, in contrast to golli isoforms. J Neurosci Res 89: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S et al. (2011) A size barrier limits protein diffusion at the cell surface to generate lipid-rich myelin-membrane sheets. Dev Cell 21: 445–456 [DOI] [PubMed] [Google Scholar]
- Fitzner D, Schneider A, Kippert A, Mobius W, Willig KI, Hell SW, Bunt G, Gaus K, Simons M (2006) Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J 25: 5037–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colello RJ, Devey LR, Imperato E, Pott U (1995) The chronology of oligodendrocyte differentiation in the rat optic nerve: evidence for a signaling step initiating myelination in the CNS. J Neurosci 15: 7665–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen LS, Chan CW, Ffrench-Constant C (2011) Translation of myelin basic protein mRNA in oligodendrocytes is regulated by integrin activation and hnRNP-K. J Cell Biol 192: 797–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Lee PR, Fields RD (2011) Control of local protein synthesis and initial events in myelination by action potentials. Science 333: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Gonsior C, Kramer-Albers EM, Stohr N, Huttelmaier S, Trotter J (2008) Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J Cell Biol 181: 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Albers EM, White R (2011) From axon-glial signalling to myelination: the integrating role of oligodendroglial Fyn kinase. Cell Mol Life Sci 68: 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R (1999) Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem 274: 34389–34395 [DOI] [PubMed] [Google Scholar]
- Carson JH, Barbarese E (2005) Systems analysis of RNA trafficking in neural cells. Biol Cell 97: 51–62 [DOI] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, D'Sa C, Carson JH, Barbarese E (2005) The microtubule-associated protein tumor overexpressed gene binds to the RNA trafficking protein heterogeneous nuclear ribonucleoprotein A2. Mol Biol Cell 16: 1938–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, Korza G, Lee JW, Carson JH, Barbarese E (2006) Heterogeneous nuclear ribonucleoprotein (hnRNP) E1 binds to hnRNP A2 and inhibits translation of A2 response element mRNAs. Mol Biol Cell 17: 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Gonsior C, Bauer NM, Kramer-Albers EM, Luhmann HJ, Trotter J (2012) Heterogeneous nuclear ribonucleoprotein (hnRNP) F is a novel component of oligodendroglial RNA transport granules contributing to regulation of myelin basic protein (MBP) synthesis. J Biol Chem 287: 1742–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M (2010) Control of oligodendroglial cell number by the miR-17-92 cluster. Development 137: 2127–2132 [DOI] [PubMed] [Google Scholar]
- Dugas JC et al. (2010) Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65: 597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH (2009) Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol 66: 843–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X et al. (2010) MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Notterpek L (2011) MicroRNAs in oligodendrocyte and schwann cell differentiation. Dev Neurosci 33: 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Jung M et al. (1995) Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci 7: 1245–1265 [DOI] [PubMed] [Google Scholar]
- Pfeiffer SE, Warrington AE, Bansal R (1993) The oligodendrocyte and its many cellular processes. Trends Cell Biol 3: 191–197 [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M (1998) Sox10, a novel transcriptional modulator in glial cells. J Neurosci 18: 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur PK, Claussen M, Koch S, Tarbashevich K, Jahn O, Pieler T (2009) Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem 284: 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ, Ffrench-Constant C (2008) Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci 9: 839–855 [DOI] [PubMed] [Google Scholar]
- Fancy SP, Zhao C, Franklin RJ (2004) Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci 27: 247–254 [DOI] [PubMed] [Google Scholar]
- Wolswijk G (1998) Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci 18: 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A et al. (2009) MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132: 3342–3352 [DOI] [PubMed] [Google Scholar]
- Junker A, Hohlfeld R, Meinl E (2011) The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol 7: 56–59 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.