Abstract
Alternative approaches complementing the existing technologies for analysis of nucleic acids and their assemblies are necessary to take on the new challenges posed by the post-genomic era. The versatility of MS in biopolymer analysis and its ability to reach beyond sequence information are the basis of ever expanding applications aimed at the elucidation of nucleic acid structure-function relationships. This Feature summarizes the current state of MS-based approaches devised to overcome the limitations of traditional techniques and to advance different facets of nucleic acids research.
The completion of the Human Genome Project, which is widely regarded as one of the most significant accomplishments at the turn of the new millennium, was fueled by the development of analytical technologies capable of performing high-throughput sequencing of nucleic acids (NAs) with high accuracy and low cost.1 In the post-genomic era, the challenge is to elucidate the function of the vast majority of the genome sequence, estimated to comprise ~98.5% of the total 3 billion base pairs, that does not code for proteins.2 The observation that well-defined 3D structures determine catalytic RNA and riboswitch biological function has led to the realization that sequence information alone is not sufficient to explain the function of noncoding elements. This realization has sharply increased the need for diversified experimental approaches to annotate the enormous body of genetic information accumulated over the years by large-scale sequencing projects.
By virtue of its versatility in biopolymer analysis and breadth of accessible information, MS is poised to play a major role in the investigation of the structure–function relationships of noncoding elements. Unique among the platforms employed for NA analysis, MS can analyze the covalent adducts and noncovalent complexes of all classes of biopolymers, which are readily recognizable from their characteristic mass signatures. This versatility offers the opportunity of pursuing the direct characterization of functional assemblies without requiring antibodies, specific staining, or radioactive/fluorescent labels. Modest sample consumption facilitates the exploration of systems that are not amenable to overexpression or amplification by reverse transcription and PCR. In addition to ligand identity and stoichiometry, MS can also offer a direct assessment of binding activity, which is critical to understand the mechanism of action of functional assemblies. This Feature provides a brief overview of the different aspects of MS technologies that make them well-suited for NA studies and summarizes cutting-edge applications that illustrate the yet unfulfilled potential in different areas of NA research.
General approaches for identification and characterization
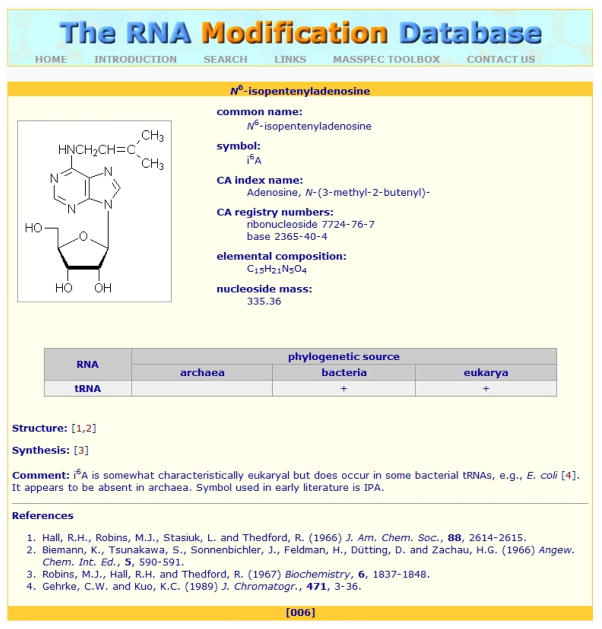
The combination of molecular mass determination and MS/MS constitutes the basis for analytical strategies that mirror very closely those employed in proteomics; MALDI and ESI can both successfully characterize these analytes.3, 4 An NA’s phosphodiester backbone affords net negative charges that make it better suited for analysis in negative-ion mode, although positive-ion data also can be obtained with somewhat lower sensitivity. The availability of endonucleases with well-defined cleavage specificity enables bottom-up approaches in which MS is used to fingerprint/map the specific hydrolytic products. The quantized nature of the fundamental units and the characteristic masses of covalent modifiers allow direct assessment of base composition5 and detection of possible nucleotide modifications.6 To date, 109 post-transcriptional modifications of RNA have been discovered, as described in the RNA Modification Database (Figure 1),7 and numerous DNA modifications, which are produced by damage and epigenetic processes, have been also reported. In addition, characteristic man-made modifications can be generated by chemicals employed to detect mass-silent nucleotides,8 to assess spatial contiguity in structural studies,9 and to achieve stable isotopic labeling for quantification purposes.10 In the absence of suitable chromophores, covalent modifications tend to elude spectroscopic detection but are immediately recognizable by MS analysis.
Figure 1.
Example of a natural post-transcriptional modification from the RNA Modification Database.7 Additional non-natural modifications are also accessible by MS technologies but off limits to conventional sequencing approaches.
Sequence information is effectively obtained by generating suitable product ladders in solution or during MS/MS analysis in the gas phase. Various reviews have extensively discussed the merits and typical applications of these approaches, and they will not be revisited here.3, 4 However, collision induced dissociation (CID) and IR multiphoton dissociation (IRMPD) generally can obtain characteristic ion series in which each signal is spaced from the next by the incremental mass of the intervening nucleotide.11 This feature not only solves any ambiguity about the identity of digestion/ladder oligonucleotides produced in solution, but also locates covalent modifications on the basis of their unique mass shifts. These MS/MS methods have a current upper limit of ~100 nucleotides;12 further work will be necessary to increase the size accessible to direct gas-phase sequencing without prior hydrolysis (i.e., top-down analysis).
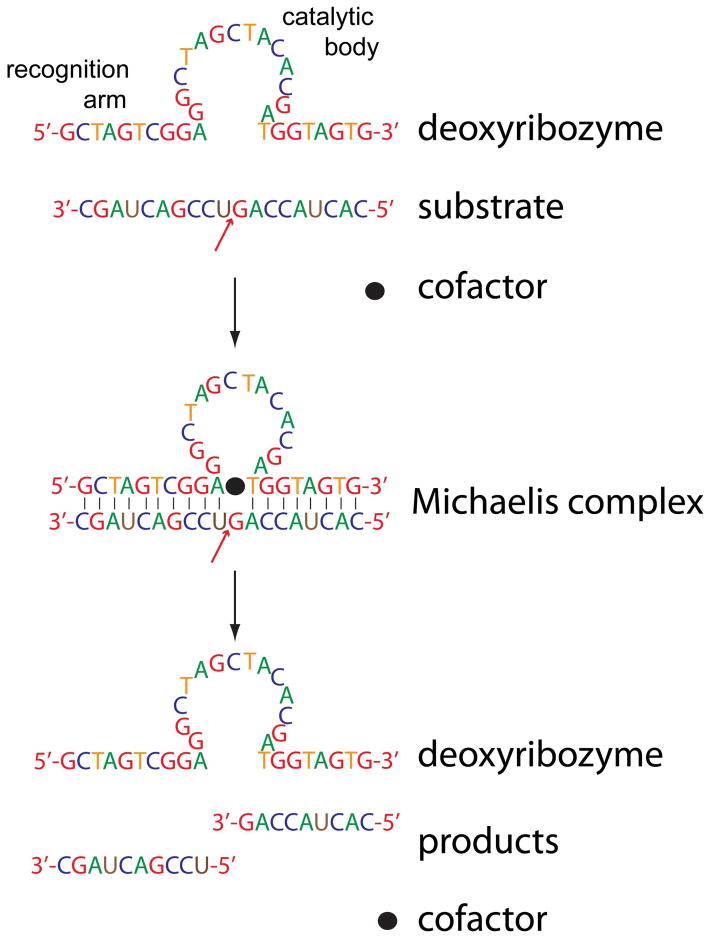
Strategies based on substrate digestion stand to greatly benefit from RNA-cleaving deoxyribozymes. These synthetic restriction enzymes consist of single-stranded DNA folded into a catalytic domain flanked by two recognition arms (Figure 2). Well-defined catalytic domains are available for each RNA nucleotide, whereas the flanking strands can be designed ad hoc to anneal with the sequences surrounding the target nucleotide. The ability to vary the recognition arms makes the enzymes’ hydrolytic activity not only residue specific, as typical for most proteases, but also sequence specific, which is unparalleled in protein analysis. Thus, deoxyribozymes can be designed ad hoc to isolate selected domains for further analysis or to probe their accessibility for structural elucidation.13 Deoxyribozymes are expected to facilitate the pursuit of species that are too large for top-down analysis but may become accessible after segmentation into targets of intermediate size (i.e., mid-down analysis).
Figure 2.
An RNA-cleaving deoxyribozyme. The red arrow indicates the target phosphodiester bond. Ad hoc recognition arms position the catalytic domain over the target.
Secondary and tertiary structure determination
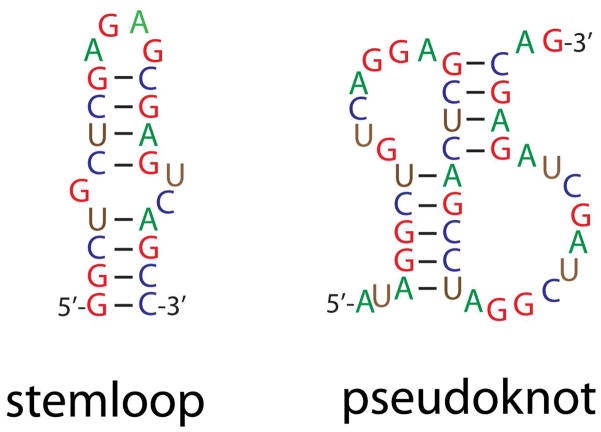
Intramolecular base pairing predominantly defines NA higher-order structure, implying that any technique capable of identifying paired regions can in principle enable structure determination. The importance of base pairing can be immediately appreciated by examining the structure of stemloop and pseudoknot domains, which represent typical secondary and tertiary structures, respectively (Figure 3). In light of this observation, the ability of ESI14 and MALDI15 to preserve intact pairing interactions suggests that one day MS technologies may become viable tools for structural determination. This prediction draws further strength from the demonstration that electron-detachment dissociation, which is achieved by irradiating anionic precursor ions with >10 eV electrons, is capable of inducing gas-phase fragmentation of the phosphodiester backbone while leaving pairing interactions undisturbed.16 Combining this non-ergodic (i.e., without energy redistribution) activation method with ergodic counterparts (i.e., CID and IRMPD) that induce pairing dissociation could help MS/MS discriminate single- versus double-stranded sequences.17 Although the possibility of gleaning structural information is clearly within reach, the potential of direct gas-phase strategies is still largely unexplored.
Figure 3.
Elements of RNA higher-order structure. Intramolecular annealing of contiguous sequences forms secondary structures, such as stemloop hairpins, and annealing of distal regions creates tertiary structures, such as pseudoknots.
More indirectly, MS is rapidly expanding its role supporting chemical/biochemical approaches to identify paired regions from their solvent accessibility or backbone flexibility. The fast rate of exchange displayed by NA functional groups has hampered the possibility of using hydrogen-deuterium exchange in solution,18 but studying nucleotides19 and oligonucleotides20 in the gas phase is feasible. In contrast, MS detection has been successfully combined with footprinting probes that assess the different accessibility21, 22 and flexibility23 exhibited by single- versus double-stranded regions. Typical solvent-accessibility probes include kethoxal (KT), which modifies N1 and N2 of unpaired guanines; dimethylsulfate (DMS), used for N1-adenine and N3-cytidine; and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide (CMCT) for N3-uridine. The main flexibility probe is N-methylisatoic anhydride, which modifies the 2′-hydroxyl group of unpaired nucleotides. Pairing patterns revealed by these probes are subsequently interpreted with the aid of algorithms that are capable of predicting NA secondary structure on the basis of thermodynamic or statistic principles.24 The MS platform eliminates any reliance on strand cleavage by probe-specific chemistries or extension of staggered primer series to generate the typical ladders necessary for electrophoretic analysis. This platform is expected to foster the introduction of new probes for functional groups involved in noncanonic pairing.
MS detection also offers an edge in the characterization of products of bifunctional probes, which electrophoretic methods traditionally cannot determine.9 For example, the identification of crosslinks produced by nitrogen mustard (NM), which can bridge N7-guanine and N3-adenine on distal regions, would require the simultaneous sequencing of both conjugated strands. Bridging residues situated within well-defined distances allow triangulation of their mutual positions in 3D space, thus complementing the information from footprinting probes. Whereas the latter can only indicate whether a given nucleotide may be involved in a base pair but cannot identify its paired counterpart, bifunctional crosslinkers clearly identify both bridged nucleotides, reveal their mutual distance, and constrain the underlying base pairing pattern. In addition, crosslinkers can also identify contacts between regions that are not necessarily complementary and that footprinting experiments would miss. This point is exemplified by the discovery of a novel GNRA (N = A, C, G, or U; R = A or G) tetraloop–receptor interaction in the HIV-1 packaging signal (Ψ-RNA); the detection of numerous NM conjugates supported this finding.25 This tertiary contact is stabilized by hydrogen bonds and stacking patterns, which are not predictable by the normal pairing rules. The validity of the crosslinking data was confirmed by introducing a key mutation that destabilized the interaction and resulted in the elimination of previously observed crosslinks.
3D structural determination
The development of ever more sophisticated computational approaches for predicting and modeling NA structure is progressively closing the resolution gap between chemical/biochemical methods and the established high-resolution techniques.26 Benefitting from these advances, the concerted use of chemical probing and MS detection has proven capable of yielding the information necessary to generate valid, all-atom 3D structures. For example, the spatial constraints of this combined approach were used to solve the structure of the ribosomal frameshifting pseudoknot of mouse mammary tumor virus.27 In this case, the monofunctional probes KT, DMS, and CMCT assessed the pairing status of each nucleotide in the construct, while the bifunctional reagent NM confirmed the pairing register and detected valuable long-range relationships. The probing data identified the best fit within a library of 3D structures generated by the MC-Sym algorithm,28 as well as restrained rounds of simulated annealing by Crystallography and NMR System.29 The energy-minimized model displayed only ~3Å root mean square deviation from the corresponding NMR structure available in the Protein Data Bank (entry 1RNK), which provided validation to the probing approach.27
This platform constitutes the basis of a general divide-and-conquer strategy for NA structure determination, for which monofunctional probes obtain the secondary structure of individual domains and bifunctional crosslinkers identify their mutual tertiary contacts. This strategy takes advantage of the highly hierarchic nature of NA folding, which proceeds through initial formation of local secondary structures, followed by the establishment of interdomain contacts that determine the overall morphology. Along these lines, an all-atom model of HIV-1 Ψ-RNA was built from the high-resolution structures of its stemloop domains—solved separately by NMR—by probing their mutual contacts in the global fold.25 Footprinting probes initially confirmed the integrity of the individual stemloops in the context of full-length Ψ-RNA, whereas crosslinkers provided the information necessary to correctly assemble their individual NMR structures into a full-fledged model. The energy-minimized structure retained the atomic resolution of the NMR data and displayed a global fold that satisfied the probing data.25
Ion mobility spectrometry (IMS)30 MS could potentially obtain direct information about the topology of NA structures, which could guide the assembly of high-resolution building blocks. This technique involves determining the mobility of ions in a moderate electric field in a gaseous environment. Ions with extended conformations are expected to undergo more collisions with the inert gas than those adopting compact conformations, which translates into different drift times along the IMS cell. Drift times can determine the collision cross-section (CCS) of the different conformers, which is a function of their global topology. In the case of NAs, CCS values obtained from the lowest charge states of stemloop, pseudoknot, and cruciform constructs matched very closely those calculated by molecular dynamics.31 These results add further proof of the stability of higher-order structures in a solvent-free environment and provide the rationale for a greater role of IMS/MS in NA structure elucidation.
Assemblies’ quaternary structure
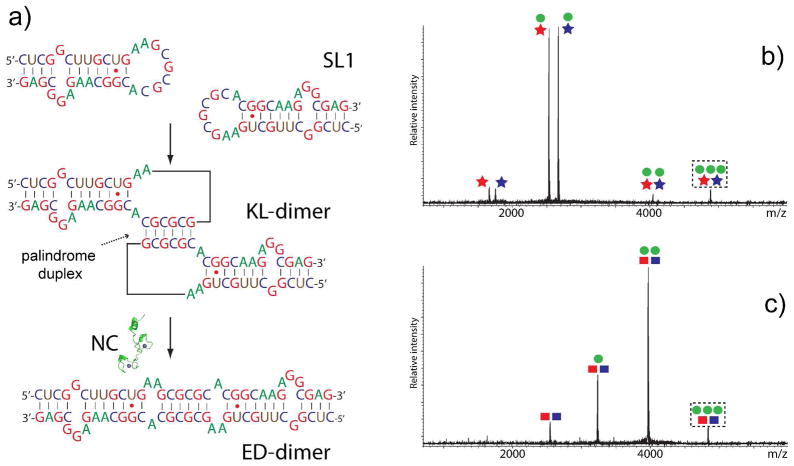
The demonstration that soft ionization techniques can transfer intact NA complexes into the gas phase has prompted the use of MS-based technologies for the investigation of their noncovalent assemblies with species of very different nature.32, 33 In addition to what molecular mass determination has to offer, gas-phase activation and IMS experiments enable the examination of NA quaternary structure. For example, MS/MS can interrogate the conformational state of dimeric complexes of SL1 RNA, which mediate genome dimerization and packaging in HIV-1.34 When two copies of SL1 interact through a highly-conserved palindromic sequence (Figure 4), they initially form a metastable kissing-loop dimer (KL), which can subsequently isomerize into a thermodynamic extended duplex (ED) mediated by HIV-1 nucleocapsid (NC) protein. Though an identical base composition/sequence prevented the recognition of the alternative forms by simple molecular mass determination, discrimination was achieved on the basis of their different gas-phase stabilities. Indeed, submitting isomer-specific mutants to gentle CID induced the dissociation of the KL-specific dimer into individual stemloops, consistent with the limited number of intermolecular base pairs displayed by its quaternary structure. In contrast, strands of the ED-specific dimer did not dissociate, even under more energetic activation conditions. Distinctive results were also obtained from the corresponding assemblies with the viral NC protein, a NA chaperone that mediates RNA structure remodeling. In this case, KL complexes still exhibited predominant dimer dissociation, whereas the ED ones displayed stepwise release of NC (Figure 4). These characteristic CID patterns elucidated the isomerization mechanism of the wildtype construct, which can form both KL and ED conformers in solution and afford NC complexes with identical stoichiometry/molecular mass.34
Figure 4.
a) Dimerization of HIV-1 SL1 into a kissing-loop (KL) complex and isomerization into an extended duplex (ED) mediated by nucleocapsid (NC) protein. Gentle CID of b) NC•KL and c) NC•ED dimer. Stars and squares identify KL- and ED-specific dimers, respectively, with blue and red colors indicating isomerization-deficient counterparts. Green circles identify NC. Precursor ions in dashed boxes possess identical masses. Adapted with permission from ref. 34.
This principle is also applicable to larger multicomponent assemblies, which have shown excellent correlation between the order of subunit dissociation at increasing collision energy and the organization of NA and protein components.35 Extensive interaction networks can be studied by the concerted application of gas-phase dissociation and either disruption of intact complexes in solution (e.g., by varying ionic strength, pH, temperature, and other conditions) or controlled complex reassembly (e.g., by varying the type and composition of subunits in solution). For example, a combined approach of this type compared the architecture of the human translation initiation factor eIF3 in free form and bound to the hepatitis C virus internal ribosome entry site (HCV-IRES) RNA.36 The MS determination of intact assembly in the absence of RNA confirmed that all 13 protein subunits were present at unit stoichiometry. Increasing collision energy provided different products with unambiguous subunit composition. At the same time, solution dissociation at increasing ionic strength produced three stable modules that were characterized individually by CID. This systematic dissection process revealed that 27 discrete subcomplexes defined the interactions within and between modules. Integrating this information with immunoprecipitation data lead to a comprehensive interaction map highlighting the significance of HCV-IRES RNA in the organization of human eIF3.36
Direct IMS analysis also has the potential to provide complementary information on quaternary organization and topology, as demonstrated by the study of trp RNA-binding attenuation protein (TRAP).37 In the presence of tryptophan substrate, the observed CCS was consistent with the characteristic ring organization shown by the corresponding crystal structure. After addition of a 53-nucleotide section of the cognate mRNA leader, the overall CCS increased by an amount consistent with the binding of RNA to the periphery of the protein ring, again in agreement with the high-resolution structure.37 A measure of the complexity of NA assemblies accessible to ion mobility techniques can be gleaned from the investigation of intact RNA viruses by gas-phase electrophoretic mobility molecular analysis, in which four icosahedral viruses had particle diameters that deviated by only 15% from the values of the respective crystal structures.38 This remarkable agreement suggests that applications aimed at studying viral particles and capsid structures have a bright future despite the challenges posed by size and sample heterogeneity.39
Mechanism of NA interactions
The ability to detect intact NA complexes offers the unique opportunity to elucidate important functional features by evaluating the determinants of their binding activity. Great attention was initially paid to the possibility of assessing the stability of NA–NA, protein–NA, and small ligand–NA complexes by activating their dissociation directly in the source or in the analyzer after mass selection. Though these gas-phase approaches offered further evidence of the preservation of base pairing and stacking interactions in a solvent-free environment, they also highlighted the challenges of obtaining quantitative determinations of dissociation constant (Kd) values, which matched those observed in solution. On the contrary, excellent correlations have been demonstrated when MS is employed to assess the partitioning between free and bound species in solution, which can lead to Kd values matching those obtained by bulk methods.40 As compared to such methods, which rely on curve fitting of spectroscopic/calorimetric data, MS analysis enables the direct assessment of coexisting binding equilibria by differentiating all species in solution. Therefore, competition schemes can be readily implemented in which multiple ligands vie for the same substrate and the observed product distributions translate into relative scales of binding affinities.41 Inverting the competition terms, multiple substrates with different features can compete for the same ligand to identify the structural determinants of binding.42 Though the simultaneous detection of all binding events in the same experiment affords virtually no bias, these types of strategies have attracted only limited interest from the broader NA community.
The type of functional groups involved, their spatial orientation, and the presence of possible cofactors define the mechanism of specific NA interactions. The interplay between such factors and the actual environmental conditions (i.e., pH, ionic strength, temperature, presence of organic solvents, etc.) determine the stoichiometry and position of binding equilibria, which can be investigated directly by MS approaches. The broad applicability to very diverse ligands is particularly advantageous for investigating the binding of small molecules that do not possess distinctive spectroscopic properties and cannot tolerate the introduction of chromophoric labels without perturbing their activity profile. For this reason, the binding modes of intercalators, minor groove binders, and many other classes of NA ligands have been successfully investigated over the years (see ref. 33 and references therein) and the results presented in reports that clearly showcase the potential of MS technologies in NA drug discovery. For example, a new class of small ligands targeting the subdomain IIA of HCV-IRES RNA was identified from a 180,000-member library by MS-based screening.43 A 29-mer stemloop mimicking the double-stranded stem of subdomain IIA competed against an unstructured 33mer control in a high-throughput competition scheme. MS-assisted SAR (structure–activity relationship) studies on the initial benzimidazole “hit” subsequently improved its initial binding properties. Systematic variation of the hit’s functional groups led to an optimized ligand with a 140-fold overall increase in binding affinity over the initial molecule.43
Analogous strategies have been developed to identify specific structural determinants of substrate interactions by monitoring the ability to establish stable complexes with molecules of well-known binding modes. For example, the structure of the HIV-1 polypurine tract (PPT), the RNA:DNA hybrid that primes the synthesis of plus-strand DNA during reverse transcription of viral RNA, was studied with classic NA binders.42 Intercalators and groove binders exhibit high affinity for helical structures with regular stacking arrangement and groove geometry, respectively, whereas aminoglycosidic antibiotics prefer anomalies such as bulging nucleotides and noncanonic base pairs, which result in highly electronegative patches and helix distortion. The data provided by the former showed no detectable deviation from a normal helical pattern, but the latter highlighted two significant anomalies that sustained specific binding. The putative sites were located by submitting the corresponding noncovalent complexes to CID,44 which identified the contact nucleotides on the basis of the inhibition of gas-phase fragmentation induced by the bound aminoglycoside. NMR analysis subsequently confirmed these findings, which contributed to a new understanding of the mechanism of PPT recognition by reverse transcriptase.
Monitoring intact functional complexes also enables the recognition of possible cofactors and the investigation of their specific roles, as exemplified by the study of the stabilizing effects of metals on drug–DNA complexes. When the binding of chromomycin A3 to duplex DNA was compared with that of the new compound UK-1 in the presence of divalent cations, a 2:1 drug:metal ratio was observed for the former, while a 1:1 ratio was detected for the latter.45 The stability of the interaction between UK-1 and a GC-rich DNA duplex was probed by CID, which provided a Ni2+ > Zn2+ > Co2+ relative stabilization scale. The development of strategies for discriminating coordinated metals that participate in the actual structure of NA assemblies versus those that form nonspecific adducts on their surfaces is expected to facilitate the investigation of metals as functional cofactors.46
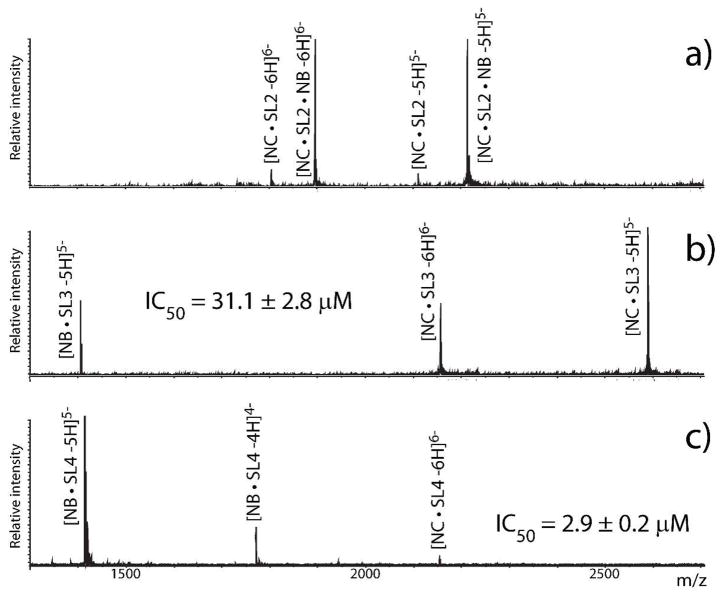
Most NA activities rely on their ability to establish productive interactions with cognate proteins. Approaches based on crosslinking, affinity capture, immunoprecipitation, and others can help identify such protein components,47, 48 while the analysis of intact assemblies can support structure elucidation, as discussed above. Additionally, investigating the effects of ligands on assembly organization/stability can provide valuable details on their inner workings. Indeed, schemes in which target complexes are treated with series of ligands have not only screened for possible inhibitors in drug discovery, but also gleaned mechanistic information. For example, when pre-formed complexes of NC with separate domains of Ψ-RNA were treated with the aminoglycoside neomycin B, those involving stemloop 3 and 4 (SL3 and SL4) showed signs of perturbation, whereas that with stemloop 2 (SL2) remained intact (Figure 5).49 IC50 (the concentration inducing 50% dissociation of initial complex) was determined with titration procedures that monitored the partitioning between NC- and neomycin B-complexes at increasing ligand concentrations. The realization that NC can bind individual Ψ-RNA domains with very distinctive mechanisms helped change the perception of its broad chaperone activities by revealing a manifest structure-specific character.
Figure 5.
Effects of neomycin B (NB) on pre-formed complexes of HIV-1 nucleocapsid (NC) with a) SL2, b) SL3, and c) SL4 (SL = stemloop). NB binds NC•SL2 without causing dissociation but displaces both NC•SL3 and NC•SL4. Corresponding IC50 values were obtained from titration procedures.
This conclusion was further supported by examining the interactions of NC with the dimeric forms of SL1 RNA. In this case, the KL- and ED-specific mutants exhibited very different binding patterns with both intercalators and minor groove binders, which are sensitive to the integrity of helical structures.50 The observed differences were attributed to the significant distortion of the palindrome duplex in the KL conformer as compared to the ED form, in which it assumes a more regular helical structure. Neither class of ligand affected the ability of SL1 dimers to bind NC, thus suggesting that protein interactions did not significantly alter their distinctive conformations. In contrast, neomycin B and its affinity for helix anomalies greatly affected NC binding. The ability of neomycin B to selectively displace NC from the hinge of the KL conformer translated into a marked inhibition of its transition into ED form. These data provided new insights into the mechanism of dimer isomerization and highlighted the importance of the structural context in determining the NC chaperone activities.34, 50 The ability of MS approaches to monitor simultaneous equilibria without fluorescent labeling of any of the species involved contributed greatly to the feasibility of these studies.
Conclusions
The recognition that sequence alone may not provide all the information necessary to understand the function of most NA species presents the opportunity for significant contributions by MS-based technologies. This brief overview shows that such technologies can reach well beyond sequence information to provide correlations between sequence, higher-order structure, and relationships with other cellular components, which are necessary to understand NA function and mechanism of activity. The benefits are already evident in some areas of NA research, though further development will be necessary in others before the largely untapped potential will come to full fruition. These techniques’ ability to study a range of subjects from covalent nucleotide modifications to large noncovalent assemblies offers the possibility of exploring directions that may have been neglected for dearth of convenient tools. Indeed, the emphasis on sequence information fueled by the availability of powerful electrophoretic techniques might have delayed the investigation of the biological significance of covalent modifications, which are off-limits to such techniques. The reliance on amplification methods that are suitable only for NA species also might have slowed the characterization of functional assemblies with proteins and other cellular components that cannot be readily amplified. For this reason, MS is primed to expand in the functional elucidation of noncoding elements. This expansion will afford a greater understanding of the biological activity of a large portion of genomic sequences preserved through evolution, which should lead to the discovery of new avenues for therapeutic intervention in human diseases and cancer.
Acknowledgments
Support from The RNA Institute and the College of Arts and Sciences of University at Albany (SUNY) and from the National Institutes of Health (GM064328-11) is gratefully acknowledged.
Biography
D. Fabris is Professor of Chemistry and Biological Sciences at the University at Albany. His research includes developing MS-based technologies for the investigation of the structure-function relationships of nucleic acids in retroviral systems. In particular, he has developed approaches that combine footprinting and crosslinking probes with high-resolution MS detection to solve the 3D structure of noncoding nucleic acids that are not directly amenable to traditional techniques. His interests also include the application of novel top-down strategies for elucidating the interactions of nucleic acids with proteins, small molecule ligands, and metals.
References
- 1.Venter JC, Smith HO, Hood L. Nature. 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- 2.Pheasant M, Mattick JS. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- 3.Nordhoff E, Kirpekar F, Roepstorff P. Mass Spectrom Rev. 1996;15:67–138. doi: 10.1002/(SICI)1098-2787(1996)15:2<67::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Limbach PA. Mass Spectrom Rev. 1996;15:297–336. doi: 10.1002/(SICI)1098-2787(1996)15:5<297::AID-MAS2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz SC, Kowalak JA, McCloskey JA. J Am Soc Mass Spectrom. 1993;4:204–209. doi: 10.1016/1044-0305(93)85082-9. [DOI] [PubMed] [Google Scholar]
- 6.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limbach PA, Crain PF, McCloskey JA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patteson KG, Rodicio LP, Limbach PA. Nucleic Acids Res. 2001;29:E49–49. doi: 10.1093/nar/29.10.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Yu ET, Kellersberger KA, Crosland E, Fabris D. J Am Soc Mass Spectrom. 2006;17:1570–1581. doi: 10.1016/j.jasms.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Meng Z, Limbach PA. Anal Chem. 2005;77:1891–1895. doi: 10.1021/ac048801y. [DOI] [PubMed] [Google Scholar]
- 11.McLuckey SA, Habibi-Goudarzi S. J Am Chem Soc. 1993;115:12085–12095. [Google Scholar]
- 12.Little DP, Aaserud DJ, Valaskovic GA, McLafferty FW. J Am Chem Soc. 1996;118:9352–9359. [Google Scholar]
- 13.Turner KB, German M, Hawkins A, Berton A, Fabris D. 14th Annual Meeting of the RNA Society; Madison (WS). May 2009. [Google Scholar]
- 14.Ganem B, Li YT, Henion JD. Tetrahedron Lett. 1993;34:1445–1448. [Google Scholar]
- 15.Lecchi P, Pannell LK. J Am Soc Mass Spectrom. 1995;6:972–975. doi: 10.1016/1044-0305(95)00524-H. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Mo J, Adamson JT, Hakansson K. Anal Chem. 2005;77:1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 17.Wilhide J, Kellersberger KA, Fabris D. Proc. of the 57th ASMS Conference on Mass Spectrometry and Allied Topics; Philadelphia. June 2009; 2009. [Google Scholar]
- 18.Johnston PD, Redfield AG. Nucleic Acids Res. 1977;4:3599–3615. doi: 10.1093/nar/4.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson JM, Greig MJ, Griffey RH, Mohan V, Laude DA. Anal Chem. 1998;70:3566–3571. doi: 10.1021/ac9805302. [DOI] [PubMed] [Google Scholar]
- 20.Hofstadler SA, Sannes-Lowery KA, Griffey RH. J Mass Spectrom. 2000;35:62–70. doi: 10.1002/(SICI)1096-9888(200001)35:1<62::AID-JMS913>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Yu E, Fabris D. J Mol Biol. 2003;330:211–223. doi: 10.1016/s0022-2836(03)00589-8. [DOI] [PubMed] [Google Scholar]
- 22.Kellersberger KA, Yu E, Kruppa GH, Young MM, Fabris D. Anal Chem. 2004;76:2438–2445. doi: 10.1021/ac0355045. [DOI] [PubMed] [Google Scholar]
- 23.Turner KB, Yi-Brunozzi HY, Brinson RG, Marino JP, Fabris D, Le Grice SF. RNA. 2009;15:1605–1613. doi: 10.1261/rna.1615409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu ET, Hawkins AE, Eaton J, Fabris D. Proc Natl Acad Sci U S A. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabris D, Yu ET. J Mass Spectrom. 2010;45:841–860. doi: 10.1002/jms.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu ET, Zhang Q, Fabris D. J Mol Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Major F, Gautheret D, Cedergren R. Proc Nat Acad Sci USA. 1993;90:9408–9412. doi: 10.1073/pnas.90.20.9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstelve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst. 1998:D54. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Clemmer DE, Jarrold MF. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 31.Baker ES, Dupuis NF, Bowers MT. J Phys Chem B. 2009;113:1722–1727. doi: 10.1021/jp807529m. [DOI] [PubMed] [Google Scholar]
- 32.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Mass Spectrom Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 33.Hofstadler SA, Griffey RH. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 34.Turner KB, Hagan NA, Fabris D. J Mol Biol. 2007;369:812–828. doi: 10.1016/j.jmb.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benesch JL. J Am Soc Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV. Proc Natl Acad Sci U S A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 38.Thomas JJ, Bothner B, Traina J, Benner WH, Siuzdak G. Spectroscopy. 2004;18:31–36. [Google Scholar]
- 39.Morton VL, Stockley PG, Stonehouse NJ, Ashcroft AE. Mass Spectrom Rev. 2008;27:575–595. doi: 10.1002/mas.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Int J Mass Spectrom Ion Proc. 2002;216:1–27. [Google Scholar]
- 41.Mei HY, Mack D, Galan AA, Halim NS, Heldsinger A, Loo JA, Moreland DW, Sannes-Lowery KA, Sharmeen L, Truong HN, Czarnik AW. Bioorg Med Chem. 1997;5:1173–1184. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 42.Turner KB, Brinson RG, Yi-Brunozzi HY, Rausch JW, Miller JT, Le Grice SFJ, Marino JP, Fabris D. Nucleic Acids Res. 2008;36:2799–2810. doi: 10.1093/nar/gkn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, Swayze EE, Griffey RH. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 44.Turner KB, Hagan NA, Kohlway A, Fabris D. J Am Soc Mass Spectrom. 2006;17:1401–1411. doi: 10.1016/j.jasms.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Nucleic Acids Res. 2001;29:e103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner KB, Monti S, Fabris D. J Am Chem Soc. 2008;30:13353–13363. doi: 10.1021/ja8045734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steen H, Jensen ON. Mass Spectrom Rev. 2002;21:163–182. doi: 10.1002/mas.10024. [DOI] [PubMed] [Google Scholar]
- 48.Rusconi F, Guillonneau F, Praseuth D. Mass Spectrom Rev. 2002;21:305–348. doi: 10.1002/mas.10036. [DOI] [PubMed] [Google Scholar]
- 49.Turner KB, Hagan NA, Fabris D. Nucl Acids Res. 2006;34:1305–1316. doi: 10.1093/nar/gkl004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner KB, Kohlway AS, Hagan NA, Fabris D. Biopolymers. 2009;91:283–296. doi: 10.1002/bip.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]