Abstract
Background
In this paper, we describe cDNA cloning of a new anionic trypsin and a collagenolytic serine protease from king crab Paralithodes camtschaticus and the elucidation of their primary structures. Constructing the phylogenetic tree of these enzymes was undertaken in order to prove the evolutionary relationship between them.
Results
The mature trypsin PC and collagenolytic protease PC contain 237 (Mcalc 24.8 kDa) and 226 amino acid residues (Mcalc 23.5 kDa), respectively. Alignments of their amino acid sequences revealed a high degree of the trypsin PC identity to the trypsin from Penaeus vannamei (approximately 70%) and of the collagenolytic protease PC identity to the collagenase from fiddler crab Uca pugilator (76%). The phylogenetic tree of these enzymes was constructed.
Conclusions
Primary structures of the two mature enzymes from P. camtschaticus were obtained and compared with those of other proteolytic proteins, including some enzymes from brachyurans. A phylogenetic analysis was also carried out. These comparisons revealed that brachyurins are closely related to their vertebrate and bacterial congeners, occupy an intermediate position between them, and their study significantly contributes to the understanding of the evolution and function of serine proteases.
Background
King crab collagenolytic serine protease PC [1] and trypsin PC [2] are brachyurins (MEROPS [3] peptidase classification: CLAN SA, family S1; NC-IUBMB enzyme classification: EC 3.4.21.32). Brachyurin is a term recommended by NC-IUBMB in 1992 for serine endopeptidases of a distinctive type found in crabs and their relatives. The name is derived from brachyurans, the phylogenetic subgroup of "true" crabs, which are a common source of these enzymes [4]. Early examples of the enzyme family include fiddler crab collagenase I [5], crayfish trypsin [6,7] and shrimp trypsin [8]. Other serine proteases were isolated from krill [9], crabs [10,11], crayfish [12], shrimps [13-17], and lobster [18]. There are at least three types of brachyurins: (Ia) the enzymes with a broad specificity, whose activities for synthetic substrates are similar to those of trypsin (Arg, Lys), chymotrypsin (Phe, Leu) and elastase (Ala, Leu) [9,16,19-21]; (Ib) broadly specific enzymes devoid of trypsin-like activity; and (II) the enzymes with a strict trypsin-like specificity (Arg, Lys) [10,13]. When preparing this article, we knew the amino acid sequences of six mature brachyurins: crab collagenase I [19,21], two crayfish trypsins [7,12], shrimp chymotrypsins I and II [22], and a shrimp trypsin [17]. The sequences encoding all the enzymes, except for crayfish trypsin, have been cloned.
In recent years, our laboratory has been engaged in the investigation of proteases from the Kamchatka king crab (P. camtschaticus) [1,2,23-25]. A number of proteases were isolated from the crab hepatopancreas. They are capable of collagen and fibrin lysis and are shown to exhibit a wound-healing activity in treatment of burns, trophic ulcers, and postoperative scars [24]. A homogeneous collagenolytic protease PC [1] and trypsin PC [2] were isolated from the king crab hepatopancreas using ion-exchange and affinity chromatographies.
In this article, we describe the construction of cDNA library from the total RNA of king crab P. camtschaticus and the isolation and sequencing of genes encoding trypsin PC and collagenolytic serine protease PC. We also compare here the primary structures of the enzymes with those of other serine proteases from invertebrate and vertebrate species. The evolution of crab trypsin and collagenase are examined by constructing a phylogenetic tree.
Results and discussion
Structural features of king crab brachyurins
The coding sequences of collagenolytic protease PC and trypsin PC were cloned and their amino acid sequences were established. An analysis of their nucleotide sequences suggests that the gene products are initially synthesized as precursor proteins, which are subsequently processed to the mature enzymes. The deduced protein sequence of collagenolytic protease PC consists of 270 residues and includes initiation Met, a 15-aa signal peptide, a 29-aa precursor peptide, and the active enzyme. The deduced protein sequence of trypsin PC comprises 266 aa and includes Met, derived from the initiation methionine codon, a 15-aa signal peptide, a 14-aa precursor peptide, and the mature enzyme. A comparison of the brachyurin propeptides additionally characterizes the enzyme group, because the propeptides have variable lengths and negligible identity. Crab collagenolytic protease PC, collagenase I, shrimp chymotrypsins I and II, and shrimp trypsin are derived from propeptides that are longer (29–44 residues) than those of the vertebrate proteases (8–15 residues). The function of these large activation domains is unclear, since they are unnecessary for the heterologous expression of proteases from vertebrates. The activation site of procollagenase PC (Val-Lys-Ser-Gln-Arg-Ile-Val-Gly-Gly) is closer to those of chymotrypsinogen (Ser-Gly-Leu-Ser-Arg-Ile-Val-Val-Gly) and proelastase (Glu-Thr-Asn-Ala-Arg-Val-Val-Gly-Gly), which are activated by trypsin, than to that of trypsinogen (Asp-Asp-Asp-Asp-Lys-Ile-Val-Gly-Gly), which is activated by enteropeptidase [26]. Interestingly, the identical activation sites of trypsin PC and shrimp trypsin (Arg-Gly-Leu-Asn-Lys-Ile-Val-Gly-Gly) are also devoid of an enteropeptidase site. This suggests that the activation cascades of the vertebrate digestive proteases have significantly diverged from those of crustaceans. Brachyurins may be self-activated, or other trypsin-like proteases in the hepatopancreas may fulfill this function.
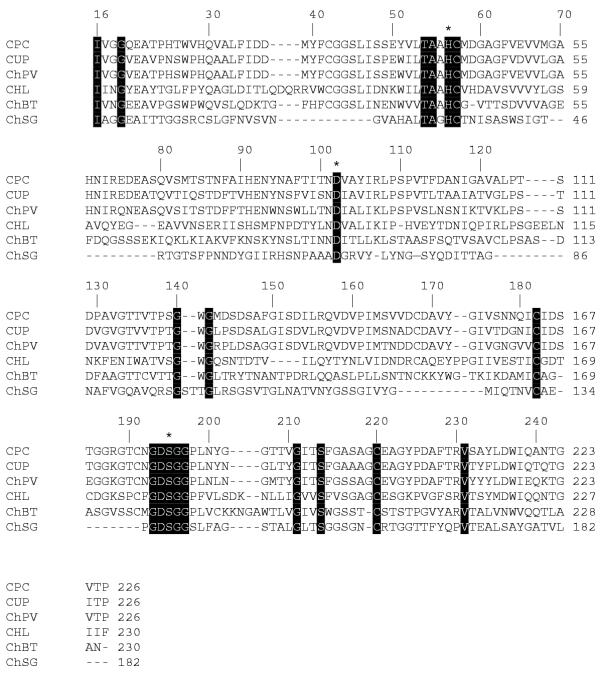
The mature collagenolytic protease PC contains 226 aa residues (which corresponds to its molecular mass of 23555 Da) and includes the active site residues His41 (57), Asp87 (102) and Ser179 (195), which are involved in the catalytic mechanisms of all serine proteases. The residue numbers in parentheses refer to topological equivalences in the classical nomenclature of chymotrypsin-like serine proteases. The sequences near these residues are highly conserved. Six half-cystinyl residues were found in king crab collagenolytic protease PC, which is comparable to six half-cystinyl residues in Uca pugilator brachyurin and ten half-cystinyl residues in chymotrypsinogen [27]. These residues probably form disulfide bonds Cys42–Cys58, Cys168–Cys182, and Cys191–Cys220, identical to three of four disulfide bonds in chymotrypsin (Cys42–Cys58, Cys168–Cys182, and Cys191–Cys220). In chymotrypsin, Ser189 forms the bottom of the substrate-binding pocket. The king crab collagenase has Gly in this position. At the upper part of the binding pocket, where chymotrypsin contains a Gly residue (Gly226), crab collagenase contains an Asp residue. An alignment of the amino acid sequences of the brachyurin collagenase from U. pugilator [28] and the collagenolytic protease PC reveals an unusual distribution of negative charges in their specific primary structures: they have a lower number of basic (six His and five Arg) than acidic residues (24) (Fig. 1). A low isoelectric point of collagenolytic protease PC (3.0) reflects this characteristic of the enzyme. Brachyurin from U. pugilator [21] also contains four His, four Arg, and only one Lys in contrast to 25 acidic residues, whereas chymotrypsin A has 16 acidic residues and 19 basic residues and its pI = 7.0.
Figure 1.
Alignment of amino acid sequences of collagenases and chymotrypsins. The completely conserved sites (13) are marked by black boxes. The catalytic triad (His, Asp, Ser) is denoted by stars above the sequences. Amino acid sequences are shown for: CPC, collagenase from P. camtschaticus (Q8WR11); CUP, collagenase from Uca pugilator (Q27824); ChPV, chymotrypsin II from Pen. vannamei (P36178); CHL, collagenase from Hypoderma lineatum (P08897); ChBT, chymotrypsin A from B. taurus (P00788); and ChSG, chymotrypsin-like protease from Streptomyces griseus (P00776). Swiss-Prot database accession numbers are given in parentheses. The numbers above the sequences follow the established chymotrypsinogen numbering system.
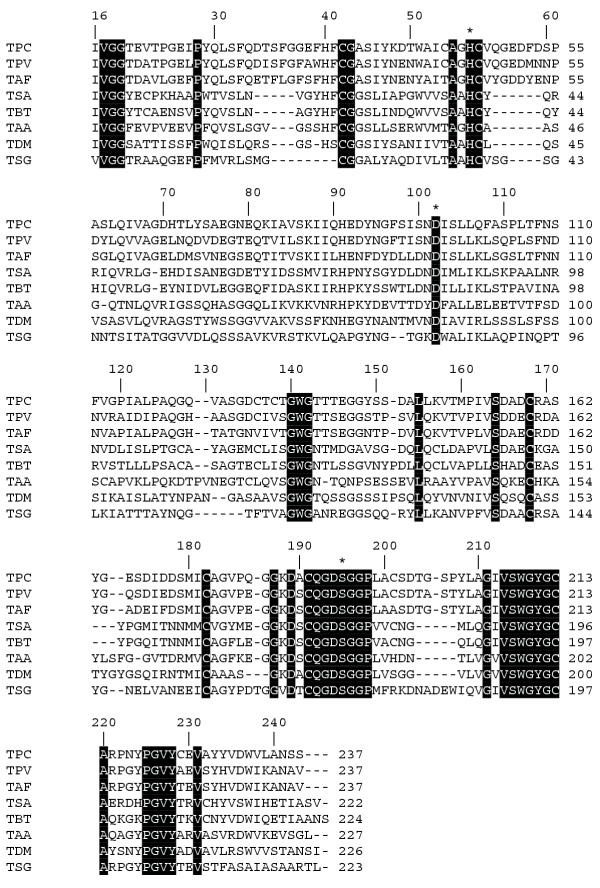
The mature trypsin PC contains 237 aa residues, which corresponds to its molecular mass of 24804 Da. The N-terminal amino sequence of the active enzyme, Ile-Val-Gly-Gly, is highly conserved in all trypsins. Multiple sequence alignments of trypsins are shown in Fig 2. They demonstrate that the residues forming the charge relay system in the active site of king crab trypsin [His41 (57), Asp87 (102) and Ser179 (195)] and the highly conserved sequences near these residues are the same in the corresponding regions of other serine proteases. Residue numbers in parentheses refer to topological equivalences in the classical nomenclature of chymotrypsin-like serine proteases. The primary (S1) binding pocket of all trypsins is strictly conserved; it includes Asp102 responsible for the interaction with the side chain of Arg or Lys (P1) residue of substrate. The amino acid residues that determine the trypsin specificity (Gly216 and Gly226) are conserved in trypsin PC as well. The active form of king crab trypsin comprises 11 Cys residues. Six Cys residues have been found in crayfish trypsin, whereas twelve Cys residues are characteristic of trypsins from vertebrate species [29]. The bridges Cys42–Cys58, Cys168–182, Cys191–Cys219, and Cys136–Cys201 are most likely present in trypsin PC. The Cys136–Cys201 disulfide bridge is a distinguishing feature of vertebrate serine proteases and is also present in shrimp trypsin [17] and in abalone, a chymotrypsin from a mollusk [30]. However, it is absent from the Pacifastacus leniusculus [7] and Astacus fluviatilis trypsins [12]. There are two additional disulfide bridges Cys22–Cys157 and Cys127–Cys232 in vertebrates [12]. The Glu70 residue, known as the calcium-binding site in bovine trypsin [31], is replaced by Asp in the crab trypsin, whereas Glu80, which also presents in calcium-binding sites of mammalian trypsins, is conserved in the crab trypsin. The calcium-binding site Glu230 of bacterial trypsin is also localized in the crab, crayfish, and shrimp trypsins but not in trypsins of vertebrates and insects [32]. The crab trypsin contains Thr in position 145, unlike bovine trypsin that contains Lys (a point of autocatalytic cleavage) in this position. Trypsin PC is resistant to autocatalytic cleavage [2], and it is interesting to emphasize that the crayfish trypsin contains Ser in this position [12]. A low isoelectric point of trypsin PC (3.0) reflects a low number of basic residues (four His, four Arg, and one Lys) and a large number of acidic residues (26) in its primary structure. Note that bovine trypsin contains three His, two Arg, and 14 Lys basic residues together with 10 acidic residues.
Figure 2.
Alignment of amino acid sequences of trypsins. The completely conserved sites (19) are marked by black boxes. The catalytic triad (His, Asp, Ser) is denoted by stars above the sequences. Amino acid sequences are shown for: TPC, trypsin from P. camtschatica (Q8WR10); TPV, trypsin from Pen. vannamei (Q27761); TAF, trypsin from Astacus fluviatilis (P00765); TSA, trypsin from Squalus acanthias (P00764); TBT, trypsin from Bos taurus (Q29463); TAA, trypsin from Aedes aegypti (P29787); TDM, trypsin from Drosophila melanogaster (P04814); and TSG, trypsin from S. griseus (P00775). Swiss-Prot database accession numbers are given in parentheses. The numbers above the sequences follow the established chymotrypsinogen numbering system.
A BLAST analysis reveals a high degree of structural identity of king crab collagenolytic protease to other brachyurins I (Fig. 1). The sequence identity (Table 1) reflects a short evolutionary distance between these enzymes. An alignment of the entire amino acid sequences of collagenase PC and trypsin PC revealed that they are more related to other crustacean enzymes than to those from vertebrates and microorganisms. For example, the sequence identity between the king crab collagenolytic serine protease and the collagenase from U. pugilator is 76%. Approximately the same correlation is characteristic of collagenase PC and the P. vannamei chymotrypsin (75%). A lesser sequence identity is found between collagenase PC and other chymotrypsins from vertebrates, insects, and microorganisms (18–29%). The sequences of trypsin PC and other trypsins of vertebrates and invertebrates are more or less correlated (Table 2). However, the sequence identity between the king crab collagenolytic protease PC and trypsin PC is no greater than that with other members of the S1 family (32%).
Table 1.
A comparison of mature collagenases and chymotrypsins. Figures indicate the identity percentage in alignment of the sequences presented in Fig. 1. The percentages are calculated using the TreeTop http://www.genebee.msu.su/services/phtree_reduced.html program
| CPC | CUP | ChPV | CHL | ChBT | ChSG | |
| CPC | 100 | 76 | 75 | 29 | 26 | 18 |
| CUP | 100 | 77 | 30 | 25 | 17 | |
| ChPV | 100 | 30 | 27 | 15 | ||
| CHL | 100 | 24 | 11 | |||
| ChBT | 100 | 14 | ||||
| ChSG | 100 |
Table 2.
A comparison of mature trypsins. Figures indicate the identity percentage in alignment of the compared sequences presented in Fig. 2. The percentages are calculated using the TreeTop http://www.genebee.msu.su/services/phtree_reduced.html program
| TPC | TPV | TAF | TAA | TDM | TSA | TBT | TSG | |
| TPC | 100 | 70 | 65 | 36 | 36 | 37 | 39 | 35 |
| TPV | 100 | 75 | 38 | 39 | 39 | 43 | 38 | |
| TAF | 100 | 38 | 36 | 39 | 41 | 36 | ||
| TAA | 100 | 39 | 39 | 39 | 31 | |||
| TDM | 100 | 33 | 36 | 31 | ||||
| TSA | 100 | 65 | 31 | |||||
| TBT | 100 | 30 | ||||||
| TSG | 100 |
A comparative structural and functional analysis indicates that these invertebrate enzymes are closely related to their analogues from vertebrates and bacteria but differ from them [11,19]. Thus, brachyurins demonstrate a high degree of structural similarity (25–36 kDa, 35–40% of amino acid sequence identity) to other members of the chymotrypsin family of serine proteases. However, these invertebrate enzymes have a lesser number of disulfide bonds than their analogues from vertebrates [12,19,22] and are particularly unstable at low pH values probably due to this reason [5,14]. Exceptionally acidic pI values are another special feature of them [5,13].
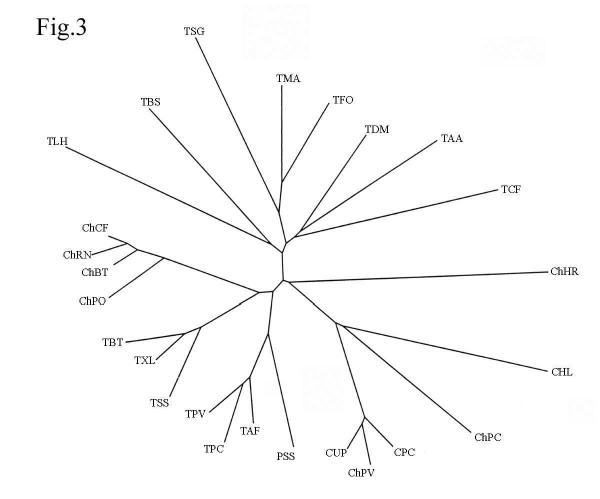
Phylogenetic tree
A phylogenetic tree, constructed in accordance with the multiple alignments, demonstrates an evolutionary equal separation of crustacean trypsins from both vertebrate and insect trypsins; they form an isolated cluster (Fig. 3). In its turn, crustacean collagenases are also equally separated from vertebrate chymotrypsins and crustacean trypsins and, together with other crustacean chymotrypsins, form a separate cluster.
Figure 3.
The unrooted phylogenetic tree of trypsins inferred from the amino acid sequence alignment (not shown). Abbreviations: ChCF, chymotrypsin from dog Canis familiaris (P04813); ChRN, chymotrypsin from rat Rattus norvegicus (P07338); ChBT, chymotrypsin from cow B. taurus (P00766); ChPO, chymotrypsin from fish Paralichthys olivaceus (Q9W7Q3); TBT, trypsin from cow B. taurus (Q29463); TXL, trypsin from frog Xenopus laevis (P19799); TSS, trypsin from fish Salmo salar (P35031); TPV, trypsin from shrimp Pen. vannamei (Q27761); TPC, trypsin from crab P. camtschaticus (Q8WR10); TAF, trypsin from crayfish Astacus fluviatilis (P00765); PSS, plasminogen activator from scolopendra Scolopendra subspinipes (O96899); CUP, collagenase from crab U. pugilator (P00771); ChPV, chymotrypsin from shrimp Pen. vannamei (P36178); CPC, collagenase from crab P. camtschaticus (Q8WR11); ChPC, chymotrypsin from insect Phaedon cochleariae (O97398); CHL, collagenase from insect Hypoderma lineatum (P08897); ChHR, chymotrypsin from mollusk Haliotis rufescens (P35003); TCF, trypsin from insect Choristoneura fumiferana (P35042); TAA, trypsin from insect A. aegypti (P29786); TDM, trypsin from insect Drosophila melanogaster (P04814); TFO, trypsin from fungus Fusarium oxysporum (P35049); TMA, trypsin from fungus Metarhizium anisopliae (Q9Y7A9); TSG, trypsin from bacterium S. griseus (P00775); TBS, trypsin from ascidium Botryllus schlosseri (O01309); and TLH, trypsin from bug Lygus hesperus (Q95P15). Primary accession numbers (Swiss-Prot database) of the protein sequences are given in parentheses. The tree was constructed as described in the Methods section.
For example, the Jones–Taylor–Thornton matrix shows that the P. camtschaticus trypsin is separated by 1.32 from Drosophila melanogaster trypsin, by 1.16 from Bos taurus trypsin, and by 0.41 from P. vannamei trypsin. King crab collagenase is 1.45 away from the king crab trypsin and 1.48 away from the bovine chymotrypsin, while the distance between the prawn chymotrypsin II and the king crab collagenase is 0.32. King crab trypsin and king crab collagenase are separated from the S. griseus trypsin by 1.57 and 1.78, respectively. Fungal trypsins are also more separated from the P. camtschaticus collagenase than from the P. camtschaticus trypsin.
Conclusions
A comparative sequence analysis of brachyurins, bacterial and vertebrate chymotrypsins, and trypsins allows us to understand the evolution of this serine protease family [12]. A closer inspection suggests that brachyurins share more sequence identity with vertebrate trypsins and chymotrypsins than with their bacterial analogues, while some important structural features, such as disulfide bonds, are conserved between the brachyurins and bacterial enzymes. Therefore, brachyurins occupy an intermediate evolutionary position between the bacterial and vertebrate trypsins and chymotrypsins. The major types of brachyurins can be distinguished on the basis of sequence similarity. The study of brachyurins significantly contributes to our understanding of the evolution of serine protease structure and function. Comparative structural and functional analyses indicate that these invertebrate enzymes are closely related to their vertebrate and bacterial analogues, but differ from them.
Methods
RNA isolation and cDNA library construction
A live king crab (P. camtschaticus) was obtained from the local market. Total RNA was isolated from 0.5 g of hepatopancreas of king crab by guanidine–phenol–chloroform extraction [33]. A cDNA library was obtained from 0.1 μg of total RNA and amplified by a SMART PCR cDNA Synthesis Kit (CLONTECH) using manufacturer's protocol. The amplified cDNA sample was 20-fold diluted with water and used in the following RACE procedures.
Isolation of the crab collagenase cDNA and trypsin cDNA
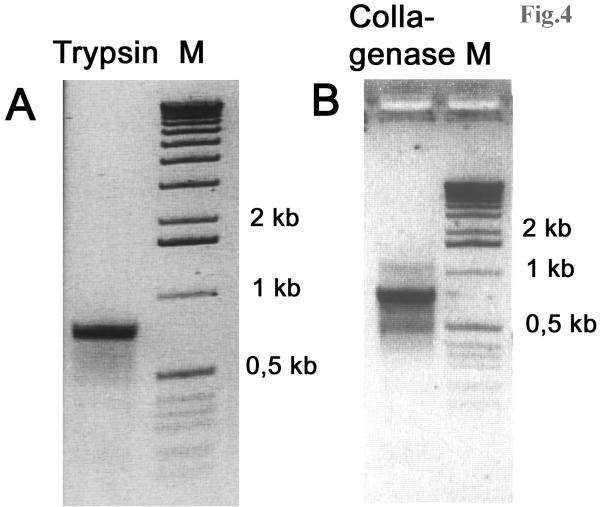
The target fragments of the cDNAs were isolated by a modified method for amplifying cDNA ends (RACE) based on step-out PCR [34]. Degenerative primers were designed to the first 10 aa residues at the N-terminus of the mature trypsin (IVGGTEVTPG) and to 9 aa of the mature collagenase (IVGGQEATP). They were 5'-GGC ACC GAG GTC ACC CCN GG and 5'-GGC GGC CAG GAG GCN ACN CC for trypsin and collagenase, respectively. The 3'-RACE PCR was carried out as follows: A 20-fold diluted amplified cDNA sample (1 μl) was added to the reaction mixture containing 1 × Advantage KlenTaq Polymerise mix (Clontech), manufacturer's 1 × reaction buffer, 200 μM dNTPs, 0.3 μM of degenerated primers (for trypsin or collagenase), and step-out primer system (0.02 μM "heel-carrier" oligo and 0.15 μM "heel-specific" oligo [34] in a total volume of 25 μl. Cycling was performed in a MJ Research PTC-200 Thermocycler in calculated mode: 25 cycles for trypsin and 26 cycles for collagenase were made using cycling profile: 95°C for 8 s, 65°C for 10 s, and 72°C for 2 min. PCR products were cloned into pT-Adv vector (Clontech) and sequenced using M13 direct and reverse universal primers by using a Beckman SEQ-2000 automated sequencer and the FS dye terminator chemistry. The primers for 5'-RACE were then designed: 5'-GTC GGA GCA GGC CAG AGG A-3' for trypsin and 5'-GGG GCC GCC AGA GTC TCC GT-3' for collagenase. PCR was carried out as described above (see Fig. 4). The putative signal peptides were determined using SignalP program [35].
Figure 4.
RT-PCR results for 5'-termini of trypsin and collagenase. A. Agarose gel electrophoresis of PCR products generated using 5'-RACE primer for trypsin and step-out primer system. B. Agarose gel electrophoresis of PCR products generated using 5'-RACE primer for collagenase and step-out primer system. About 1 ng of the first strand cDNA was used as a starting material for PCR reaction. Cycling was performed in a MJ Research PTC-200 Thermocycler in calculated mode: 25 cycles for trypsin and 26 cycles for collagenase were made using cycling profile: 95°C for 8 s, 65°C for 10 s, and 72°C for 2 min.
The GenBank accession numbers for two sequences determined in this study are AF461035 and AF461036 for collagenolytic serine protease PC and trypsin PC, respectively.
Alignment of amino acid sequences and a phylogenetic analysis of the crab collagenase and trypsin
The primary structures of enzymes used for the alignment were taken from Swiss-Prot database. Multiple sequence alignments were performed using ClustalW program with manual verification [36]. To construct the phylogenetic tree on the basis of the multiple sequence alignment, a pairwise distance matrix was set up by the Protdist program within the PHYLIP package [37]. The construction of the unrooted phylogenetic tree was performed by the Bionj program [38] according to the aforementioned matrix. The unrooted phylogenetic tree was drawn by TreeView program [39].
List of abbreviations
Collagenolytic serine protease PC, colladenolytic serine protease from Paralithodes camtschaticus; Trypsin PC, trypsin from Paralithodes camschaticus; and aa, number of amino acid residues.
Authors' contributions
DR carried out cloning and sequencing; YK carried out the phylogenetic tree construction and sequence alignments; GR conceived of the study, and participated in its design and coordination. All authors participated in writing this article and read and approved the final manuscript.
Acknowledgments
Acknowledgments
This work was supported by the Russian Foundation for Basic Research, project no. 02-04-48699.
Contributor Information
Galina N Rudenskaya, Email: gnruden@genebee.msu.ru.
Yuri A Kislitsin, Email: kislitsinyalex@hotmail.com.
Denis V Rebrikov, Email: den@ibch.ru.
References
- Rudenskaya GN, Isaev VA, Stepanov VM, Dunaevsky YaE, Baratova LA, Kalebina TS, Nurminskaya MV. Isolation and properties of serine proteinase PC of the kamchatka crab Paralithodes camchatica, a proteolytic enzyme of broad specificity. Biochemistry (Moscow) 1996;61:804–814. [PubMed] [Google Scholar]
- Rudenskaya GN, Isaev VA, Kalebina TS, Stepanov VM, Mal'tsev KV, Shvets SV, Luk'anova NA, Kislitsin YuA, Miroshnikov AI. Isolation and properties of trypsin PC from the king crab Paralithodes camtschatica. Bioorg Khim (Russian) 1998;24:112–118. [PubMed] [Google Scholar]
- Barret AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. Academic Press. 1998.
- Brusca RC, Brusca GJ. Invertebrates. Sunderland, MA:Sinauer Associates. 1990. pp. 595–666.
- Eisen AZ, Henderson KO, Jeffrey JJ, Bradshaw RA. A collagenolytic protease from the hepatopancreas of the fiddler crab, Uca pugilator, purification and properties. Biochemistry. 1973;12:1814–1822. doi: 10.1021/bi00733a024. [DOI] [PubMed] [Google Scholar]
- Zwilling R, Pfleiderer G, Sonneborn H, Kraft V, Stucky I. The evolution of the endopeptidases: V. Common and different traits of bovine and crayfish trypsin. Comp Biochem Physiol. 1969;28:1275–1287. doi: 10.1016/0010-406X(69)90566-0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Cortes P, Cerenius L, Garcia-Carreno F, Soderhall K. Trypsin from Pacifastacus leniusculus hepatopancreas : purification and cDNA cloning of the synthesized zymogen. Biol Chem. 1999;380:499–501. doi: 10.1515/BC.1999.065. [DOI] [PubMed] [Google Scholar]
- Gates RJ, Travis J. Isolation and comparative properties of shrimp trypsin. Biochemistry. 1969;8:4483–4489. doi: 10.1021/bi00839a039. [DOI] [PubMed] [Google Scholar]
- Turkiewicz M, Galas E, Kalinowska H. Collagenolytic serine protease from Euphausia superba dana (antarctic krill) Comp Biochem Physiol. 1991;99B:359–371. doi: 10.1016/0305-0491(91)90056-j. [DOI] [PubMed] [Google Scholar]
- Grant GA, Sacchettini JC, Welgus HG. A collagenolytic serine protease with trypsin-like specificity from the fiddler crab Uca pugilator. Biochemistry. 1983;22:2228–2233. doi: 10.1021/bi00271a019. [DOI] [PubMed] [Google Scholar]
- Klimova OA, Chebotarev YuV. Collagenic complex of proteases from hepatopancreas kamtchatka crab. Bull Exp Biol Med (Russia) 1999;128:308–313. [PubMed] [Google Scholar]
- Titani K, Sasagawa T, Woodbury RG, Ericsson LH, Dorsam H, Kraemer M, Neurath H, Zwilling R. Amino acid sequence of crayfish (Astacus fluviatilis) trypsin If. Biochemistry. 1983;22:1459–1465. doi: 10.1021/bi00275a021. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Liu HC, Tsai IH. The midgut trypsins of shrimp (Penaeus monodon): high efficiency toward native protein substrates including collagens. Biol Chem Hoppe-Seyler. 1990;371:851–859. doi: 10.1515/bchm3.1990.371.2.851. [DOI] [PubMed] [Google Scholar]
- Tsai IH, Chuang KI, Chuang JL. Chymotrypsins in digestive tracts of crustacean decapods (shrimps) Comp Biochem Physiol. 1986;85B:235–239. [Google Scholar]
- Tsai IH, Lu PJ, Chuang JL. The midgut chymotrypsins of shrimps (Penaeus monodon, Penaeus japonicus and Penaeus penicillatus) Biochim Biophys Acta. 1991;1080:59–67. doi: 10.1016/0167-4838(91)90112-D. [DOI] [PubMed] [Google Scholar]
- Van Wormhoudt A, Le Chevalier P, Sellos D. Purification, biochemical characterization and N-terminal sequence of a serine-protease with chymotrypsic and collagenolytic activities in a tropical shrimp, Penaeus vannamei (Crustacea, Decapoda) Comp Biochem Physiol. 1992;103B:675–680. doi: 10.1016/0305-0491(92)90389-9. [DOI] [PubMed] [Google Scholar]
- Klein B, Le MG, Sellos D, Van Wormhoudt A. Molecular cloning and sequencing of trypsin cDNAs from Penaeus vannamei (Crustacea, Decapoda): use in assessing gene expression during the moult cycle. Int J Biochem Cell Biol. 1996;28:551–563. doi: 10.1016/1357-2725(95)00169-7. [DOI] [PubMed] [Google Scholar]
- Johnston D, Hermans JM., Yellowlees D. Isolation and characterization of a trypsin from the slipper lobster, Thenus orientalis (Lund) Arch Biochem Biophys. 1995;324:35–40. doi: 10.1006/abbi.1995.9933. [DOI] [PubMed] [Google Scholar]
- Grant GA, Henderson KO, Eisen AZ, Bradshaw RA. Amino acid sequence of a collagenolytic protease from the hepatopancreas of the fiddler crab, Uca pugilator. Biochemistry. 1980;19:4653–4659. doi: 10.1021/bi00561a018. [DOI] [PubMed] [Google Scholar]
- Tsu CA, Perona JJ, Fletterick RJ, Craik CS. Structural basis for broad substrate specificity of the fiddler crab collagenolytic serine protease 1. Biochemistry. 1997;36:5393–5412. doi: 10.1021/bi961753u. [DOI] [PubMed] [Google Scholar]
- Tsu CA, Craik CS. Substrate recognition by recombinant serine collagenase 1 from Uca pugilator. J Biol Chem. 1996;271:11563–11570. doi: 10.1074/jbc.271.19.11563. [DOI] [PubMed] [Google Scholar]
- Sellos D, Van Wormhoudt A. Molecular cloning of a cDNA that encodes a serine protease with chymotryptic and collagenolytic activities in the hepatopancreas of the shrimp Penaeus vanamei (Crustacea, Decapoda) FEBS Lett. 1992;309:219–224. doi: 10.1016/0014-5793(92)80777-E. [DOI] [PubMed] [Google Scholar]
- Rudenskaya GN, Kupenko OG, Isaev VA, Stepanov VM, Dunaevsky YaE. Isolation and characterization of a carboxypeptidase from a kamchatka crab Paralithodes camtshatica. Bioorg Khim (Russian) 1995;21:249–255. [PubMed] [Google Scholar]
- Rudenskaya GN, Isaev VA, Shmoylov AM, Karabasova MA, Shvets SV, Miroshnikov AI, Brusov AB. Preparation of proteolytic enzymes from Kamchatka crab Paralithodes camchatica hepatopancreas and their application. Appl Biochem Biothech. 2000;88:175–183. doi: 10.1385/ABAB:88:1-3:175. [DOI] [Google Scholar]
- Rudenskaya GN, Shmoylov AM, Isaev VA, Ksenofontov AV, Shvets SV. Aminopeptidase PC from the hepatopancreas of the Kamchatka crab Paralithodes camchatica. Biochemistry (Moscow) 2000;65:164–170. [PubMed] [Google Scholar]
- Light A, Janska H. Enterokinase (enteropeptidase) comparative aspects. Trends Biochem Sci. 1989;14:110–112. doi: 10.1016/0968-0004(89)90133-3. [DOI] [PubMed] [Google Scholar]
- Brown JR, Hartley BS. Location of disulphide bridges by diagonal paper electrophoresis: the disulphide bridges of bovine chymotrypsinogen A. Biochem J. 1966;101:214–228. doi: 10.1042/bj1010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi M, Nakamura Y, Ogawa M, Yamamoto T, Nishide T, Mori T, Matsubara K. Cloning, characterization and nucleotide sequences of two cDNAs encoding human pancreatic trypsinogens. Gene. 1986;41:305–310. doi: 10.1016/0378-1119(86)90111-3. [DOI] [PubMed] [Google Scholar]
- Groppe JC, Morse DE. Molluscan chymotrypsin-like protease: structure, localization, and substrate specificity. Arch Biochem Biophys. 1986;305:159–169. doi: 10.1006/abbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- Rypniewski WR, Perrakis A, Vorgias CE, Wilson KS. Evolutionary divergence and conservation of trypsin. Protein Eng. 1994;7:57–64. doi: 10.1093/protein/7.1.57. [DOI] [PubMed] [Google Scholar]
- Muller H, Crampton A, della Torre, Sinden R, Crisanti A. Members of a trypsin gene family in Anopheles gambiae are induced in the gut by blood meal. EMBO J. 1993;12:2891–2900. doi: 10.1002/j.1460-2075.1993.tb05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signal P. p. V1.1.http://www.cbs.dtu.dk/services/SignalP/.
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressivemultiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) version 3.6. Distributed by author, Department of Genetics, University of Washington, Seattle. 1993.
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- http://taxonomy.zoology.gla.ac.uk/rod/treeview.html