Summary
GATA-1 and its cofactor FOG-1 are required for the differentiation of erythrocytes and megakaryocytes. In contrast, mast cell development requires GATA-1 and the absence of FOG-1. Through genome-wide comparison of the chromatin occupancy of GATA-1 and a naturally occurring mutant that cannot bind FOG-1 (GATA-1V205G), we reveal that FOG-1 intricately regulates the chromatin occupancy of GATA-1. We identified GATA1-selective and GATA-1V205G-selective binding sites and show that GATA-1, in the absence of FOG-1, occupies GATA-1V205G-selective sites, but not GATA1-selective sites. By integrating ChIP-seq and gene expression data, we discovered that GATA-1V205G binds and activates mast cell-specific genes via GATA-1V205G-selective sites. We further show that exogenous expression of FOG-1 in mast cells leads to displacement of GATA-1 from mast cell-specific genes and causes their downregulation. Together these findings establish a novel mechanism of gene regulation whereby a non-DNA binding cofactor directly modulates the occupancy of a transcription factor to control lineage specification.
Introduction
Cell lineage fate decisions are driven in large part by transcription factors that activate gene regulatory networks of one lineage while simultaneously repressing those of other lineages (Stathopoulos and Levine, 2005). Transcription factors achieve their specific activities by interactions with other transcription factors, cofactors, basal transcriptional machinery, and chromatin modifying enzymes. In the hematopoietic system, many lineage-specific transcription factors have been identified, and the complex interplay among them is central to the production of the broad diversity of cell types derived from the hematopoietic stem cell (Dore and Crispino, 2011; Orkin and Zon, 2008).
GATA-1 is a zinc finger transcription factor that drives the differentiation of megakaryocytes and erythrocytes and also is required for mast cell and eosinophil development (Crispino, 2005; Ferreira et al., 2005). The essential GATA-1 co-factor, Friend of GATA-1 (FOG-1), is likewise required for the differentiation of megakaryocytes and erythrocytes (Chang et al., 2002; Tsang et al., 1998; Tsang et al., 1997), but antagonizes GATA-1 mediated eosinophil and mast cell development (Cantor et al., 2008; Querfurth et al., 2000). FOG-1 contains nine zinc finger domains, but does not bind DNA directly (Fox et al., 1999). Rather, it interacts with the N-terminal zinc finger of GATA-1 and regulates GATA-1 transcriptional activity in diverse ways. For example, FOG-1 mediates both activation and repression of GATA-1 target genes (Fox et al., 1999; Grass et al., 2003; Letting et al., 2004), is required for GATA-1-driven chromatin looping (Vakoc et al., 2005), and facilitates GATA-1 chromatin occupancy at some sites (Letting et al., 2004; Pal et al., 2004). Furthermore, FOG-1 recruits the Nucleosome Remodeling/histone Deacetylase (NuRD) complex to both GATA-1 repressed and activated genes, and NuRD activity is required for lineage specification (Gao et al., 2010; Gregory et al., 2010; Hong et al., 2005; Miccio et al., 2010). Several recent studies have revealed a role for the FOG-NuRD complex in antagonizing mast cell differentiation in favor of erythromegakaryocytic fate (Cantor et al., 2008; Maeda et al., 2006; Sugiyama et al., 2008). FOG-1 is thought to interfere with mast cell development by repressing GATA-1 mast cell target genes.
The recent advent of next generation sequencing technology has provided the ability to study transcription factor occupancy on a genome scale. Despite a wealth of new data identifying the precise binding sites of dozens of transcription factors, binding site selection by these proteins remains a poorly understood phenomenon. In part, this process likely involves a permissive chromatin environment, such as nucleosome free region or a region of “open” chromatin marked by H3K4-methylation and H3 acetylation (Li et al., 2007). Other transcription factors or co-regulator complexes play a role in “reading” the chromatin landscape or preparing the site for binding (Guccione et al., 2006).
Mutations in the N-terminal zinc finger of GATA1 are found in rare congenital blood diseases. Specifically, GATA1 mutations that disrupt binding to FOG-1 have been found in patients with congenital dyserythropoietic anemia/thrombocytopenia (Ciovacco et al., 2008). These mutations include the V205M substitution, which is associated with defects in both red blood cell and platelet formation (Nichols et al., 2000). Although the presumption has been that loss of the interaction with FOG-1 alters GATA-1 transcriptional regulation due to lost recruitment of co-regulator complexes, we suspected that the mutation would also affect the chromatin occupancy of GATA-1 in vivo. Here, by comparing genome-wide chromatin occupancy of wild-type GATA-1 with that of GATA-1V205G (Crispino et al., 1999), we reveal intricate regulation of GATA-1 chromatin occupancy by FOG-1 and show that this modulation is critical for its control of hematopoietic lineage specification.
Results
Altered chromatin occupancy of a GATA-1 mutant that fails to interact with FOG-1
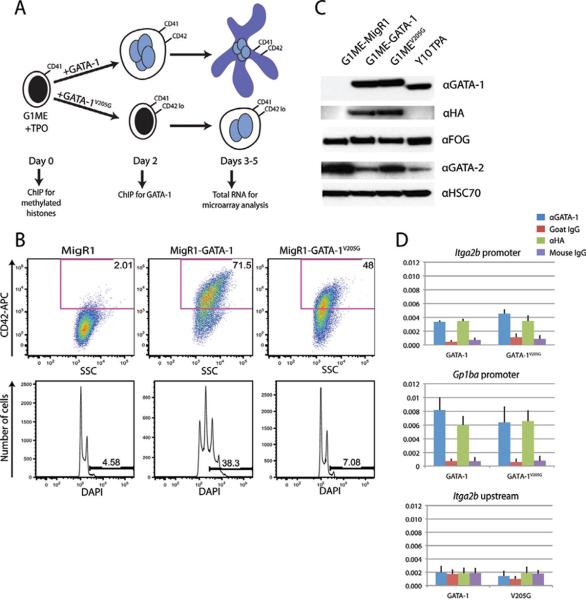
GATA-1 depends upon interaction with FOG-1 during terminal differentiation of both erythroid cells and megakaryocytes (Crispino et al., 1999; Nichols et al., 2000). To test the necessity of the GATA-1:FOG-1 interaction for GATA-1 chromatin binding, we expressed wild-type or the V205G mutant of GATA-1 in G1ME cells, a Gata1-null cell line that approximates the megakaryocyte-erythroid progenitor (Stachura et al., 2006). In the presence of thrombopoietin, cells reconstituted with wild-type GATA-1 undergo terminal megakaryocyte differentiation, including increased cell size and DNA content and expression of the CD42 cell surface marker (Stachura and Fig 1A,B). In contrast, cells expressing GATA-1V205G showed a marked deficiency in differentiation (Fig 1A,B), consistent with the phenotypes of mouse and humans harboring FOG-1 non-interacting mutations of GATA-1 (Chang et al., 2002; Nichols et al., 2000). Western blot analysis revealed that both GATA-1 and GATA-1V205G are expressed at a level approximately equal to the expression of endogenous GATA-1 in the Y10 megakaryocytic cell line after 48h treatment with TPA (Fig 1C). Given the essential requirement for FOG-1 in transcriptional regulation by GATA-1, we predicted that loss of the interaction might affect the ability of GATA-1 to bind chromatin in vivo. Chromatin immunoprecipitation (ChIP) with antibodies against either the GATA-1 protein or its HA tag revealed that enrichment at the Itga2b and Gp1ba promoters was equivalent between wild-type GATA-1 and GATA-1V205G (Fig 1D). This indicates that GATA-1 and GATA-1V205G can be immunoprecipitated from chromatin with relatively equal efficiency using either antibody.
Figure 1. The GATA-1 FOG-1 interaction is required for megakaryocytic differentiation.
(A) Schematic representation of megakaryocytic differentiation of G1ME cells after transduction with GATA-1 or V205G-expressing retroviruses. (B) FACS analysis of CD42 expression and DNA content (DAPI) 48h post infection with empty, GATA-1, or V205G-expressing MigR1 retrovirus. (C) Western blot of nuclear extracts from GFP+ G1ME cells 48h post-infection with empty, GATA-1, or GATA1V205G-expressing retrovirus and Y10 cells treated with TPA for 48h. (D) ChIP for GATA-1 and GATA-1V205G in G1ME cells using anti-GATA-1 and anti-HA-tag antibodies at the promoters of Itga2b and Gp1ba and a negative control region upstream of Itga2b. Means ± SD are depicted.
Genome wide analysis reveals GATA1-selective and GATA1V205G-selective binding sites
We performed ChIP-Seq for GATA-1 and GATA-1V205G in reconstituted G1ME cells cultured in TPO. Using anti-HA ChIP samples, we obtained nearly 20 million unique mappable reads for each factor and identified 12,736 and 11,696 binding sites enriched over input for GATA-1 and V205G, respectively. (Table S1). Each peak was matched to the nearest transcription start site (TSS), yielding 7,912 and 7,312 bound genes for GATA-1 and GATA-1V205G, respectively.
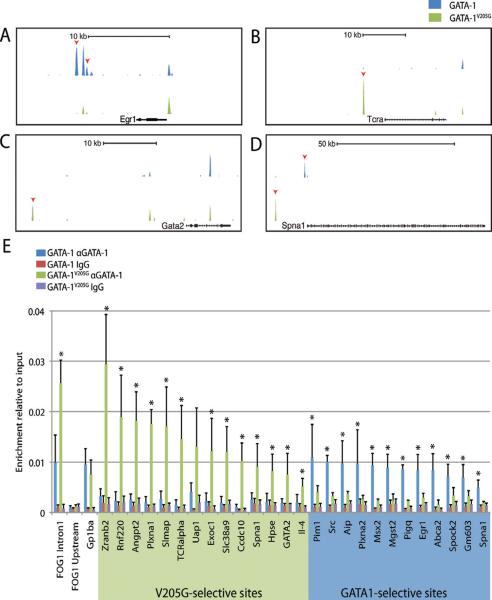
In order to identify peaks that are differentially bound by the two factors, we identified binding sites where one factor was enriched over the other factor by a p-value<1E-8 and where no peak was called for the other factor in the initial analysis. This approach identified 1421 GATA1-selective peaks and 1513 GATA1V205G-selective peaks (Fig 2A–D). To confirm this result, we performed qPCR analysis on GATA-1 and GATA-1V205G anti-HA ChIP material at ten GATA1-selective sites and ten GATA1V205G-selective sites. We found that all of the GATA1V205G selective peaks displayed at least a 2.5-fold increase in enrichment compared to GATA-1 (data not shown). Moreover, 9/10 of the GATA1-selective sites validated by the same standard (data not shown). In order to rule out differential antibody sensitivity as the cause for differences in chromatin binding, we repeated the ChIP-PCRs at the validated sites with an anti-GATA-1 antibody and found similar differences (Fig 2E).
Figure 2. GATA1 and GATA-1V205G bind distinct sites in chromatin.
(A–D) UCSC Genome Browser depiction of GATA1-selective and GATA-1V205G -selective peaks at representative loci. Red arrows indicate sites of differential binding. (E) ChIP-qPCR with a GATA1-specific antibody for GATA-1 and GATA-1V205G at a panel of validated GATA-1V205G -selective and GATA1-selective sites. Means ± SD are depicted. Asterisks mark sites where enrichment is significantly different.
We also compared binding to several other sites located at genes of interest, including a GATA1V205G-selective peak 24kb upstream of Gata2 (Fig 2D), and both a GATA1-selective and a GATA1V205G-selective peak upstream of the Spna1 gene, which encodes the erythroid membrane protein α-spectrin (Fig 2E). We found that these sites were also validated by the same standard (Fig 2F). The results of this qPCR validation confirm that there exist sites where the inability of GATA-1 to interact with FOG-1 can either increase or decrease its affinity for chromatin.
GATA1V205G-selective binding sites are distinct in genomic location and chromatin conformation
To understand how FOG-1 alters GATA-1 binding, we studied the differences among the full set of GATA-1 binding sites and our GATA1-selective and GATA1V205G-selective binding sites. We mapped the location of all peaks from the GATA-1 and GATA-1V205G datasets and fit each location into one of five categories: upstream (2–100kb), promoter (<2kb upstream), intragenic (exon or intron), downstream (<100kb), or gene desert (>100kb) (Fig S1B). This analysis revealed that about half of the peaks for GATA-1 and GATA-1V205G were located at promoters (10% and 8%) or within genes (37% and 36%). About one-third of the peaks lie upstream of the assigned TSS (32% and 34%), and relatively few lie downstream of the closest gene (16% and 16%) or in gene deserts (5% and 6%). We compared these data to a randomly generated set of peaks and found that the GATA peaks are enriched at locations where gene regulatory elements tend to be found (promoter, intragenic, and upstream regions) and depleted in gene desert regions (Fig S1B).
We next compared the distribution of the full set of GATA-1 peaks to the GATA1-selective and GATA1V205G-selective binding sites. This analysis revealed that the GATA1-selective peaks were distributed similarly to the full set of GATA-1 peaks (compare Fig S1C and Fig S1B). In contrast, the GATA1V205G-selective peaks displayed a reduced frequency at promoter elements (10% vs 2%, p=2.94E-20) and increased frequencies in gene desert regions (5% vs 10%, p=1.57E-18) (Fig S1C). Strikingly, the GATA1V205G-selective peaks were no more likely to be found at a promoter than the randomly generated set of peaks (3% vs 2%, p=0.14). This analysis indicates that the GATA1V205G-selective sites are more likely to be distal elements than GATA1-selective and FOG-independent GATA-1 binding sites.
The chromatin environment, including nucleosome occupancy and histone modifications, is thought to be a major determinant of binding by transcription factors and their co-regulators. Previous studies have shown that GATA-1 prefers to bind to sites with an “open” chromatin conformation, characterized by histone acetylation and H3K4-methylation (Wozniak et al., 2007; Wu et al., 2011). To examine the role of the chromatin environment in the regulation of GATA-1 binding by FOG-1, we overlapped the primary sets of GATA-1 and GATA-1V205G peaks with a histone H3 trimethyl-lysine 4 (H3K4me3) ChIP-Seq dataset from proliferating G1ME cells (Dore et al., 2012). This analysis revealed that 33% of the total GATA-1 binding sites and 27% of the total GATA-1V205G binding sites lie at sites of H3K4me3 enrichment (Table S2). We then analyzed the GATA1-selective and GATA1V205G-selective binding sites and found that 28% of the GATA1-selective sites overlap with H3K4me3 peaks. In contrast, only 10% of the GATA1V205G-selective sites displayed H3K4me3 enrichment. This indicates that the GATA1V205G-selective sites are much less likely (p=5.37E-57) to have an “open” chromatin conformation before binding than normal GATA-1 binding sites. Together with the altered genomic distribution of the GATA1V205G-selective sites, these data suggest that GATA-1V205G binds chromatin promiscuously in that it finds additional binding sites that are unnecessary for normal megakaryocyte differentiation.
GATA1V205G-selective binding sites are distinct in motif composition
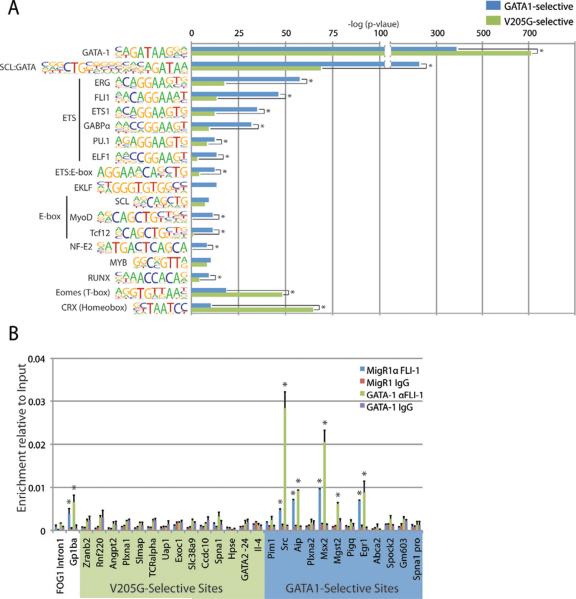
GATA-1 interacts with a myriad of transcriptional co-regulators alone or through FOG-1 (Lowry and Mackay, 2006). We predicted that these interacting factors might play a crucial role in regulation of GATA-1 binding site selection. To test this hypothesis, we performed de novo and informed motif finding on the full peak sets, as well as the GATA1-selective and GATA1V205G-selective binding sites. Unbiased motif finding using DREME revealed a perfect WGATAA motif enriched in both the GATA-1 and GATA-1V205G peak sets (Fig S2A). The second-most commonly discovered motif in both peak sets was an Ets-like motif with the core sequence AGGAA (Fig S2A). Previous studies have shown that GATA-1 and several Ets proteins, including GABPA, ETS1, and FLI1, co-regulate target genes in megakaryocytes (Deveaux et al., 1996; Eisbacher et al., 2003; Pang et al., 2006; Tijssen et al., 2011; Wang et al., 2002). The co-occurrence of their motifs at a high percentage of GATA-1 binding sites suggests a functional relationship between GATA-1 and Ets-family transcription factors in megakaryocyte differentiation. Next, using MEME for de novo motif discovery in the GATA1-selective and GATA1V205G-selective binding sites, we identified enrichment of the WGATAA motif, an Ets motif, an E-box motif, and a CACCC box motif within the GATA1-selective sites (Fig S2B). In contrast, at the GATA1V205G-selective sites, we observed enrichment of the WGATAA motif, an AP2-like motif, an E-box-like motif, and TG and CA dinucleotide repeat motifs. Strikingly, Ets motifs were not enriched among the GATA1V205G-selective sites.
To identify statistically significant differences in motif enrichment, we used an informed motif finding program with experimentally validated transcription factor binding motifs. This analysis revealed that Ets motifs are strongly enriched (Erg motif, p=1E-37) in the GATA1-selective sites relative to the GATA1V205G-selective sites (Fig 3A). Several other classes of motifs were also enriched, including the GATA:SCL combined motif (p=1E-35), an E-box (MYOD, p=1E-23), and the NF-E2 motif (p=1E-13). The inverse analysis revealed that the WGATAA motif is significantly more enriched in the GATA1V205G-selective sites relative to the GATA1-selective sites (p=1E-55). This result indicates that the GATA1V205G-selective sites are highly enriched for a WGATAA consensus GATA-1 binding motif and that if anything, these sites are more likely to contain the WGATAA motif than the GATA1-selective sites. In addition, a Homeobox motif (p=1E-26) and a T-box motif (p=1E-8), two motifs that previously had no association with GATA-1 function, were significantly enriched in the GATA1V205G-selective sites. Taken together, these data indicate that GATA-1V205G binds chromatin at sites that contain a distinct milieu of transcriptional regulators and chromatin conformation.
Figure 3. GATA1V205G-selective sites are devoid of ETS protein binding sites.
(A) Informed motif analysis of the ChIP-seq datasets. The enrichment of selected motifs in each peak set was plotted as the −log(p-value). Asterisks mark motifs that are significantly enriched in one peak set relative to the other (p<.01). (B) ChIP for FLI-1 in control and GATA1-transduced G1ME cells at the GATA-1V205G and GATA1-specific sites. Means ± SD are depicted. Asterisks mark sites where enrichment is significantly greater than control IgG.
The ETS protein FLI-1 occupies GATA1-selective sites but not GATA1V205G-selective sites
Due to the strong enrichment in ETS motifs at GATA-1 selective sites, we sought to determine if FLI-1, an ETS protein with known roles in megakaryocyte development, binds at these sites. We performed ChIP on FLI-1 in control and GATA1-expressing G1ME cells and found that FLI-1 significantly occupies 5/12 GATA1-selective sites and 0/14 GATA1V205G-selective sites (Figure 3B). These data confirm that the GATA1V205G-selective sites are devoid of ETS proteins and are thus distinct from normal GATA-1 binding sites. Furthermore, it is likely that FLI-1 and other ETS proteins likely contribute to the GATA1-selective occupancy of these sites.
Wild-type GATA-1 binds to GATA1V205G-selective sites, but not GATA1-selective sites in the absence of FOG-1
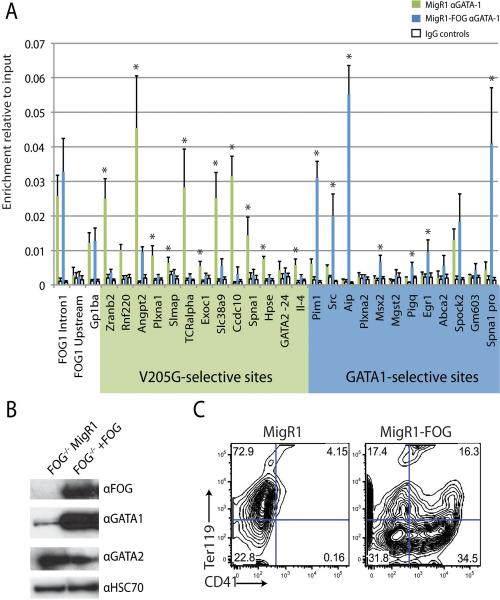
If FOG-1 indeed regulates the chromatin occupancy of GATA-1, we would expect that in the absence of FOG-1, GATA-1 would fail to bind GATA1-selective sites but would bind strongly to GATA1V205G-selective sites. To test this hypothesis, we assayed binding of endogenous wild-type GATA-1 in FOG-1 deficient hematopoietic cells (Cantor et al., 2002). As predicted, wild-type GATA-1 occupancy closely resembled that of GATA-1V205G in this context (Fig 4A). Specifically, GATA-1 bound 12/13 GATA1V205G-selective sites and failed to bind 9/12 GATA1-selective sites. Next, we sought to determine if reconstitution of FOG-1 expression could revert GATA-1 occupancy to the wild-type pattern (Fig 4A). We transduced the cells with control and FOG1-expressing MigR1 retrovirus and found that GATA-1 occupancy was significantly reduced at all 12 of the bound GATA1V205G-selective sites and significantly increased at 8/12 GATA1-selective binding sites. Western blot analysis confirmed expression of FOG-1 in the reconstituted, but not control infected cells (Fig 4B). Interestingly, the western blot also revealed that restoration of FOG-1 expression increased the expression of GATA-1. Despite this upregulation, GATA-1 was still efficiently displaced from the GATA1V205G-selective sites. Of note, the change in occupancy caused by reconstitution of FOG-1 was not a result of terminal differentiation of the cells, as although FOG-1 reconstituted cells expressed lower levels of Ter119 and higher levels of CD41, they did not become polyploid and only marginally up-regulated CD42 (Fig 4C and data not shown). Taken together, these data verify that the abnormal binding behavior of GATA-1V205G is a direct result of its inability to associate with FOG-1.
Figure 4. Ectopic expression of FOG-1 in FOG-deficient cells displaces wild-type GATA-1 from GATA-1V205G-selective binding sites.
(A) FOG-1 deficient hematopoietic cells were infected with empty or FOG-1 and the occupancy of endogenous GATA-1 was assayed at a panel of GATA-1V205G-selective and GATA1-selective sites by ChIP-qPCR. Means ± SD are depicted. (B) Western blot analysis on nuclear extracts from sorted GFP+ FOG-1 deficient cells infected with empty or FOG-1 MigR1 expressing retrovirus. (C) FACS analysis for Ter119 and CD41 expression on infected FOG-1 deficient cells 3 days post-infection.
Gene expression analysis reveals that GATA-1V205G aberrantly regulates GATA-1 target genes
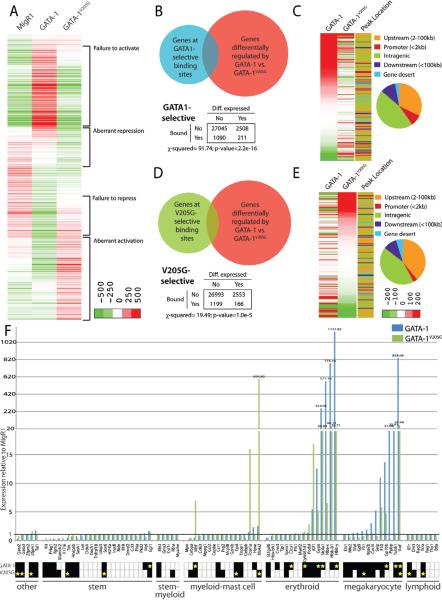
To determine how altered GATA-1 chromatin occupancy affects gene expression, we examined global gene expression by microarray in G1ME cells 72h post-transduction with GATA1, GATA1V205G, or a control virus. Between the 3 conditions, we identified a total of 5123 differentially expressed genes (fold change>1.5, p-value<0.05), of which 2720 were differentially expressed in the GATA1V205G-expressing cells relative to the GATA-1-expressing cells (Fig 5A). We overlapped the genes differentially expressed by each factor with the genes that are bound by each factor in the full ChIP-Seq datasets and found statistically significant overlap (p-value<2.2E-16) (Fig S3A, C). There was no tendency towards activation or repression in the direct target genes of either factor (Fig S3B, D).
Figure 5. Global gene expression analysis reveals that FOG-1 modulates GATA-1 chromatin occupancy to maintain lineage-specific gene expression.
(A) Heatmap of 5123 differentially expressed genes among sorted GFP+ G1ME cells infected with MigR1 GATA-1, or GATA1V205G-expressing retrovirus. The genes were divided into groups based on hierarchical clustering. (B) Overlap of the genes closest to the GATA1-selective binding sites with the genes differentially expressed by GATA-1 relative to GATA-1V205G. Chi-square analysis reveals that the differentially bound genes are enriched for differentially expressed genes (p<2.2E-16). (C) Heatmap of the genes that overlap in panel B. The location of each peak relative to the TSS is plotted in an adjacent column and in a pie chart. (D) Overlap of the genes closest to the GATA-1V205G-selective binding sites with the genes differentially expressed by GATA-1 relative to GATA-1V205G. Chi-square analysis reveals that the differentially bound genes are enriched for differentially expressed genes (p<2.2E-16). (C) Heatmap of the genes that overlap in panel B. The location of each peak relative to the TSS is plotted in an adjacent column and in a pie chart. (D) Overlap of the genes closest to the GATA-1V205G-selective binding sites with the genes differentially expressed by GATA-1 relative to GATA-1V205G. Chi-square analysis reveals that the differentially bound genes are enriched for differentially expressed gene (p=1.0E-5). (E) Heatmap of the genes that overlap in panel D. The location of each peak relative to the TSS is plotted in an adjacent column and in a pie chart. (F) The expression of genes for multiple hematopoietic cell types is plotted for GATA-1 and GATA1V205G-expressing cells relative to control cells. The presence of a ChIP-Seq peak for GATA-1 or GATA-1V205G at each gene is included in the boxes below the graph: black box indicates that the gene is bound by the factor, while a yellow star indicates that a GATA1-selective or GATA1V205G-selective binding site is present.
Using hierarchical clustering, we identified four major groups of differentially expressed genes (Fig 5A). The groups were defined as genes that GATA-1V205G (1) fails to activate, (2) aberrantly represses, (3) fails to repress, or (4) aberrantly activates. We completed gene ontology analysis on these groups and found results consistent with the failed differentiation of GATA1V205G-expressing cells (Fig S4). Notably, the “fails to activate” and “aberrant repression” groups are enriched for classes of genes that are associated with megakaryocytic differentiation, such as platelet aggregation and cytoskeletal rearrangement, and the “fails to repress” and “aberrant activation” are enriched for cell cycle progression and DNA damage response genes.
FOG-1 mediated modulation of chromatin occupancy alters hematopoietic gene expression programs
Having identified sites where FOG-1 regulates GATA-1 chromatin occupancy, we set out to determine if this activity altered the expression of nearby genes. To do this, we overlapped the genes assigned to the GATA1-selective and GATA1V205G-selective sites with the 2720 genes differentially expressed in the GATA-1V205G cells relative to GATA-1 (Fig 5B, D). This analysis identified 211 differentially expressed genes located near GATA1-selective sites and 166 differentially expressed genes near GATA1V205G-selective sites. These numbers represent a significant enrichment of the differentially bound genes for differentially expressed genes by chi-square analysis (Fig 5B, D). We used hierarchical clustering to make heatmaps of the differentially bound and expressed genes. This revealed that 68% of the differentially expressed genes with a GATA1-selective binding site are activated, indicating a preference for gene activation by GATA-1 from these sites (Fig 5C). Among the differentially regulated genes that have a GATA1V205G-selective binding site, 110 are significantly changed compared to MigR1 control and 63 of these are activated (57%) (Fig 5E).
In order to understand how the modulation of GATA-1 chromatin occupancy alters hematopoietic gene expression, we next analyzed the expression and occupancy status of gene sets associated with various hematopoietic cell types including stem cells, myeloid progenitors, mature myeloid and mast cells, erythroid cells, megakaryocytes, and lymphocytes (Fig 5F). Consistent with the differentiation of the cells to mature megakaryocytes, GATA-1 directly induces robust activation of nearly all of the megakaryocyte signature genes. Additionally, the GATA-1 expressing cells exhibit expression of a subset of erythroid genes, most notably the genes encoding hemoglobin subunits, despite the culture of the G1ME cells in thrombopoietin and not in erythropoietin. As expected, this analysis revealed that GATA1V205G-expressing cells are deficient in expression of genes in the megakaryocyte and erythrocyte differentiation programs. By inspection of GATA-1 and GATA-1V205G chromatin occupancy at these genes, we found that many have FOG-regulated GATA-1 binding sites. Notably, there are GATA1-selective binding sites at the megakaryocytic genes Gp1bb, Tubb1, and Vwf, three genes associated with megakaryocyte and platelet function. There are also GATA1-selective sites at several erythroid genes, some of which were previously identified, such as the globin genes and Slc4a1 (Band3).
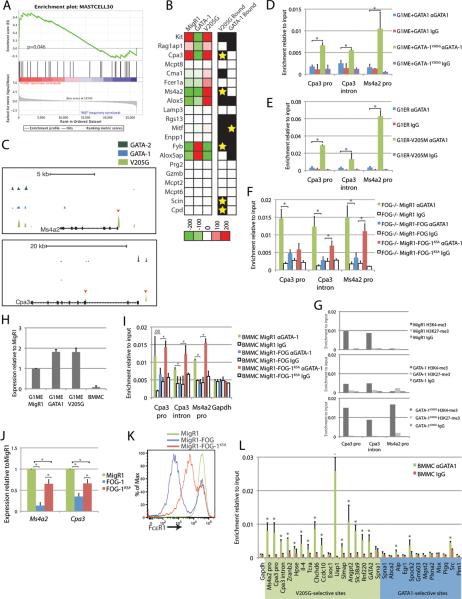
While the failure of GATA-1V205G to induce erythromegakaryocytic differentiation programs was expected, the most remarkable finding from this analysis was that GATA-1V205G strongly activates the expression of genes in the myeloid-mast cell group including Mitf, which encodes a transcription factor with known roles in mast cell development, and Fcer1a and Ms4a2, which encode the two chains of the Fc-epsilon receptor 1, a marker of mast cells. This aberrant activation of mast cell genes is reminiscent of the expansion of mast cells in mice harboring FOG-1 non-binding mutations in GATA-1 and GATA-2, FOG-1 knockout mice, and FOG-1 NuRD-binding mutations (Cantor et al., 2008; Gao et al., 2010; Gregory et al., 2010). Furthermore, the Ms4a2 promoter contains a GATA1V205G-selective binding site, indicating that the activation of this gene is direct. We performed Gene Set Enrichment Analysis (GSEA) on a set of mast cell-expressed genes previously defined from human primary mast cells (Nakajima et al., 2001). This analysis revealed significant enrichment of mast cell gene expression in the GATA-1V205G-transduced cells but not in the GATA1-transduced or the control cells (p-value=0.046, Fig 6A). Together, our findings suggest that FOG-1 antagonizes GATA-1 function in mast cells not by suppressing GATA-1 target gene expression, but rather by directly inhibiting chromatin binding of GATA-1 at mast cell gene regulatory elements.
Figure 6. FOG-1 prohibits GATA-1 occupancy from mast cell genes to maintain lineage fidelity.
(A) GSEA for a set of mast cell-specific genes on the GATA-1V205G samples relative to the MigR1 and GATA-1 samples. (B) Heatmap of expression of mast cell genes from the microarray. The presence of a ChIP-Seq peak for GATA-1 or GATA-1V205G at each gene is included in the boxes beside the graph: black box indicates that the gene is bound by the factor, while a yellow star indicates that a GATA1-selective or GATA-1V205G-selective binding site is present. (C) UCSC Genome Browser depiction of ChIP-Seq data at the Ms4a2 and Cpa3 loci. GATA-2 ChIP-Seq data are from uninfected G1ME cells. The red arrows indicate the location of GATA-1V205G-selective binding sites. (D) ChIP using a GATA1-specific antibody for GATA-1 and GATA-1V205G in G1ME cells. Primers amplify the GATA-1V205G-selective binding sites depicted in panel B. (E) ChIP for GATA-1 in G1ER and G1ER-V205M cells induced with β-estradiol for 24h. (F) ChIP for GATA-1 in sorted GFP+ FOG-1 deficient cells 2 days post-infection with empty, FOG-1, or FOG-1K5A MigR1 expressing retrovirus. (G) ChIP for H3K4-me3 and H3K27-me3 at GATA-1V205G-selective sites at Cpa3 and Ms4a2. (H) qPCR analysis for expression of the Zfpm1 gene (FOG-1) in G1ME cells 3 days post-infection with empty, GATA-1, or GATA-1V205G-expressing and in BMMCs. (I) ChIP for GATA-1 in sorted GFP+ BMMCs 2 days post infection with empty, FOG-1, or FOG-1K5A MigR1 expressing retrovirus. (J) qPCR analysis of gene expression for the indicated genes in sorted GFP+ BMMCs 3 days post infection with empty, FOG-1, or FOG-1K5A MigR1 expressing retrovirus. (K) FACS analysis of FcεR1 expression on GFP+ BMMCs 3 days post infection with empty, FOG-1, or FOG-1K5A MigR1 expressing retrovirus. (L) ChIP for GATA-1 in BMMCs at a panel of validated GATA1V205G-selective and GATA1-selective sites.
FOG-1 prohibits GATA-1 occupancy at mast cell genes to maintain lineage fidelity
We next examined the expression and GATA factor occupancy of a set of mast cell genes culled from the literature (Fig 6B). This revealed that, in addition to Ms4a2, GATA-1V205G also selectively binds and regulates the promoter of the mast cell specific carboxypeptidase Cpa3, a known GATA-1 target gene (Tripic et al., 2009). Additionally, there are GATA1V205G-selective binding sites at other mast cell genes including Fyb, Scin, and Cpd.
To test whether FOG-1 prohibits GATA-1 binding at these genes, we performed ChIPs for GATA-1 and GATA-1V205G at the Ms4a2 and Cpa3 promoters and intron 3 of Cpa3 in G1ME and G1ER cells, a related GATA1-null cell line with strictly erythroid differentiation potential (Fig 6C–E). We also probed enrichment for the endogenous GATA-1 in the FOG-1 deficient cell line transduced with control or FOG1-expressing retrovirus (Fig 6F). This analysis revealed that the GATA-1:FOG-1 interaction dramatically reduces GATA-1 chromatin binding at these sites in all three cell line models. Furthermore, since FOG-1 requires its interaction with the NuRD repressor to repress mast cell-specific gene expression, we used a mutant of FOG-1 (FOG-1K5A) that is deficient for the interaction with the NuRD repressor to reveal that this interaction is required for FOG-1 to prohibit occupancy at Cpa3 intron 3 and the Ms4a2 promoter (Fig 6F). In order to confirm that the reduced enrichment by wild-type GATA-1 at these sites was not caused by masking of the GATA-1 epitope by FOG-1, we completed ChIPs for FOG-1 in control-infected and GATA1-expressing G1ME cells and found enrichment for FOG-1 at 3 control sites, but not at the Ms4a2 or Cpa3 promoters (Fig S5).
Next, we examined the changes in the chromatin environment induced by expression of GATA-1 or GATA-1V205G. We performed ChIP for tri-methylated H3K4 and H3K27 in MigR1, GATA-1, and GATA1V205G-expressing G1ME cells 3 days post-infection (Figure 6G). This analysis revealed that GATA-1V205G induces robust tri-methylation of H3K4 at the Ms4a2 promoter, consistent with its direct activation of the gene. In contrast, there is no increase in H3K4-me3 at this promoter in GATA1-expressing cells. At Cpa3, the promoter and intron are bound by H3K4-me3 in control cells, and this signal is maintained in GATA1V205G-expressing cells, consistent with the continued elevated expression of this gene. In contrast, expression of GATA-1 reduces the H3K4-me3 at this locus by about half. This is likely caused by reduced occupancy by GATA-2 in this context since its expression is decreased.
Next, we tested whether exogenous FOG-1 expression could prohibit GATA-1 occupancy at mast cell genes in primary mast cells. We generated BMMCs from mouse bone marrow cells and confirmed that these cells express dramatically less FOG-1 than erythromegakaryocytic cells by qPCR (Figure 6H). We then infected these cells with retroviruses expressing wild-type and K5A-mutant FOG-1, and performed ChIP on sorted GFP+ cells. We found that GATA-1 bound to the Cpa3 and Ms4a2 sites in the control-infected BMMCs, but upon overexpression of FOG-1, GATA-1 binding was lost (Fig 6I). In contrast, FOG-1K5A was unable to displace GATA-1, consistent with a requirement for NuRD complex in FOG-mediated repression of these genes. The FOG-NuRD-mediated changes in chromatin occupancy coincided with reduced expression of both genes (Fig 6J) and reduced staining for FcεR1 on the cell surface (Fig 6K).
Lastly, in order to determine if the global occupancy pattern of GATA-1 in mast cells is similar to that of GATA-1V205G in megakaryocytes, we examined enrichment for endogenous GATA-1 in BMMCs at the same panel of GATA1V205G-selective and GATA1-selective sites. We found that 12 of 14 GATA1V205G-selective sites were bound in mast cells whereas only 3 of 12 GATA1-selective sites were bound (Fig 6L). Together, these data lead us to conclude that FOG-1 actively restricts GATA-1 chromatin binding in megakaryocytes to prevent activation of the mast cell gene expression program and maintain megakaryocyte identity.
Discussion
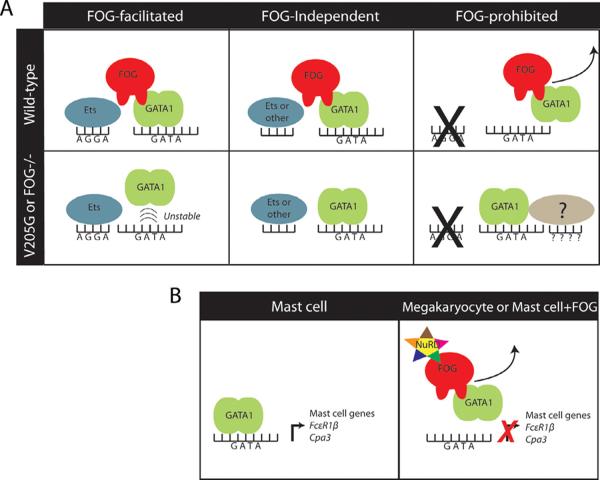
In this study, we discovered that FOG-1 regulates GATA-1 transcriptional activity not merely by recruiting co-activators and co-repressors, but also by directly modulating chromatin occupancy. We have identified three classes of GATA-1 binding sites: GATA1-selective, GATA1V205G-selective, and sites that are bound by both factors. We consider these sites to be 1) FOG-facilitated, 2) FOG-prohibited, and 3) FOG-independent (Fig 7A). At the FOG-facilitated sites, GATA-1 requires FOG-1 to either achieve or stabilize binding. We predict that this difference is mediate by interactions of FOG-1 with other chromatin factors that increase the stability of GATA-1 at the sites, such as ETS family proteins. This model is supported by our detection of a high proportion of FOG-facilitated sites that contain ETS motifs. Indeed, several previous studies have shown that GATA-1, FOG-1, and the ETS protein FLI-1 exist in transcriptional complexes at megakaryocyte genes (Huang et al., 2009; Pang et al., 2006; Wang et al., 2002; Woo et al., 2008). One study found that FOG-1 was required for activation of the αIIb promoter by GATA-1 and FLI-1, suggesting that FOG-1 is a vital participant in this complex (Wang et al., 2002). It is enticing to speculate that FLI-1 physically interacts with GATA-1/FOG-1 pairs to promote their occupancy at GATA-ETS elements. There is one report of a direct physical interaction between FLI-1 and GATA-1(Eisbacher et al., 2003), but we and others have been unable to confirm this interaction in co-immunoprecipitation experiments on the endogenous proteins. This study also indicated that FLI-1 facilitated GATA-1 binding to DNA in an EMSA assay. Perhaps then the interaction between FLI-1 and GATA-1 is a transient one that occurs on chromatin in the presence of FOG-1. The tri-fold interaction between these proteins might provide stability that allows for continuous occupancy of a regulatory element.
Figure 7. FOG-1 modulates GATA-1 chromatin occupancy in a context-dependent manner.
(A) An illustration of the three types of GATA-1 binding sites as they relate to dependence on FOG-1. (B) An illustration of FOG-1 prohibiting GATA-1 occupancy at mast cell genes.
At the FOG-prohibited sites, FOG-1 normally acts to prevent wild-type GATA-1 occupancy. . In the context of a V205 mutation or in cells that do not express FOG, FOG-less GATA-1 proteins are permitted to occupy these sites. We predict that GATA-FOG complexes fail to occupy these sites due to interactions with transcriptional co-regulators or other chromatin proteins that are repelled by FOG-1. The identity of this FOG-repelling factor is yet to be determined, though the enrichment of T-box and Homeobox motifs at the sites provides direction for future research. Since our data show that FOG-1 prohibits GATA-1 occupancy at these sites in several different cell systems, the factors or chromatin conditions required for this regulation are likely expressed broadly. Finally, GATA-1 and GATA-1V205G both bind significantly to the FOG-independent sites, which represent the majority of GATA-1 binding sites.
The promiscuous binding behavior of GATA-1V205G reveals that FOG-1, despite its inability to bind DNA directly, actively regulates GATA-1 chromatin occupancy. Global gene expression analysis shows that FOG-mediated regulation of chromatin occupancy is required for lineage-appropriate gene expression and thus represents a bona fide mode of transcriptional regulation. Several studies have revealed a requirement for the GATA-1:FOG-1:NuRD axis in antagonizing mast cell fate in favor of erythromegakaryocytic fate. First, mice bearing a Gata1V205G knock-in mutation show increased mast cell potential in the fetal liver and aberrant expression of mast cell genes in erythroid and megakaryocytic cells (Cantor et al., 2008). Second, mouse models of the FOG-1R3K5A knock-in mutation, which abrogates the FOG-1:NuRD interaction, display a similar phenotype (Gao et al., 2010; Gregory et al., 2010). Finally, exogenous expression of FOG-1 in mast cell progenitors induces erythromegakaryocytic features. Although the prevailing model has been that GATA-1 actively represses mast cell gene expression in erythromegakaryocytic cells by binding chromatin and recruiting FOG-1 and NuRD, our data reveal that FOG-1 actually prevents GATA-1 from binding and activating these mast-cell specific genes.
Through ChIP assays in multiple systems, we discovered that GATA-1 binds mast cell genes when there is no interaction with FOG-1, such as in mast cells, in FOG-1 deficient cells, or when GATA-1 harbors a FOG non-interacting mutation. When the interaction with FOG-1 is restored, however, GATA-1 is displaced from the mast cell promoters and the genes are no longer expressed (Fig 7B). These data reveal that FOG-1 prohibits mast cell-specific gene expression by directly inhibiting GATA-1 chromatin occupancy at select sites.
GATA and FOG family members are conserved throughout vertebrates and are required for the development of diverse tissues, including the hematopoietic system and the cardiac system of flies, frogs, mice, and humans. Based on these parallels, it is enticing to speculate that FOG-mediated modulation of chromatin occupancy is a conserved mechanism of lineage fate regulation throughout metazoans.
Materials and Methods
Cell Culture
G1ME cells were maintained as described (Stachura et al., 2006) in 1% TPO-conditioned media. FOG−/− cells were maintained as described (Cantor et al., 2002). BMMCs were produced by harvesting whole bone marrow from 6–8 week old C57BL/6 mice and culturing for 4 weeks in IMDM (Invitrogen) containing15% FBS, 2mM L-glutamine, Pen/strep, 10ng/ml murine recombinant IL-3 (Peprotech), and 20ng/ml murine recombinant SCF (Peprotech). The purity of BMMCs was assessed by toluidine blue staining and FACS analysis with anti-FcεR1 and anti-cKit antibodies.
Chromatin immunoprecipitation and sequencing
ChIP was performed as described previously(Forsberg et al., 2000). For FOG-1 ChIPs, the cells were crosslinked in 1.5mM EGS for 30 minutes at room temperature before formaldehyde crosslinking. For ChIP-Seq, 50E6 cells were infected with MigR1-GATA-1 or MigR1-GATA-1GATA-1V205G and were harvested 48 hours later. An anti-HA tag antibody was used for the ChIP-Seq and in the validation of peaks. An anti-GATA-1 antibody was used for all other ChIP-qPCR experiments. Three biological replicate ChIP samples for each factor and three input samples were processed as described previously (Johnson et al., 2007) and sequenced on a Genome Analyzer II or IIx (Illumina). Sequence tags were mapped to the mouse genome (mm9) using the Illumina pipeline.
ChIP-Seq Binding Site Identification
Peaks of enrichment compared to input were indentified as described previously (Dore et al., 2012; Zhang Y, 2008). Each peak was assigned to the gene with the nearest TSS using the ChIP-Seq Tool Set(Blahnik et al., 2010). The overlaps of peaks were determined basewise with the ChIP-Seq Tool Set. The selective peak sets were identified by running MACS with the other factor as the `control', p-value<10−8, and mfold value of 15. The resulting peak sets were then overlapped with the peaks from the initial analysis, and those that existed in both peak sets were called `selective'. Control peak sets were generated using a custom Perl script that randomly assigns a new start position from the same chromosome as each experimentally identified peak, with the constraint that the new start position is chosen from the locations of input tags mapped to that chromosome. The length of the original peak is maintained in the newly generated control peak.
Motif Analysis
MEME (Bailey et al., 2009) was used for de novo motif finding for peak sets smaller than 2000 peaks. 200bp of genomic sequence surrounding each peak summit was submitted to MEME using the `zoops' option and the max motif width set to 10. For larger peak sets, DREME was used (Bailey, 2011 Homer findmotifs program was used on default parameters (Heinz et al., 2010). To determine statistically significant differences in motif enrichment, one peak set was used as the treatment and the other was used as the control.
Gene Expression Analysis
G1ME cells were transduced with MigR1 retroviruses and the GFP+ cells were sorted 68h later. The cells were allowed to recover in culture for 4h, and then the total RNA was extracted using RNeasy Plus mini columns (Qiagen) following manufacturer's protocol. The samples were hybridized to Illumina MouseWG-6 v2.0 Expression BeadChips. The background was subtracted and the data was quantile normalized to remove batch effects. The probe level data was analyzed using GeneSpring software (Agilent Technologies). Probes with signals below 20.5 were eliminated from the analysis as `absent'. Unpaired t-tests were performed for the remaining 16380 probes with asymptomatic p-value calculation. Probes with fold-change>1.5 and p-value<0.05 were considered differentially expressed. To make heatmaps, the genes were centered and then hierarchically clustered by centroid linkage using the software program Gene Cluster 3.0 (de Hoon et al., 2004), and then the Java program TreeView was used to make and edit the heatmap (Saldanha, 2004).
Statistical Analysis
All statistical analysis was completed using StatPlus:mac software program (AnalystSoft, Inc.). For quantitative assays, treatment groups were reported as mean ± SD and compared using the unpaired Student t test. To test for independence of groups based on categorical data, Chi-square analysis with the Yates Correction for large sample sizes was used. Statistical significance was established at p less than or equal to 0.05, unless otherwise noted.
Data Access
The ChIP-Seq and microarray data used in this paper can be accessed from the Gene Expression Omnibus (GEO) using accession numbers GSE35708 and GSE31331.
Supplementary Material
Highlights
Novel example of regulation of chromatin occupancy by a non DNA binding cofactor
FOG-1 prohibits GATA-1 from binding to mast cell genes to maintain lineage fidelity
GATA-FOG regulation of cell fate in other systems likely requires same mechanism
Acknowledgments
The authors thank Ari Melnick and the Epigenomics Core at Weill Cornell Medical College. This work was supported by the Chicago Center for Systems Biology (P50 GM081892) and in part by a grant from the NCI (R01 CA101774).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions T.M.C. performed the experiments. T.M.C. and L.C.D. analyzed the data. J.D.C. helped design and interpret the study. T.M.C. and J.D.C. wrote the paper.
References
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahnik KR, Dou L, O'Geen H, McPhillips T, Xu X, Cao AR, Iyengar S, Nicolet CM, Ludascher B, Korf I, et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Research. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. The Journal of Experimental Medicine. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciovacco WA, Raskind WH, Kacena MA. Human phenotypes associated with GATA-1 mutations. Gene. 2008;427:1–6. doi: 10.1016/j.gene.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD. GATA1 in normal and malignant hematopoiesis. Seminars in Cell & Developmental Biology. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of Altered Specificity Mutants to Probe a Specific Protein-Protein Interaction in Differentiation: the GATA-1:FOG Complex. Molecular Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Deveaux S, Filipe A, Lemarchandel V, Ghysdael J, Romeo P, Mignotte V. Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood. 1996;87:4678–4685. [PubMed] [Google Scholar]
- Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012 doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118:231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, Chong BH. Protein-Protein Interaction between Fli-1 and GATA-1 Mediates Synergistic Expression of Megakaryocyte-Specific Genes through Cooperative DNA Binding. Mol Cell Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 Function, a Paradigm for Transcription Factors in Hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the Œ≤-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC. FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J. 2010;29:457–468. doi: 10.1038/emboj.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proceedings of the National Academy of Sciences. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory GD, Miccio A, Bersenev A, Wang Y, Hong W, Zhang Z, Poncz M, Tong W, Blobel GA. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood. 2010;115:2156–2166. doi: 10.1182/blood-2009-10-251280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen Y-Y, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yu M, Akie TE, Moran TB, Woo AJ, Tu N, Waldon Z, Lin YY, Steen H, Cantor AB. Differentiation-Dependent Interactions between RUNX-1 and FLI-1 during Megakaryocyte Development. Molecular and Cellular Biology. 2009;29:4103–4115. doi: 10.1128/MCB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen Y-Y, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lowry JA, Mackay JP. GATA-1: One protein, many partners. The International Journal of Biochemistry & Cell Biology. 2006;38:6–11. doi: 10.1016/j.biocel.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Maeda K, Nishiyama C, Tokura T, Nakano H, Kanada S, Nishiyama M, Okumura K, Ogawa H. FOG-1 represses GATA-1-dependent Fc,àäRI Œ≤-chain transcription: transcriptional mechanism of mast-cell-specific gene expression in mice. Blood. 2006;108:262–269. doi: 10.1182/blood-2005-07-2878. [DOI] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Matsumoto K, Suto H, Tanaka K, Ebisawa M, Tomita H, Yuki K, Katsunuma T, Akasawa A, Hashida R, et al. Gene expression screening of human mast cells and eosinophils using high-density oligonucleotide probe arrays: abundant expression of major basic protein in mast cells. Blood. 2001;98:1127–1134. doi: 10.1182/blood.v98.4.1127. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang L, Xue H-H, Szalai G, Wang X, Wang Y, Watson DK, Leonard WJ, Blobel GA, Poncz M. Maturation stage¬ñspecific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood. 2006;108:2198–2206. doi: 10.1182/blood-2006-04-019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPE and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes & Development. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview,Äîextensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107:87–97. doi: 10.1182/blood-2005-07-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic Regulatory Networks and Animal Development. Developmental Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Sugiyama D, Tanaka M, Kitajima K, Zheng J, Yen H, Murotani T, Yamatodani A, Nakano T. Differential context-dependent effects of friend of GATA-1 (FOG-1) on mast-cell development and differentiation. Blood. 2008;111:1924–1932. doi: 10.1182/blood-2007-08-104489. [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide Analysis of Simultaneous GATA1/2, RUNX1, FLI1, and SCL Binding in Megakaryocytes Identifies Hematopoietic Regulators. Developmental Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes & Development. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a Multitype Zinc Finger Protein, Acts as a Cofactor for Transcription Factor GATA-1 in Erythroid and Megakaryocytic Differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among Distant Regulatory Elements at the [beta]-Globin Locus Requires GATA-1 and FOG-1. Molecular cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wang X, Crispino JD, Letting DL, Nakazawa M, Poncz M, Blobel GA. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 2002;21:5225–5234. doi: 10.1093/emboj/cdf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo AJ, Moran TB, Schindler YL, Choe S-K, Langer NB, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB. Identification of ZBP-89 as a Novel GATA-1-Associated Transcription Factor Involved in Megakaryocytic and Erythroid Development. Molecular and Cellular Biology. 2008;28:2675–2689. doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RJ, Boyer ME, Grass JA, Lee Y, Bresnick EH. Context-dependent GATA Factor Function. Journal of Biological Chemistry. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen K-B, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Researc. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y LT, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.