Abstract
The rare translocation t(8;14)(q11.2;q32) has been described in patients with B-cell acute lymphoblastic leukemia (ALL), particularly patients with Down Syndrome (DS).
We describe patients with t(8;14)(q11.2;q32) that were identified by the Children's Oncology Group (COG) ALL cytogenetics database, expanding our previous report of 10 patients with this translocation. Twenty-two such patients were treated with COG protocols. All patients had B-cell ALL and 7 (31.8%) had DS. None of the children with DS had an event, thus these patients had a superior estimated 5-year event-free survival (EFS) compared to non-DS patients (100% vs. 50.1 ± 17.7%; p=0.04). Only one patient (4.5%) had a concomitant Philadelphia chromosome t(9;22)(q34;q11.2). The cytogenetics data of two additional patients, who were not eligible for COG protocols, are also included in this report.
In conclusion, ALL patients with the recurring translocation t(8;14)(q11.2;q32) have B-cell phenotype and a high percentage have DS. Children with DS and t(8;14)(q11.2;q34) have improved event-free survival using standard COG therapy compared to non-DS patients. We did not find an increased number of patients with a concomitant Philadelphia chromosome in this population.
Keywords: Acute Lymphoblastic Leukemia, B-cell, translocation, Down Syndrome
Introduction
Recurrent chromosome translocations play a critical role in the pathogenesis of ALL, and many translocations have important prognostic significance. The rare translocation t(8;14)(q11.2;q32) has been described in ALL (1, 2) and has been associated with Down syndrome (DS), although the mechanism of this association is not clear (1-6).
We previously described the clinical features of 10 ALL patients with t(8;14)(q11.2;q32) and estimated the frequency of this translocation to be 0.2% of children with B-cell ALL (1). The Mitelman database reports that t(8;14)(q11.2;q32) has a prevalence of 0.7% in pediatric ALL (7). We now expand the cytogenetic, clinical and outcome data to include 22 patients with this translocation. We confirm the high proportion of DS in this group and the association with B-cell phenotype. We also demonstrate a remarkably good outcome of the DS subgroup using standard Children's Oncology Group (COG) therapy.
Methods
The COG ALL cytogenetic database was reviewed to identify patients with t(8;14)(q11.2a32). Demographic, treatment and survival data were accessed from the COG clinical database. Ten of the patients have been reported previously (1). All 22 patients were diagnosed and treated at COG institutions on frontline trials. All studies and treatment protocols were approved by the local institution's Investigational Review Board (IRB), and all patients and/or their parent/guardian signed an informed consent.
Diagnosis
The diagnosis of ALL was based on morphologic, cytochemical, and immunologic features of the cells, as previously described (8). Immunophenotyping was performed at local centers using standard techniques and/or centrally in the COG ALL Reference Laboratory by indirect immunofluorescence and flow cytometry.
Cytogenetic Analysis
Analyses of bone marrow or unstimulated peripheral blood specimens obtained prior to initiation of remission induction therapy were evaluated at COG Institutional Laboratories according to standard protocols. Criteria for clonality were based on guidelines as defined by the International System for Cytogenetic Nomenclature (i.e., 2 or more metaphase spreads with identical structural or additional chromosomes or 3 or more metaphases with identical chromosome loss) (9). Members of the COG cytogenetics committees reviewed the cytogenetic reports and representational karyotypes for each abnormal clone. Patients were eligible if, after review, an abnormal clone with t(8;14)(q11.2;q32) was identified.
Statistical Methods
The outcomes evaluated were event-free survival (EFS), calculated as the time from entry on a therapeutic trial to first event or date of last follow-up, where an event was defined as induction failure, relapse at any site, secondary malignancy, or death, and overall survival (OS), calculated as the time from entry on a therapeutic trial to death or date of last follow-up. Patients who did not experience an event were censored as of the date of last contact. Event-free survival curves were constructed using the Kaplan-Meier life table method (10). Standard errors of the estimates were determined by the method of Peto and Peto (11). The log-rank test was used for comparison of survival curves between groups.
Results
Twenty-two patients diagnosed with ALL between 2/24/1987 and 4/11/2007 and treated on COG protocols were found to have the t(8;14)(q11.2;q32) (Table 1). Two additional ALL patients with DS-ALL and t(8;14)(q11.2;q32) were not included in the clinical data because they were retrospectively found to be ineligible for COG therapeutic trials. Epidemiologic, clinical, and treatment data are summarized in Table 2. All patients had B-cell ALL; 9 were males and 13 were females. The median age at diagnosis was 10.7 years (range: 2.7 – 17.1 years). Median diagnostic white blood cell count (WBC) was 9,950 (range: 700-172,000 × 103/mm3). Seven of the 22 patients were designated as NCI standard-risk and 15 were designated as high-risk.
Table 1.
Patient Data
| Patient number | Sex | Syndrome | Phenotype | Age at Diagnosis (years) | Presenting WBC | NCI Risk | COG Treatment Protocol | Early Response |
|---|---|---|---|---|---|---|---|---|
| 1 | Male | Down | B-Cell | 11.47 | 11,000 | High risk | 9406 | M3-SER |
| 2 | Male | Down | B-Cell | 8.95 | 29,300 | Standard risk | 9905 | M3-SER |
| 3 | Female | Down | B-Cell | 8.19 | 66,400 | High risk | 1961 | M1-RER |
| 4 | Female | Down | B-Cell | 8.35 | 8,600 | Standard risk | 9405 | M3-SER |
| 5 | Male | Down | B-Cell | 9.35 | 57,800 | High risk | 9900 | M3-SER |
| 6 | Female | Down | B-Cell | 16.97 | 2,875 | High risk | 8602 | M3-SER |
| 7 | Female | Down | B-Cell | 8.87 | 37,300 | Standard risk | 9905 | M3-SER |
| 8 | Female | None | B-Cell | 4.11 | 34,300 | Standard risk | 1991 | M1-RER |
| 9 | Female | None | B-Cell | 15.09 | 6,700 | High risk | 1882 | M1-RER |
| 10 | Female | None | B-Cell | 8.85 | 7,000 | Standard risk | 8699 | M3-SER |
| 11 | Male | None | B-Cell | 17.11 | 4,000 | High risk | 8602 | M3-SER |
| 12 | Female | None | B-Cell | 12.95 | 126,000 | High risk | 8602 | M3-SER |
| 13 | Male | None | B-Cell | 10.14 | 23,800 | High risk | 9406 | M3-SER |
| 14 | Male | None1 | B-Cell | 4.00 | 20,300 | Standard risk | 9203 | M3-SER |
| 15 | Male | None | B-Cell | 2.69 | 12,000 | Standard risk | AALL0331 | M1-RER |
| 16 | Female | None | B-Cell | 11.12 | 8,900 | High risk | 9900 | M3-SER |
| 17 | Male | None | B-Cell | 12.78 | 700 | High risk | AALL0232 | M1-RER |
| 18 | Female | None | B-Cell | 9.95 | 172,000 | High risk | 8698 | M3-SER |
| 19 | Female | Turner | B-Cell | 17.06 | 2,700 | High risk | 1882 | M1-RER |
| 20 | Female | None | B-Cell | 16.46 | 8,900 | High risk | 1882 | M1-RER |
| 21 | Male | None | B-Cell | 16.45 | 8,000 | High risk | 8602 | M3-SER |
| 22 | Female | None2 | B-Cell | 17.12 | 2,900 | High risk | AALL0232 | M2-SER |
Abbreviations: M1 = 0-5% blasts in the bone marrow; M2: 5-25% blasts in the in bone marrow; M3 = > 25% blasts in the in bone marrow; RER = rapid early response; SER = slow early response; WBC = white blood cell count.
Patient with spina bifida.
Patient with small kidney and aniridia.
Table 2.
Summary Demographic Data
| Patients | n = 22 | |
|---|---|---|
| Gender | Males | 9 (40.9%) |
| Females | 13 (59.1%) | |
| Age at diagnosis (Years) | median = 10.6 (range 2.7 -17.1) | |
| WBC at diagnosis (per mm3) | median = 9,950 (range 700-172,000) | |
| NCI Risk Group | Standard | 7 (31.8%) |
| High | 15 (68.2%) | |
| ALL Phenotype | B-cell | 22 (100%) |
| Constitutional Chromosomal Abnormality | Down | 7 (31.8%) |
| Turner* | 1 (4.5%) | |
| Normal | 14 (63.6%) | |
Phenotypic Turner syndrome with mosaic constitutional r(Y)(p11.2q11.2)
Additional Cytogenetic Abnormalities
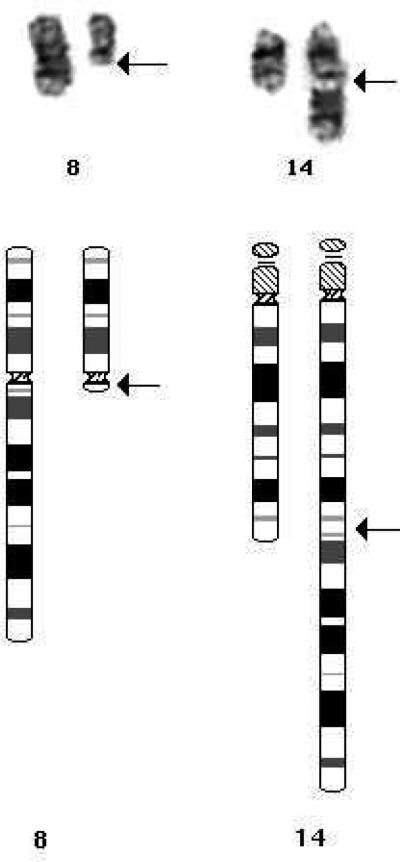
A representative G-banded t(8;14)(q11.2;q32) and diagrammatic representation of the translocation is shown in figure 1. Cytogenetics data are shown in table 3 and include the two excluded patients. The most common additional cytogenetic abnormality was trisomy 21 (Table 3). Seven of the 10 cases with trisomy 21 were constitutional DS. In total 7/22 or 32% patients had DS, confirming previous reports that the number of t(8;14)(q11.2;q32) who have DS is much higher than the roughly 3% rate of DS among children with ALL (4, 12). Interestingly, three additional patients had acquired +21, but the significance of this is not known. One patient in this cohort, who lacked DS, had a Turner syndrome phenotype and a mosaic constitutional r(Y)(p11.2q11.2); thus, 8 of the 22 patients (36%) with t(8;14)(q11.2;q32) had constitutional cytogenetic abnormalities. Secondary abnormalities included additional X (n=5); additional 5 (n=3); and a Philadelphia chromosome (n=1). Two cases had a second der(14)t(8;14) and two had loss of the der(8)t(8;14), consistent with the der(14)t(8;14) as the significant abnormality. Five cases or 23% had 9p deletions compared with 11% in the overall ALL population (13), and two cases or 9% had abnormalities of 13q compared with 2% in the overall population (14). Numerically, 12 cases were pseudodiploid, 8 cases were low hyperdiploid, one was high hyperdiploid and one was pseudo-tetraploid. Of the seven DS patients six or 86% were pseudodiploid with five or 71% having abnormalities in addition to the t(8;14). Of the non-DS patients the t(8;14) was the only cytogenetic abnormality in one patient, while all the other non-DS patients had secondary abnormalities in addition to the t(8;14).
Figure 1.
G-banded t(8;14)(q11.2;q32) and diagrammatic representation of the translocation.
Table 3.
Cytogenetic Data
| CONSTITUTIONAL +21 | |
| 1* | 47,XY,t(8;14)(q11.2;q32),+21c[7]/47,XY,+21c[2] |
| 2 | 47,XY,t(8;14)(q11.2;q32),+21c[12]/47,XY,+21c[8] |
| 3 | 47,XX,der(14)t(8;14)(q11.2;q32),+21c[135] |
| 4* | 47,XX,t(8;14)(q11.2;q32),del(9)(p13p22),+21c[9]/47,XX,+21c[6] |
| 5 | 47,XY,t(8;14)(q11.2;q32),add(9)(p13),+21c[6]/47,XY,+21c[4] |
| 6* | 47,XX,t(8;14)(q11.2;q32),+21c[9]/47,idem,add(16)(q24)[9]/47,XX,+21c[2] |
| 7 | 48,XX,+X,t(8;14)(q11.2;q32),+21c[17] |
| ACQUIRED +21 | |
| 8 | 47,XX,t(8;14)(q11.2;q32),+21[16]/46,XX[4] |
| 9 | 47,XX,t(8;14)(q11.2;q32),del(13)(q14),+?21[18]/46,XX[2] |
| 10* | 56,XX,+X,der(3)t(3;8)(p27;q13),+5,+7,+8,t(8;14)(q11.2;q32),+10,+14,+18,+21,+21,+22[6]/46,XX[12] |
| NON-TRISOMY 21 | |
| 11* | 46,XY,t(8;14)(q11.2;q32)[22]/46,XY[1] |
| 12* | 46,XX,t(8;14)(q11.2;q32),i(9)(q10)[20] |
| 13* | 46,XY,t(1;5)(p32;q31),t(8;14)(q11.2;q32)[20] |
| 14* | 46,XY,t(8;14)(11.2;q32),t(9;22)(q34;q11.2),add(17)(p13)[6]/46,X,idem,der(X)t(X;8)(q28;q11.2)[12]/46,XY[3] |
| 15 | 46,XY,t(8;14)(q11.2;q32),add(9)(p22)[7]/46,XY[13] |
| 16 | 46,XX,t(8;14)(q11.2;q32)[2]/45,idem,-der(8)t(8;14)[5]/46,XX[13] |
| 17 | 47,XY,+X,t(8;14)(q11.2;q32)[9]/46,XY[20] |
| 18* | 47,XX,t(8;14)(q11.2;q32),del(13)(q14),+der(14)t(8;14)[14]/46,XX[6] |
| 19 | 47,X,r(Y)(p11.2q11.2)c,+5,t(8;14)(q11.2;q32),der(21)t(1;21)(q12;q22)[9]/46,X,r(Y)(p11.2q11.2)c[11] |
| 20 | 48,XX,+2,t(8;14)(q11.2;q32),+mar[10]/47,idem,-20[6]/46,XX[1] |
| 21* | 50,XY,+X,+4,+5,t(8;14)(q11.2;q32),+der(14)t(8;14)[15]/46,XY[6] |
| 22 | 92,XXXX,+X,t(8;14)(q11.2;q32)x2,-9,del(9)(p13p22)x2[cp9]/46,XX[1] |
| EXCLUDED | |
| E1 | 48,XX,+X,t(8;14)(q11.2;q32),+21c[5]/47,XX,+21c[17] |
| E2 | 47,XX,t(8;14)(q11.2;q32),+21c[6]/47,XX,+21c[17] |
Cytogenetic results of all 22 patients that are included in the clinical analysis (patients 1-22) and the 2 excluded patients who were not treated on Children's Oncology Group therapeutic trials (patients E1 and E2).
Previously reported by Kaleem et al. (1)
Therapy and Outcome
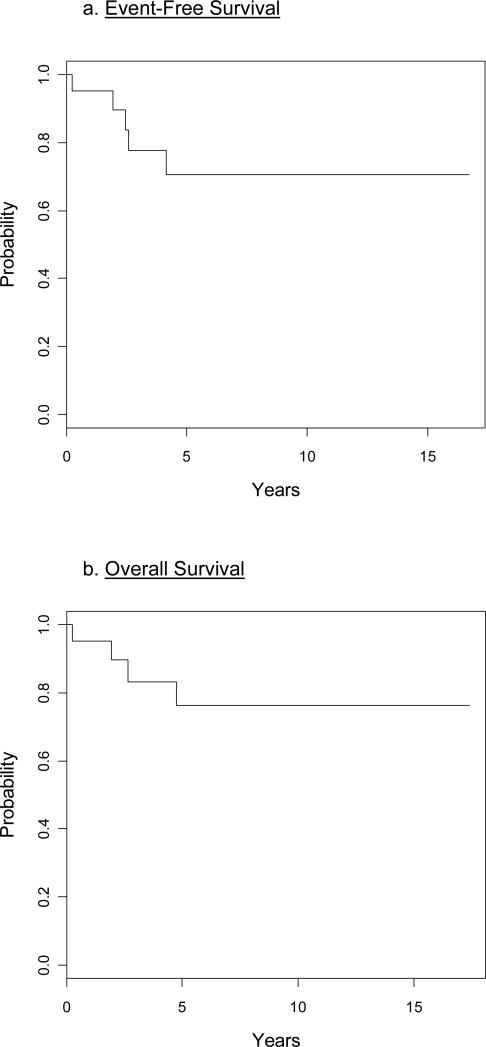
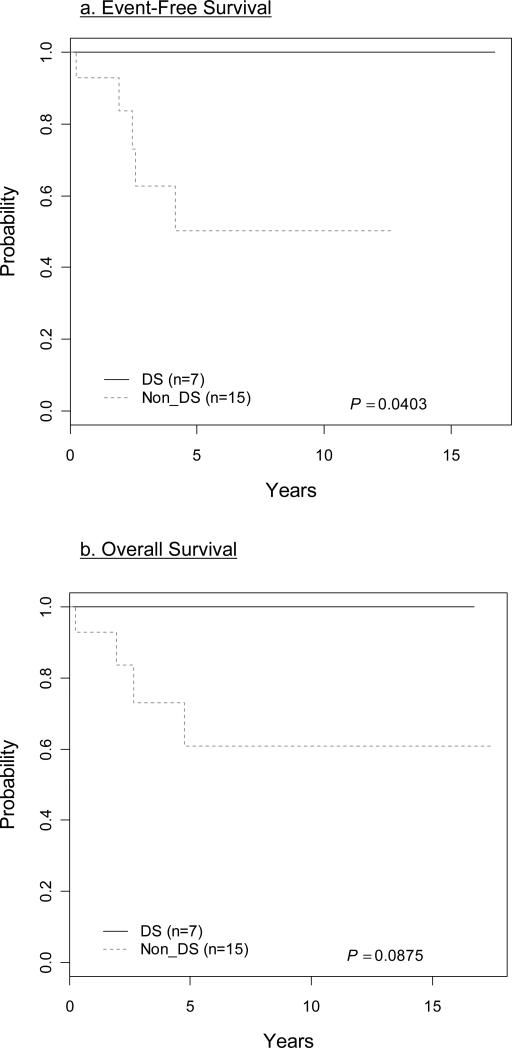
Patients were treated on COG protocols. A rapid early response (RER) as determined by a reduction of day 7 or day 14 bone marrow blasts to < 5%, was found in 7 of 22 (32%) patients (Table 1). All 22 (100%) patients achieved complete remission at the end of induction therapy. The 5-year EFS was 70.6 ±14.4% and the overall survival was 76.3 ±13.1% (Fig 2). None of the children with DS had an event and these patients had a superior estimated 5-year EFS (100%) compared to non-DS patients (50.1±17.7%; p=0.04) (Fig. 3a). Overall survival was also superior for DS patients but did not reach conventional levels of statistical significance: 100% for DS versus 60.9±17.0% for non-DS patients (p=0.088) (Fig 3b). There was no significant difference in the age at diagnosis or WBC at presentation between DS and non-DS patients with a t(8;14)(q11.2;q32).
Figure 2.
Twenty two patients with t(8;14)(q11.2;q32) had 5 years event-free-survival of 70.6 ±14.4% (a) and 5 years overall-survival of 76.3 ±13.1% (b).
Figure 3.
Children with Down Syndrome (DS) compared to non-DS. a. 5-year event-free-survival = 100% (DS) vs. 50.1±17.7% (non-DS); p=0.04. b. Overall survival = 100% (DS) vs. 60.9±17.0% (non-DS); p=0.088.
Discussion
We describe twenty-two childhood ALL patients with the recurring translocation t(8;14)(q11.2;q32) enrolled on COG protocols between 2/24/1987 and 4/11/2007. Consistent with prior reports, all patients with this translocation had B-cell ALL (1, 15).
The translocation t(8;14)(q11.2;q32) results in fusion of the chromosome 8 CEBPD (delta) gene to the IGH (immunoglobulin heavy chain) gene (16). As pointed out by Dryer et al. chromosomal translocations involving the immunoglobulin loci are extremely common in mature B-cell malignancies such as Burkitt lymphoma (17). In contrast, in B-cell ALL translocations involving the immunoglobulin loci are very uncommon with 2-3% estimated frequency (17). The most frequent IGH translocation involves the cytokine receptor CRLF2 (on chromosome Xp22 or Yp11), followed by the ID4 gene (on chromosome 6p22.3) and the CEBP family (CCAAT enhancer binding protein) translocations (17). CEBP is a family with 5 members of multifunctional basic leucine zipper (bZIP) transcription factors. In t(8;14)(q11.2;q32) CEBPD (a member of the CEBP family) is deregulated because of transcriptional enhancers within the IGH locus (16, 17). The mechanism by which deregulated CEBP gene expression transforms B-cell precursors into neoplastic cells is not yet known (16).
About one-third of the patients with t(8;14)(q11.2;q32) identified in our study had Down syndrome, an incidence that is much higher than the approximate 3% overall rate of DS in childhood ALL (4, 12). Lundin et al. report a similar high 27% rate of DS among patients with t(8;14)(q11.2;q32) in their recent review (15). In patients with DS and ALL this translocation was the second most common balanced aberration (2.8%) as reported by the International Berlin-Frankfurt-Munster (iBFM) cytogenetics register (4). Several recent studies have shown that about half of children with DS-ALL have high level expression of CRLF2, due to either a cryptic interstitial deletion of the pseudoautosomal region1 (PAR1) of X/Y that causes P2RY8-CRLF2 fusion, or a cryptic translocation between IGH and CRLF2, and about half of these patients have concomitant JAK2 point mutations (18, 19). The mechanism of CRLF2 activation by fusion to IGH is much like the activation of CEBP by IGH fusion in DS patients with ALL and the t(8;14)(q11.2;q32). Both cases (activation of CRLF2 and CEBP) are much more frequent in DS ALL than in childhood ALL as a whole. At this time the mechanism of the association between trisomy 21 and these translocations is not clear. Nor is it known whether activating JAK2 mutations are present in patients with t(8;14)(q11.2;q32).
In this series only 1 of 22 patients (a non-DS patient) had concomitant t(9;22)(q34;q11.2) – the Philadelphia chromosome. This is in contrast to the high (30%) prevalence of a Philadelphia chromosome in children with t(8;14)(q11.2;q32) reported by others (15). Similarly, more females have this translocation in our series in contrast to the male preponderance included in that report (15).
In B-cell ALL most patients (68.7%) can be classified as NCI standard-risk (8). In contrast, in t(8;14)(q11.2;q32) ALL only 31.8% were standard-risk (p=0.0002). The EFS of the non-DS t(8;14)(q11.2;q32) patients (50.1±17.7%) was low, more in line with the low 8-year EFS (56.5% to 64.9%) reported for high-risk B-cell ALL in COG studies from the same era (8). These patients were treated on protocols spanning several decades, during which consistent improvement in survival was achieved.
Recent outcome of ALL patients with DS is reportedly similar to non-DS ALL patients, using risk-adapted therapies, after adjusting for biological differences (12, 20). This is in contrast to AML in which patients with DS have improved outcome compared to non-DS patients (21). Lundin et al., in their literature review of patients with t(8;14)(q11.2;q32), show slightly worse median survival in DS compared to non-DS (15). In contrast, in our COG patients with t(8;14)(q11.2;q32) the DS patients seem to have an improved outcome versus non-DS patients. In fact, all seven DS patients have survived thus far with no event (follow up range: 2.6 - 16.7 years). Age and presenting WBC were similar in the DS and the non-DS groups. Thus the NCI-risk group cannot explain the improved outcome of DS patients on frontline COG protocols. Our results require further confirmation from other cooperative group studies.
In conclusion, the rare recurring translocation t(8;14)(q11.2;q32) in childhood ALL is associated with B-cell phenotype and often with high-risk features. About 30% of patients with this translocation have constitutional DS and an outcome superior to non-DS patients.
Acknowledgements
This work was supported in part by grants to the COG including the COG Chair's Grant CA098543 and CA98413 supporting the COG Statistical Center. SPH is the Ergen Family Chair in Pediatric Cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaleem Z, Shuster JJ, Carroll AJ, et al. Acute lymphoblastic leukemia with an unusual t(8;14)(q11.2;q32): a Pediatric Oncology Group Study. Leukemia. 2000;14:238–40. doi: 10.1038/sj.leu.2401675. [DOI] [PubMed] [Google Scholar]
- 2.Moore S, Suttle J, Bain S, et al. Acute lymphoblastic leukemia characterized by t(8;14)(q11.2;q32). Cancer Genet Cytogenet. 2003;141:1–4. doi: 10.1016/s0165-4608(02)00643-x. [DOI] [PubMed] [Google Scholar]
- 3.Byatt SA, Cheung KL, Lillington DM, et al. Three further cases of t(8;14)(q11.2;q32) in acute lymphoblastic leukemia. Leukemia. 2001;15:1304–5. doi: 10.1038/sj.leu.2402166. [DOI] [PubMed] [Google Scholar]
- 4.Forestier E, Izraeli S, Beverloo B, et al. Cytogenetic features of acute lymphoblastic and myeloid leukemias in pediatric patients with Down syndrome: an iBFM-SG study. Blood. 2008;111:1575–83. doi: 10.1182/blood-2007-09-114231. [DOI] [PubMed] [Google Scholar]
- 5.Lee AC, Chan LC, Kwong KW. Down syndrome, acute lymphoblastic leukemia, and t(8;14)(q11;q32). Cancer Genet Cytogenet. 1996;88:92. doi: 10.1016/0165-4608(95)00285-5. [DOI] [PubMed] [Google Scholar]
- 6.Secker-Walker LM, Hawkins JM, Prentice HG, et al. Two Down syndrome patients with an acquired translocation, t(8;14)(q11;q32), in early B-lineage acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1993;70:148–50. doi: 10.1016/0165-4608(93)90189-s. [DOI] [PubMed] [Google Scholar]
- 7.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (2012) 2012 Available from: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 8.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG). Blood. 2007;109:926–35. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ISCN . An International System for Human Cytogenetic Nomenclature 2005. S. Karger; Basel: 2005. [Google Scholar]
- 10.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Joural of the American Statistical Association. 1958;53:457–481. [Google Scholar]
- 11.Peto R, Peto J. Asymptotically efficient rank invariant test procedure. J Royal Stat Soc. 1972;135:185–198. [Google Scholar]
- 12.Maloney K, Carroll WL, Carroll A, et al. Comparison of the biology of Down syndrome (DS) acute lymphoblastic leukemia (ALL) and non-DS ALL: Children's Oncology Group (COG) study P9900. J Clin Oncol. 2008;26 Abstr 10003. [Google Scholar]
- 13.Heerema NA, Sather HN, Sensel MG, et al. Association of chromosome arm 9p abnormalities with adverse risk in childhood acute lymphoblastic leukemia: A report from the Children's Cancer Group. Blood. 1999;94:1537–44. [PubMed] [Google Scholar]
- 14.Heerema NA, Sather HN, Sensel MG, et al. Abnormalities of chromosome bands 13q12 to 13q14 in childhood acute lymphoblastic leukemia. J Clin Oncol. 2000;18:3837–44. doi: 10.1200/JCO.2000.18.22.3837. [DOI] [PubMed] [Google Scholar]
- 15.Lundin C, Heldrup J, Ahlgren T, et al. B-cell precursor t(8;14)(q11;q32)-positive acute lymphoblastic leukemia in children is strongly associated with Down syndrome or with a concomitant Philadelphia chromosome. Eur J Haematol. 2009;82:46–53. doi: 10.1111/j.1600-0609.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 16.Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109:3451–61. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 17.Dyer MJ, Akasaka T, Capasso M, et al. Immunoglobulin heavy chain locus chromosomal translocations in B-cell precursor acute lymphoblastic leukemia: rare clinical curios or potent genetic drivers? Blood. 2010;115:1490–9. doi: 10.1182/blood-2009-09-235986. [DOI] [PubMed] [Google Scholar]
- 18.Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–17. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 19.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–6. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitlock JA. Down syndrome and acute lymphoblastic leukaemia. Br J Haematol. 2006;135:595–602. doi: 10.1111/j.1365-2141.2006.06337.x. [DOI] [PubMed] [Google Scholar]
- 21.Gamis AS. Acute myeloid leukemia and Down syndrome evolution of modern therapy--state of the art review. Pediatr Blood Cancer. 2005;44:13–20. doi: 10.1002/pbc.20207. [DOI] [PubMed] [Google Scholar]