Abstract
Protein kinase C (PKC) ε, a member of the novel PKC family, plays key roles in mitogenesis and survival in normal and cancer cells. PKCε is frequently overexpressed in epithelial cancers, particularly in lung cancer. Using a shRNA approach, here we established that PKCε is required for non-small cell lung carcinoma (NSCLC) growth in vitro as well as tumor growth when inoculated into athymic mice. Moreover, sustained delivery of a PKCε selective inhibitor peptide, εV1-2, reduced xenograft growth in mice. Both RNAi depletion and pharmacological inhibition of PKCε caused a marked elevation in the number of apoptotic cells in NSCLC tumors. PKCε-depleted NSCLC cells show elevated expression of pro-apoptotic proteins of the Bcl-2 family, caspase recruitment domain (CARD)-containing proteins, and TNF ligands/receptor superfamily members. Moreover, a Gene Set Enrichment Analysis (GSEA) revealed that a vast majority of the genes changed in PKCε-depleted cells were also deregulated in human NSCLC. Our results strongly suggest that PKCε is required for NSCLC cell survival and maintenance of NSCLC tumor growth. Therefore, PKCε may represent an attractive therapeutic target for NSCLC.
Keywords: PKCε, non-small cell lung carcinoma, tumorigenesis, cell survival, apoptotic genes
Introduction
Protein kinase C (PKC) isozymes comprise 3 classes of serine-threonine kinases (“classical” cPKCα, β, γ, “novel” nPKCs δ, ε, η, θ, and “atypical” aPKCζ and ι/λ) that play important roles in the control of cellular growth, survival, differentiation and transformation. Their cooperation with oncogenic stimuli such as Ras, Myc and Fos, is well established (Barr et al., 1991; Han et al., 1995; Hsiao et al., 1989). Despite the extensive knowledge on PKC signaling, there are still major gaps in our understanding of the striking functional specificity displayed by PKC isozymes. Depending on the cellular context, individual members of the DAG/phorbol ester regulated cPKC and nPKC classes modulate cellular responses either in cooperative or antagonistic manners. Studies have shown that PKCα and PKCδ generally inhibit cell cycle progression or promote apoptotic and senescent responses, whereas PKCε mediates mitogenic responses (Black, 2000; Brodie & Blumberg, 2003; Caino et al., 2009; Nakagawa et al., 2005; Oliva et al., 2008; Slupsky et al., 2007). In addition, activation of PKCε inhibits apoptotic responses triggered by a number of stimuli through both Akt-dependent and Akt-independent mechanisms (Basu & Sivaprasad, 2007; Lu et al., 2006; McJilton et al., 2003; Okhrimenko et al., 2005). The diversity and heterogeneity in PKC isozyme function in distinct cell types may largely explain the limited therapeutic success of PKC inhibitors which in most cases fail to discriminate between PKC isozymes.
The expression of PKC isozymes and their effectors is altered in human cancer. One of the most frequent alterations in tumors of epithelial origin is the overexpression of PKCε, particularly in lung, prostate, and breast cancer. PKCε up-regulation in tumors was defined as a prognostic marker for recurrence and patient survival (Gorin & Pan, 2009; Griner & Kazanietz, 2007). Ectopic expression of PKCε in non-transformed epithelial cells leads to either growth advantage or a transformed phenotype (Mischak et al., 1993; Perletti et al., 1996). A recent study revealed that transgenic overexpression of PKCε in the mouse prostate confers a preneoplastic phenotype (Benavides et al., 2011). In the mouse skin, transgenic overexpression of PKCε leads to metastatic squamous carcinoma (Jansen et al., 2001). Thus, despite cell type differences, it became clear that PKCε may have prominent roles in cancer initiation and progression (Gorin & Pan, 2009).
Lung cancer is one of the malignancies in which the role of PKC isozymes is still poorly understood. Studies from our laboratory revealed that PKCα and PKCδ promote anti-mitogenic responses in non-small cell lung carcinoma (NSCLC) cells (Caino et al., 2009; Nakagawa et al., 2005; Oliva et al., 2008). A study by Xiao and coworkers showed that expression of a dominant-negative PKCε mutant inhibits the progression of NSCLC cells through the cell cycle, suggesting that PKCε may have an important role in lung cancer development (Bae et al., 2007). Moreover, PKCε inhibition resulted in a significant amplification of the cytotoxic activity of TRAIL in A549 cells and increased their apoptotic responsiveness (Farber et al., 2004). Notably, more than 90% of human NSCLC tumors display elevated PKCε levels (Bae et al., 2007). Several oncogenic stimuli implicated in lung cancer development, including the epidermal growth factor (EGFR) or K-Ras, signal through the second messenger DAG, a key step for PKCε activation (Parker & Murray-Rust, 2004; Toker, 1998). It is therefore conceivable that PKCε plays a role in the maintenance of the tumorigenic phenotype in lung cancer.
In this study we present strong evidence that PKCε is required for the growth of NSCLC cells in vitro and tumorigenicity in nude mice. Moreover, we identified novel apoptosis-related transcriptional targets for PKCε that correlate with disease status in NSCLC.
Results and discussion
PKCε depletion impairs anchorage-dependent and anchorage-independent NSCLC cell growth
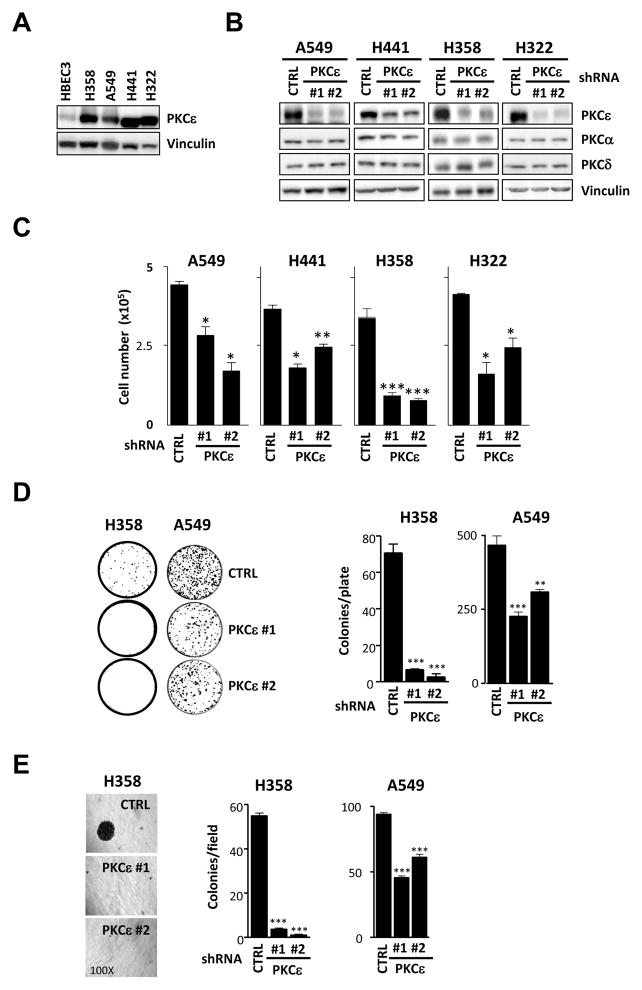
Overexpression of PKCε, a member of the novel PKC family, is a hallmark of human epithelial cancers particularly in NSCLC tumors (Bae et al., 2007; Griner & Kazanietz, 2007). We therefore speculated that this kinase plays a role in the maintenance of the malignant phenotype in NSCLC cells. Analysis of PKCε expression in 4 different human NSCLC cells (H358, A549, H441 and H322) revealed a remarkable overexpression of PKCε relative to immortalized non-tumorigenic (HBEC3) cells (Fig. 1A), which fits with observations in human lung cancer specimens. To establish a role of PKCε in growth and tumorigenesis, we used a RNAi silencing approach. Each of the four NSCLC cell lines was infected with PKCε shRNA lentiviruses or a non-target shRNA lentivirus and stable pools selected with puromycin. Two different sequences (ε#1 and ε#2) were used in all cases to minimize off-target effects. Expression of PKCε was reduced more than 75% by either sequence without any noticeable change in the levels of the other DAG-responsive PKCs present in these cells (PKCα and PKCδ) (Fig. 1B). Cell proliferation was significantly reduced in all four NSCLC cell lines in which PKCε was stably depleted (Fig. 1C). Moreover, assays of colony formation in liquid and semisolid medium revealed that both anchorage-independent and anchorage-dependent growth were impaired in PKCε-depleted H358 cells relative to control cells. Similar results were observed in A549 cells (Fig. 1D and 1E). These data suggest that PKCε may be important in NSCLC growth.
Fig. 1. PKCε is required for the growth of NSCLC cells.
A) PKCε expression was analyzed by Western blot in immortalized non-tumorigenic (HBEC3) and NSCLC-derived cell lines (H358, H441, H322 and A549). Cells lines were obtained from ATCC and grown as recommended by the provider. An anti-PKCε antibody (Santa Cruz) was used at a 1:1000 dilution. B) Cells were infected with shRNA lentiviruses for PKCε (MISSION shRNA Lentiviral Transduction particles, Sigma, NM_005400 ε#1, clone ID x-741s1c1; ε#2, clone ID x-375s1c1) followed by selection with puromycin (1–2 μg/ml). MISSION non-target shRNA Lentiviral Transduction particles (Sigma, SHC002V) were used as control (CTRL). Expression of PKCε in NSCLC stable cell lines is depicted. C) Cells (5 ×104) were seeded in 12-well plates, allowed to grow in 2% FBS and counted 48 h later using a hemocytometer. D) For liquid colony formation assays cells were plated in 100 mm plates (100 cells/plate for H358 and 1000 cells/plate for A549). Medium was replaced twice a week and after 15 days colonies were stained with 0.7% methylene blue in 50% ethanol. E) To evaluate anchorage-independent growth 3 ×103 cells were plated in 0.35% agar over a 0.5% agar layer. After 10 days the plates were stained with MTS. For each well, the number of colonies was counted in 5 different fields and averaged. In all cases, data are expressed as mean ± S.E.M. of 3 individual experiments. *, p<0.05; **, p<0.01; ***, p<0.001.
PKCε depletion inhibits NSCLC xenograft growth
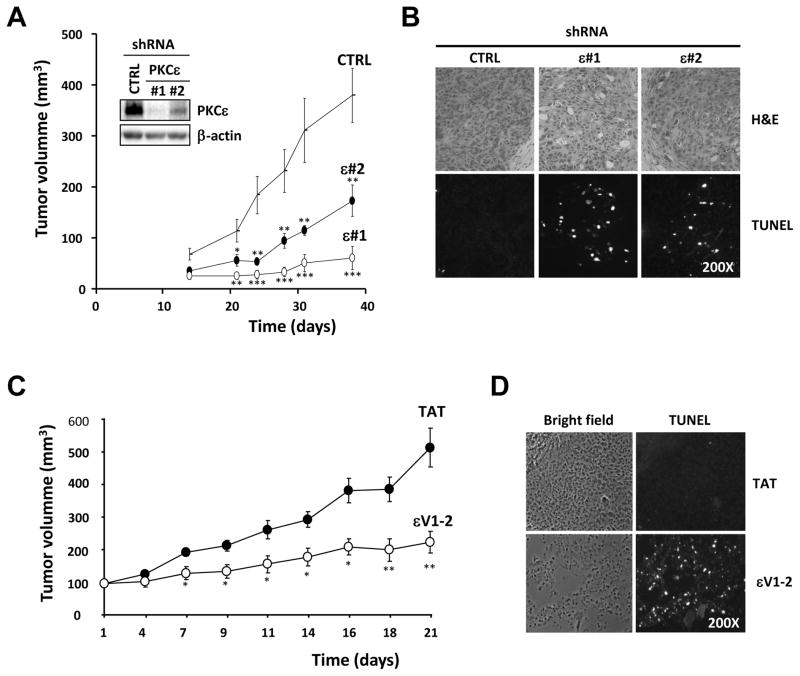
In order to assess the effect of PKCε depletion on the tumorigenic potential of NSCLC cells in vivo, we injected s.c. into athymic nude mice H358 cell lines stably expressing either ε#1 shRNA, ε#2 shRNA, or control shRNA. Inoculation of control H358 cells led to the formation of tumors with a latency of ~2 weeks. Notably, tumor growth of PKCε-depleted cells was remarkably lower compared with control NSCLC cells (Fig. 2A). Inhibition of tumor growth was more evident in NSCLC cells expressing shRNA ε#1, which shows near complete depletion of PKCε. Immunohistochemical analysis of xenografts 15 days after inoculation showed a marked induction of cell death in PKCε-depleted cells, as evidenced by a large number of TUNEL positive cells (Fig. 2B).
Fig. 2. PKCε is required for NSCLC tumor growth in athymic nude mice.
A) H358 cells expressing shRNA control (CTRL) or PKCε (ε#1 and ε#2) at 80% confluency were resuspended in serum-free medium, and then 0.1 ml containing 5 ×106 cells were injected s.c. into the flank of male athymic nude-Foxn1nu mice (Harlan Laboratories). The width and length of tumors were measured with a caliper at different times, and tumor volume calculated as Vol=π x width2 x length/6. Inset, PKCε levels at the day of inoculation. Data are expressed as mean ± S.E.M. (n=10). *, p<0.05; **, p<0.01; ***, p<0.001. A second experiment gave similar results. B) Tumors were removed and processed for immunohistochemistry, 15 days post-inoculation. Upper panels, H&E staining, lower panels, TUNEL labeling. C) H358 cells were injected s.c. in the flank of athymic mice (5 ×106 cells/mice). When tumors reached ~100 mm3 (~ 20 days post-inoculation) animals were randomized into two groups and subject to treatment with either control carrier peptide (TAT) or εV1-2 with TAT (18 mg/kg/day). Peptide delivery was achieved by weekly subcutaneous implantation of osmotic minipumps in the opposite flank. Tumor volume was expressed as the mean ± S.E.M. (n=8). *, p<0.05; **, p<0.01. A second experiment gave similar results. D) Tumors were removed 15 days after the beginning of treatment and stained for TUNEL.
Pharmacological inhibition of PKCε impairs NSCLC tumor growth
PKC isozymes have been extensively studied as therapeutic targets, and several modulators of PKC activity have been examined in clinical trials for multiple malignancies (Barry & Kazanietz, 2001; Goekjian & Jirousek, 2001; Serova et al., 2006). Surprisingly, despite the established relevance of PKCε in mitogenesis and survival there are no studies to date examining PKCε as a potential therapeutic target for lung cancer. This may be partly due to the lack of kinase inhibitors with selectivity towards PKCε. Indeed, most ATP-binding site directed inhibitors fail to discriminate between PKC isozymes and/or have additional kinases as targets (Bain et al., 2003; Bain et al., 2007; Davies et al., 2000; Mochly-Rosen & Kauvar, 1998). We decided to take advantage of εV1-2, a peptide that specifically inhibits PKCε translocation without affecting the activation of other PKC isozymes (Begley et al., 2004; Felber et al., 2007). This PKCε inhibitor has been successfully delivered as a TAT-fused protein into cellular models as well as into mice and rats (Budas et al., 2007; Gray et al., 1997; Inagaki et al., 2005; Koyanagi et al., 2007).
H358 xenografts were implanted s.c. in athymic mice and when tumors reached approximately 100 mm3 animals were randomized into two groups that received either εV1-2 conjugated to TAT or control TAT (18 mg/kg/day) by osmotic minipumps. Previous studies have found that administration of εV1-2 for 5 weeks at 20 mg/kg/day does not cause cytotoxic or systemic effects to mice (Koyanagi et al., 2007). Osmotic minipumps were replaced every 7 days and tumor growth was followed for 3 weeks. As shown in Fig. 2C, delivery of εV1-2 into nude mice greatly reduced H358 xenograft growth. Immunohistochemical analysis showed a strong induction of cell death (TUNEL positive cells) in tumors from mice that received εV1-2. On the other hand, there were essentially no TUNEL-positive cells in tumors from animals that received the control TAT peptide (Fig. 2D). These results strongly argue for a role for PKCε in NSCLC tumor cell survival.
Identification of PKCε target genes related to survival in NSCLC cells and tumors
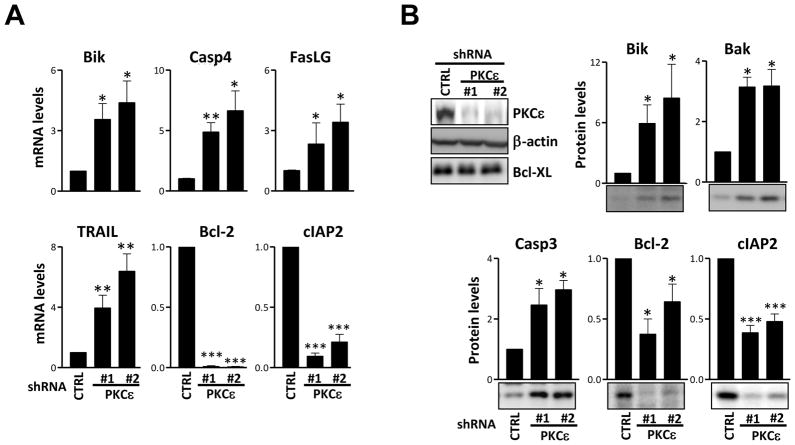
In a recent genome wide analysis we established that individual members of the PKC family differentially modulate gene expression. PKCε-regulated gene sets related to cell cycle, K-Ras oncogenesis, and transformation were identified (Caino et al., 2011). Given that PKCε loss-of-function induces cell death in NSCLC xenografts we sought to determine whether PKCε regulates the expression of apoptotic/survival genes. Using a Human Apoptosis array (QIAGEN) we compared the mRNA levels of key apoptosis-related genes in control vs. PKCε-depleted cells. A 1.5-fold cut-off value (PKCε/control) was used. This analysis revealed that PKCε depletion increased mRNA levels of pro-apoptotic proteins Bak1, Bcl2A1, Bcl2L10, Bik and HRK (Table 1). PKCε depletion was associated with a significant decrease in the expression of the pro-survival protein Bcl-2 and the inhibitor of apoptosis cIAP-2 (Fig. 3A and data not shown). In addition, PKCε-depleted cells expressed higher levels of caspase recruitment domain (CARD)-containing proteins (Bag4, CARD8) and caspases (CASP2, CASP3, CASP4 and CASP6) relative to control cells. Several ligands and receptors of the tumor necrosis factor superfamily were also elevated as a consequence of PKCε depletion (Table 1). In order to validate the results from the array, 11 genes were selected. Fig. 3A shows that in all cases the changes observed in the array could be recapitulated using qPCR. Several of these genes were further validated by Western blot (Fig. 3B). These results suggest that PKCε plays an important role in the modulation of genes involved in cell survival and raise the possibility that they contribute to the susceptibility to apoptosis upon PKCε depletion.
Table 1. Expression of PKCε-target genes in human NSCLC cells and specimens.
Simultaneous detection of apoptosis-related genes and 2 housekeeping genes by qPCR was carried out using RT2 Profiler PCR Array plates (QIAGEN) and a RT2 SYBR Green/5-carboxy-X-rhodamine (ROX) qPCR master mix. Data were normalized to GAPDH and β-actin housekeeping genes, and the relative levels of mRNA calculated according to the ΔCt method. Genes that changed expression > 1.5-fold as a consequence of PKCε depletion were selected for further analysis. Tumor cells, H358 cells expressing shRNA control (CTRL) or shRNA for PKCε (ε#1 and ε#2) were assayed for expression of key apoptosis-related genes. Human samples, data publicly available from the Oncomine repository. Lung adenocarcinomas (LAC) were compared against normal lung tissues (NL). Data for each gene were filtered for down-regulation in lung adenocarcinoma (LAC) vs. normal lung (NL) tissue with a p<0.05. Whenever possible, multiple studies containing information for each gene were compared by meta-analysis and an associated p value was determined. p values for single-gene (Single) or meta-analysis (Meta) are shown. To establish if PKCε-regulated genes are altered in lung cancer, we carried out a Gene Set Enrichment Analysis (GSEA), as previously described (Subramanian et al., 2005). A GSEA comparing LAC (n=139) vs. NL (n=17) was run for the defined PKCε-gene set using the complete dataset for the Bhattacharjee Study (Bhattacharjee et al., 2001). Enrichment in NL is indicated by “Yes”. ns, not significant.
| Gene Symbol | Tumor cells
|
Human samples
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTRL | PKCε-depleted | p-value | Oncomine p-values | GSEA (Yes/No) | |||||||
| mean | S.D. | n | mean | S.D. | n | Single gene | Meta-analysis | ||||
| Up-regulated | |||||||||||
| BAG3 | 1.01 | 0.09 | 4 | 1.63 | 0.13 | 4 | 0.0035 | * | 0.05 | N/A | No |
| BAG4 | 1.00 | 0.04 | 4 | 2.05 | 0.02 | 4 | p<0.0001 | *** | ns | N/A | No |
| BAK1 | 1.00 | 0.03 | 4 | 2.33 | 0.52 | 4 | 0.0219 | * | 0.04 | 0.04 | No |
| BCL10 | 1.01 | 0.11 | 4 | 3.10 | 0.70 | 4 | 0.013 | * | ns | N/A | No |
| BCL2A1 | 1.10 | 0.32 | 4 | 3.59 | 0.01 | 4 | p<0.0001 | *** | 6.23 ×10−5 | 0.02 | No |
| BCL2L10 | 1.07 | 0.27 | 4 | 2.99 | 0.86 | 4 | 0.0386 | * | 0.018 | N/A | No |
| BCL2L11 | 1.01 | 0.09 | 4 | 1.63 | 0.19 | 4 | 0.0124 | * | ns | N/A | No |
| BIK | 1.10 | 0.32 | 4 | 8.76 | 0.24 | 4 | p<0.0001 | *** | ns | N/A | No |
| BIRC2 | 1.09 | 0.31 | 4 | 1.53 | 0.07 | 4 | 0.1038 | ns | 1.42 ×10−4 | 3.4 ×10−4 | Yes |
| BIRC4 | 1.01 | 0.08 | 4 | 1.56 | 0.04 | 4 | 0.0004 | *** | ns | N/A | No |
| BIRC8 | 1.34 | 0.63 | 4 | 5.35 | 2.55 | 4 | 0.0889 | ns | N/A | N/A | No |
| BNIP3 | 1.01 | 0.10 | 4 | 2.44 | 0.29 | 4 | 0.0017 | ** | ns | N/A | No |
| BNIP3L | 1.02 | 0.16 | 4 | 1.99 | 0.15 | 4 | 0.0022 | ** | 1.58 ×10−5 | 0.004 | Yes |
| BRAF | 1.01 | 0.08 | 4 | 3.34 | 0.35 | 4 | 0.0003 | *** | ns | N/A | No |
| NOD1 | 1.00 | 0.06 | 4 | 1.53 | 0.19 | 4 | 0.0193 | * | ns | N/A | No |
| CASP1 | 1.06 | 0.25 | 4 | 6.47 | 2.82 | 4 | 0.0524 | ns | 1.18 ×10−5 | 1.74 ×10−8 | Yes |
| CASP2 | 1.01 | 0.10 | 4 | 1.59 | 0.35 | 4 | 0.0808 | ns | 0.008 | N/A | No |
| CASP3 | 1.00 | 0.06 | 4 | 2.80 | 0.21 | 4 | p<0.0001 | *** | ns | N/A | No |
| CASP4 | 1.09 | 0.31 | 4 | 7.05 | 1.02 | 4 | 0.0007 | *** | 6.22 ×10−6 | 0.005 | Yes |
| CASP5 | 1.00 | 0.03 | 4 | 1.69 | 0.51 | 4 | 0.1136 | ns | 3.13 ×10−4 | 0.015 | No |
| CASP6 | 1.02 | 0.15 | 4 | 2.91 | 0.58 | 4 | 0.0095 | ** | ns | N/A | No |
| CD40 | 1.22 | 0.50 | 4 | 9.33 | 0.53 | 4 | p<0.0001 | *** | 1.17 ×10−4 | 0.019 | Yes |
| CD40LG | 1.00 | 0.00 | 4 | 1.66 | 0.51 | 4 | 0.1238 | ns | 0.011 | N/A | No |
| CIDEA | 1.34 | 0.63 | 4 | 3.00 | 0.80 | 4 | 0.0768 | ns | ns | N/A | No |
| CIDEB | 1.00 | 0.06 | 4 | 1.56 | 0.09 | 4 | 0.0011 | *** | 0.049 | N/A | No |
| DFFA | 1.00 | 0.03 | 4 | 1.58 | 0.01 | 4 | p<0.0001 | *** | 0.031 | N/A | No |
| FAS | 1.00 | 0.05 | 4 | 1.63 | 0.05 | 4 | p<0.0001 | *** | 1.05 ×10−7 | 6.32 ×10−4 | Yes |
| FASLG | 1.00 | 0.06 | 4 | 4.26 | 0.43 | 4 | 0.0001 | *** | 0.028 | 0.038 | No |
| GADD45A | 1.00 | 0.04 | 4 | 3.44 | 0.01 | 4 | p<0.0001 | *** | ns | N/A | No |
| HRK | 1.00 | 0.01 | 4 | 2.67 | 0.87 | 4 | 0.0513 | ns | 0.035 | N/A | No |
| LTA | 1.18 | 0.44 | 4 | 5.02 | 0.20 | 4 | 0.0001 | *** | 0.006 | N/A | No |
| MCL1 | 1.04 | 0.21 | 4 | 2.66 | 0.13 | 4 | 0.0003 | *** | 2.36 ×10−6 | 9.52 ×10−4 | Yes |
| NOL3 | 1.07 | 0.27 | 4 | 2.95 | 0.06 | 4 | 0.0002 | *** | ns | N/A | No |
| TNF | 1.21 | 0.49 | 4 | 2.23 | 1.17 | 4 | 0.2261 | ns | 9.79 ×10−5 | 0.006 | Yes |
| TNFRSF10A | 1.00 | 0.06 | 4 | 1.86 | 0.44 | 4 | 0.0509 | ns | N/A | N/A | No |
| TNFRSF21 | 1.00 | 0.06 | 4 | 2.33 | 0.04 | 4 | p<0.0001 | *** | ns | N/A | No |
| TNFRSF25 | 1.07 | 0.27 | 4 | 7.22 | 1.96 | 4 | 0.0105 | * | 0.016 | N/A | No |
| TNFSF10 | 1.16 | 0.42 | 4 | 4.82 | 1.04 | 4 | 0.0085 | ** | 1.05 ×10−11 | 2.17 ×10−4 | Yes |
| TNFSF8 | 1.00 | 0.04 | 4 | 2.42 | 0.36 | 4 | 0.0038 | ** | 0.01 | 0.013 | No |
| TP53BP2 | 1.00 | 0.01 | 4 | 1.55 | 0.08 | 4 | 0.0002 | *** | 0.001 | 0.001 | No |
| TP73 | 1.01 | 0.10 | 4 | 2.21 | 0.51 | 4 | 0.0296 | * | ns | N/A | No |
| TRADD | 1.00 | 0.02 | 4 | 1.65 | 0.02 | 4 | p<0.0001 | *** | 7.26 ×10−5 | N/A | No |
| TRAF4 | 1.00 | 0.03 | 4 | 2.60 | 0.57 | 4 | 0.0152 | * | ns | N/A | No |
| B2M | 1.02 | 0.15 | 4 | 2.55 | 0.01 | 4 | p<0.0001 | *** | 4.1 ×10−5 | 2.91 ×10−4 | Yes |
| Down-regulated | |||||||||||
| BCL2 | 1.16 | 0.42 | 4 | 0.09 | 0.04 | 4 | 0.0216 | * | 0.013 | 0.029 | N/A |
| BIRC3 | 1.10 | 0.33 | 4 | 0.18 | 0.04 | 4 | 0.0163 | * | 6.57 ×10−4 | 0.017 | N/A |
| CARD6 | 1.00 | 0.01 | 4 | 0.60 | 0.16 | 4 | 0.0214 | * | N/A | N/A | N/A |
| IGF1R | 1.01 | 0.09 | 4 | 0.64 | 0.04 | 4 | 0.004 | ** | 0.021 | N/A | N/A |
| PYCARD | 1.00 | 0.07 | 4 | 0.01 | 0.00 | 4 | p<0.0001 | *** | ns | N/A | N/A |
| TNFRSF11B | 1.09 | 0.31 | 4 | 0.53 | 0.20 | 4 | 0.0907 | ns | ns | N/A | N/A |
| TNFRSF1A | 1.00 | 0.06 | 4 | 0.69 | 0.22 | 4 | 0.1076 | ns | ns | N/A | N/A |
| CD27 | 1.11 | 0.34 | 4 | 0.30 | 0.17 | 4 | 0.0381 | * | 2.83 ×10−4 | 6.39 ×10−4 | N/A |
| TNFRS9 | 1.12 | 0.36 | 4 | 0.55 | 0.14 | 4 | 0.0929 | ns | 2.34 ×10−4 | 2.34 ×10−4 | N/A |
| CD70 | 1.05 | 0.22 | 4 | 0.10 | 0.02 | 4 | 0.0025 | ** | 0.002 | 0.012 | N/A |
| RPL13A | 1.00 | 0.01 | 4 | 0.61 | 0.21 | 4 | 0.0559 | ns | 0.003 | 0.002 | N/A |
Fig. 3. Validation of representative PKCε target genes in NSCLC cells.
H358 cells expressing shRNA control (CTRL) or shRNA for PKCε (ε#1 and ε#2) were assayed for expression of key apoptosis-related genes. A) For qPCR, RNA was isolated using the QIAGEN RNeasy kit and reverse transcribed to cDNA using random hexamers and the TaqMan Reverse Transcription kit (Applied Biosystems). Real-time PCR assays were performed in a 7300 ABI PCR System (Applied Biosystems), using TaqMan Gene expression assays and TaqMan Universal master mix. Human 18S rRNA was used as an endogenous control for normalization. The relative levels of mRNA compared to control were calculated according to the ΔCt method. B) Western blots and the corresponding densitometric analyses were carried out essentially as previously described (Oliva et al., 2008). The following primary antibodies were used: anti-Bik, anti-Bak, anti-caspase 3, anti-cIAP2, anti-Bcl2, anti-Bcl-XL (Cell Signaling, 1:1000 dilution), and anti-β-actin (Sigma, 1:50,000 dilution). Densitometric analysis of 3 independent experiments is shown. Data are expressed as the mean ± S.E.M. (n=3). *, p<0.05; **, p<0.01.
To further investigate the relevance of PKCε target genes in NSCLC, we carried out a comprehensive analysis of publicly available human lung cancer datasets. A comparison of PKCε mRNA levels in human lung adenocarcinomas (LAC) vs. normal lung (NL) tissue using the Oncomine repository revealed marked up-regulation in LAC (fold change=2.5, p<0.05, Beer Lung study). Remarkably, the vast majority of the genes with expression altered in PKCε-depleted cells were deregulated in LAC (Table 1 and Tables S1–S2). As expected, pro-apoptotic genes up-regulated in PKCε-depleted cells were down-regulated in LAC, whereas pro-survival genes down-regulated in PKCε-depleted cells were up-regulated in LAC. Furthermore, a meta-analysis from all available studies for each gene revealed that expression changes in LAC vs. NL were statistically significant for 27 of the PKCε-regulated genes (21 genes down-regulated and 6 genes up-regulated in LAC) (Table 1). Based on these results we defined a set for genes regulated negatively by PKCε that passed the threshold of statistical significance p<0.05 that we named “PKCε-gene set”.
In order to gain biological insight from these gene expression studies and determining a potential correlation between the expression of PKCε-regulated genes and disease status, we carried out a Gene Set Enrichment Analysis (GSEA) using as a query the above defined PKCε-gene set. Interestingly, a significant correlation was found between down-regulation of PKCε-regulated genes and LAC samples (p<1 ×10−8 and FDR <0.001). Among the genes negatively correlated with LAC (enrichment score, ES<0) we found FAS, CASP4, CD40, BIRC2, CASP1, TRAIL (TNFSF10), MCL1 and BNIP3L (Table 1, GSEA column). In summary, this analysis identified novel apoptosis-related PKCε transcriptional targets associated with NSCLC, suggesting an important role for PKCε in the inhibition of pro-apoptotic genes.
Final conclusions
Our studies clearly show that overexpression of PKCε is a key driving force in the growth and tumorigenicity of NSCLC cells. PKCε depletion or pharmacological inhibition reduced tumor growth and enhanced tumor cell death, as demonstrated also in cell models in vitro. A key mechanism by which PKCε may sustain tumorigenicity of NSCLC cells is via modulation of the expression of apoptotic genes. A point that illustrates the relevance of our findings is that the majority of the PKCε apoptotic target genes are down-regulated in human NSCLC samples. Moreover, current unpublished studies from our laboratory suggest that PKCε is a key modulator of transcriptional networks of genes that dictate cell fate (M.C.C. and M.G.K., unpublished observations). Conceivably, targeting PKCε may reinstate altered transcriptional patterns displayed by NSCLC cells that contribute to disease development.
One of the outstanding questions raised by this study is the utility of PKCε as a therapeutic target. Our results suggest that PKCε is required for survival of tumor cells and that PKCε depletion of inhibition is sufficient to inhibit tumor growth of lung cancer cells. Three major issues are raised by these results that required ample investigation. The first is the mechanism that leads to PKCε up-regulation in lung cancer (or other epithelial cancers). Preliminary data from our laboratory suggest that deregulation occurs at a transcriptional level and possibly through enhanced protein stability (Wang H.B., Lu H., and M.G.K., unpublished observations). Second, it remains to be determined whether PKCε plays a role in the initiation of lung cancer and whether its overexpression cooperates with other oncogenic alterations characteristic of the disease. Lastly, it would be important to establish whether overexpression of PKCε also involves its hyperactivation. Conceivable, oncogenic alterations that occur in lung cancer lead to enhanced generation of DAG, the physiological PKCε activator. For example, mutant K-Ras is known to elevate cellular DAG levels (Fleischman et al., 1986; Matyas & Fishman, 1989; Preiss et al., 1986). EGFR, which is overexpressed/hyperactivated in 50–90% of human lung tumors (Sekido et al., 2003), couples to phospholipase Cγ, a key enzyme responsible for DAG generation (Paez et al., 2004; Pao et al., 2004; Parker & Murray-Rust, 2004; Rusch et al., 1993; Toker, 1998). Moreover, EGFR signals to PDK-1, a PIP3-dependent kinase required for the maturation of PKCε (Parker & Murray-Rust, 2004; Toker, 1998). Both K-Ras and EGFR are established drivers of survival signals (Lemmon & Schlessinger, 2010; Malumbres & Barbacid, 2003), but the implication of PKCε in these responses remains to be determined. Whether other common oncogenic alterations in lung cancer, such as bFGF and VEGF overexpression, or PI3K, p53 and Pten mutations (Sekido et al., 2003), lead to PKCε activation, needs to be established. Our data therefore warrant the analysis of signals leading to PKCε activation and up-regulation in lung cancer cells. Dissecting these mechanisms may help determining the potential of PKCε as a therapeutic target for different subsets of lung cancers or establish new regimens of combined therapy.
Supplementary Material
References
- Bae KM, Wang H, Jiang G, Chen MG, Lu L, Xiao L. Cancer Res. 2007;67:6053–63. doi: 10.1158/0008-5472.CAN-06-4037. [DOI] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr LF, Mabry M, Nelkin BD, Tyler G, May WS, Baylin SB. Cancer Res. 1991;51:5514–9. [PubMed] [Google Scholar]
- Barry OP, Kazanietz MG. Curr Pharm Des. 2001;7:1725–44. doi: 10.2174/1381612013397041. [DOI] [PubMed] [Google Scholar]
- Basu A, Sivaprasad U. Cell Signal. 2007;19:1633–42. doi: 10.1016/j.cellsig.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley R, Liron T, Baryza J, Mochly-Rosen D. Biochem Biophys Res Commun. 2004;318:949–54. doi: 10.1016/j.bbrc.2004.04.121. [DOI] [PubMed] [Google Scholar]
- Benavides F, Blando J, Perez CJ, Garg R, Conti CJ, DiGiovanni J, Kazanietz MG. Cell Cycle. 2011;10:268–77. doi: 10.4161/cc.10.2.14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JD. Front Biosci. 2000;5:D406–23. doi: 10.2741/black. [DOI] [PubMed] [Google Scholar]
- Brodie C, Blumberg PM. Apoptosis. 2003;8:19–27. doi: 10.1023/a:1021640817208. [DOI] [PubMed] [Google Scholar]
- Budas GR, Koyanagi T, Churchill EN, Mochly-Rosen D. Biochem Soc Trans. 2007;35:1021–6. doi: 10.1042/BST0351021. [DOI] [PubMed] [Google Scholar]
- Caino MC, Meshki J, Kazanietz MG. Apoptosis. 2009;14:392–408. doi: 10.1007/s10495-009-0316-z. [DOI] [PubMed] [Google Scholar]
- Caino MC, von Burstin VA, Lopez-Haber C, Kazanietz MG. J Biol Chem. 2011;286:11254–64. doi: 10.1074/jbc.M110.194332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber M, Sonnemann J, Beck JF. Pathol Oncol Res. 2007;13:295–301. doi: 10.1007/BF02940308. [DOI] [PubMed] [Google Scholar]
- Fleischman LF, Chahwala SB, Cantley L. Science. 1986;231:407–10. doi: 10.1126/science.3001936. [DOI] [PubMed] [Google Scholar]
- Goekjian PG, Jirousek MR. Expert Opin Investig Drugs. 2001;10:2117–40. doi: 10.1517/13543784.10.12.2117. [DOI] [PubMed] [Google Scholar]
- Gorin MA, Pan Q. Mol Cancer. 2009;8:9. doi: 10.1186/1476-4598-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MO, Karliner JS, Mochly-Rosen D. J Biol Chem. 1997;272:30945–51. doi: 10.1074/jbc.272.49.30945. [DOI] [PubMed] [Google Scholar]
- Griner EM, Kazanietz MG. Nat Rev Cancer. 2007;7:281. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- Han EK, Cacace AM, Sgambato A, Weinstein IB. Carcinogenesis. 1995;16:2423–8. doi: 10.1093/carcin/16.10.2423. [DOI] [PubMed] [Google Scholar]
- Hsiao WL, Housey GM, Johnson MD, Weinstein IB. Mol Cell Biol. 1989;9:2641–7. doi: 10.1128/mcb.9.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki K, Begley R, Ikeno F, Mochly-Rosen D. Circulation. 2005;111:44–50. doi: 10.1161/01.CIR.0000151614.22282.F1. [DOI] [PubMed] [Google Scholar]
- Jansen AP, Verwiebe EG, Dreckschmidt NE, Wheeler DL, Oberley TD, Verma AK. Cancer Res. 2001;61:808–12. [PubMed] [Google Scholar]
- Koyanagi T, Noguchi K, Ootani A, Inagaki K, Robbins RC, Mochly-Rosen D. J Mol Cell Cardiol. 2007;43:517–22. doi: 10.1016/j.yjmcc.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Huang J, Basu A. J Biol Chem. 2006;281:22799–22807. doi: 10.1074/jbc.M603390200. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Matyas GR, Fishman PH. Cell Signal. 1989;1:395–404. doi: 10.1016/0898-6568(89)90058-2. [DOI] [PubMed] [Google Scholar]
- McJilton MA, Van Sikes C, Wescott GG, Wu D, Foreman TL, Gregory CW, Weidner DA, Harris Ford O, Morgan Lasater A, Mohler JL, Terrian DM. Oncogene. 2003;22:7958–68. doi: 10.1038/sj.onc.1206795. [DOI] [PubMed] [Google Scholar]
- Mischak H, Goodnight JA, Kolch W, Martiny-Baron G, Schaechtle C, Kazanietz MG, Blumberg PM, Pierce JH, Mushinski JF. J Biol Chem. 1993;268:6090–6096. [PubMed] [Google Scholar]
- Mochly-Rosen D, Kauvar LM. Adv Pharmacol. 1998;44:91–145. doi: 10.1016/s1054-3589(08)60126-x. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Oliva JL, Kothapalli D, Fournier A, Assoian RK, Kazanietz MG. J Biol Chem. 2005;280:33926–34. doi: 10.1074/jbc.M505748200. [DOI] [PubMed] [Google Scholar]
- Okhrimenko H, Lu W, Xiang C, Hamburger N, Kazimirsky G, Brodie C. Cancer Res. 2005;65:7301–9. doi: 10.1158/0008-5472.CAN-05-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva JL, Caino MC, Senderowicz AM, Kazanietz MG. J Biol Chem. 2008;283:5466–76. doi: 10.1074/jbc.M707576200. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker PJ, Murray-Rust J. J Cell Sci. 2004;117:131–2. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- Perletti GP, Folini M, Lin HC, Mischak H, Piccinini F, Tashjian AH., Jr Oncogene. 1996;12:847–54. [PubMed] [Google Scholar]
- Preiss J, Loomis CR, Bishop WR, Stein R, Niedel JE, Bell RM. J Biol Chem. 1986;261:8597–600. [PubMed] [Google Scholar]
- Rusch V, Baselga J, Cordon-Cardo C, Orazem J, Zaman M, Hoda S, McIntosh J, Kurie J, Dmitrovsky E. Cancer Res. 1993;53:2379–85. [PubMed] [Google Scholar]
- Sekido Y, Fong KM, Minna JD. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- Serova M, Ghoul A, Benhadji KA, Cvitkovic E, Faivre S, Calvo F, Lokiec F, Raymond E. Semin Oncol. 2006;33:466–78. doi: 10.1053/j.seminoncol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Slupsky JR, Kamiguti AS, Harris RJ, Cawley JC, Zuzel M. Am J Pathol. 2007;170:745–54. doi: 10.2353/ajpath.2007.060557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. Front Biosci. 1998;3:D1134–47. doi: 10.2741/a350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.