Abstract
Background
Telomerase is a ribonucleoprotein enzyme that synthesises telomeres after cell division and maintains chromosomal stability leading to cellular immortalisation. hTERT (human telomerase reverse transcriptase) is the rate-limiting determinant of telomerase reactivation. Telomerase has been associated with negative prognostic indicators in some studies. The present study aims to detect any correlation between hTERT and the negative prognostic indicators VEGF and PCNA by quantitatively measuring the mRNA expression of these genes in human breast cancer and in adjacent non-cancerous tissue (ANCT).
Materials and methods
RNA was extracted from 38 breast carcinomas and 40 ANCT. hTERT and VEGF165, VEGF189 and PCNA mRNA expressions were estimated by reverse transcriptase-PCR (RT-PCR) and Taqman methodology.
Results
The level of expression of VEGF-165 and PCNA was significantly higher in carcinoma tissue than ANCT (p = 0.02). The ratio of VEGF165/189 expression was significantly higher in breast carcinoma than ANCT (p = 0.025). hTERT mRNA expression correlated with VEGF-189 mRNA (p = 0.008) and VEGF165 (p = 0.07).
Conclusions
hTERT mRNA expression is associated with the expression of the VEGF189 and 165 isoforms. This could explain the poorer prognosis reported in breast tumours expressing high levels of hTERT. The relative expression of the VEGF isoforms is significantly different in breast tumour to ANCT, and this may be important in breast carcinogenesis.
Keywords: hTERT, breast, cancer, VEGF, PCNA
Introduction
Telomerase is an RNA dependant DNA polymerase, the function of which is to synthesise the repetitive nucleotide sequence (TTAGGG in humans) forming the telomeres at the end of chromosomes 1. During cell division, DNA polymerase is unable to fully replicate the ends of linear DNA and therefore genetic material is lost from chromosomal ends. Telomeres form caps on the chromosomes that prevent fusion of chromosomal ends and are dispensable without compromising gene expression. Without telomerase activity, each round of nuclear division results in shortening of the telomeres and reaching a critical length seems to trigger off cellular apoptosis 1. Telomerase activity however appears to stabilise telomeres and thus leads to cellular immortality [1-3]. Telomerase is active in 80–90% of malignant tissues and many immortal cell lines but not most normal somatic cells [4]. Of the telomerase subunits, the reverse transcriptase (hTERT) is the determinant of enzyme activity [5-7]. We have recently demonstrated a significant correlation between telomerase activity and hTERT mRNA expression in human breast cancer [8]. Investigators have shown high levels of hTERT expression to be linked to a worse prognosis in Wilm's tumour [9], acute myelogenous leukaemia [10] and non-small cell lung carcinoma [11]. In breast carcinoma, high hTERT expression detected by quantitative RT-PCR was found to have a statistically significant correlation with disease relapse [12]. We have shown that hTERT mRNA is over-expressed in the majority of the breast carcinomas that we have studied [13]. The level of expression was much higher in breast cancer specimens than in ANCT and fibroadenomas [14]. However, apart from a weak correlation between hTERT levels and tumour size and progesterone receptor negativity, we have not detected any correlation with the standard prognostic indicators for breast carcinoma [14]. These findings could be explained if hTERT expression were linked to a negative prognostic indicator not routinely assessed by histopathological examination.
Angiogenesis in carcinomas, including breast carcinoma is an important development in maintaining tumour growth and promoting metastasis [15]. Vascular endothelial growth factor (VEGF) is a cytokine that stimulates the proliferation and migration of vascular endothelial cells, key steps in angiogenesis [16]. A number of structurally related peptides have now been discovered, named VEGF-B, VEGF-C, VEGF-D and PIGF, with VEGF now being renamed VEGF-A .15. VEGF-A is expressed in a wide range of malignancies including breast carcinoma [17]. The level of VEGF mRNA has been found to be higher in breast tumour cells than in normal breast tissue [18] and higher levels of VEGF protein are associated with increased microvascular density and disease relapse [19,20]. The VEGF-A gene is made up of 8 exons [21] and differential splicing results in at least six isoforms named VEGF-121, -145, -165, -183, -189, and -206 according to the number of amino acids [22].
Proliferating cell nuclear antigen (PCNA) is a protein that localises to the nucleus and is involved in DNA synthesis and replication [23]. PCNA levels increase in cells undergoing proliferation and increased PCNA expression has been identified as an independent prognostic indicator in breast carcinoma [24].
The aim of this study was to quantitatively measure the expression of hTERT, VEGF and PCNA mRNAs in human breast cancer and in adjacent non-cancerous tissue (ANCT) 1 cm away from the tumour in order to detect any correlation of these negative prognostic indicators with hTERT.
Materials and Methods
Samples
Brent Medical Ethics Committee provided ethical approval for the use of tissue specimens for RNA extraction and analysis. Following patient consent, samples of tissue from breast tumour and macroscopically normal tissue 1 cm away from the tumour were taken from 40 women undergoing surgery for breast cancer. Tissue was frozen in liquid nitrogen within 30 minutes of removal and was then stored at -70° Celsius until use.
RNA Extraction
Total cellular RNA was extracted from specimens of breast tissue carcinomas and matched adjacent non-cancerous tissue using the RNeasy Mini isolation kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. For cancerous tissue between 10 and 20 mg of tissue was used. However, because ANCT tended to have a high fat content, difficulty was experienced in obtaining sufficient RNA concentration for analysis. Therefore, for ANCT the extraction technique was modified as described by Vidal et al [25] to include higher amounts of lysis buffer and repeated lysis steps, and using between 100 and 200 mg of tissue. At the time of separation of tissue for RNA extraction, a frozen section was taken from the tissue immediately adjacent to allow confirmation of histological features. Extraction of RNA was carried out in a laboratory separate to the one used for RT-PCR. Quantification of the RNA following treatment with DNase (Promega) was carried out in triplicate using the RiboGreen reagent (Molecular Probes Europe BV) according to the manufacturer's protocol.
RT-PCR
TaqMan RT-PCR was performed in duplicate for each sample using the ABI PRISM 7700 Sequence Detector (Perkin-Elmer Applied Biosystems) and the TaqMan EZ RT-PCR Core Reagents Kit (Perkin-Elmer Applied Biosystems). The RT-PCR mix was set up in an extraction hood in a laboratory distant from the one in which RNA extraction was carried out. The RT-PCR was performed using the forward and reverse oligonucleotide primers and TaqMan probes given in Table 1.
Table 1.
Sequences of TaqMan Primers and Probes for RT-PCR
| Gene | Forward | Reverse | Probe |
| hTERT | 5'GAC AGT TGC AAA GCA TTG GA3' | 5'ACC TCT GCT TCC GAC AGC TC3' | 5'AGC TGC ACC CTC TTC AAG TGC TGT CTG A3' |
| VEGF-165 | 5'TGT GAA TGC AGA CCA AAG AAA GA3' | 5'GCT TTC TCC GCT CTG AGC AA | 5'AGA GCA AGA CAA GAC AAG AAA ATC CCT GTG GGC3' |
| VEGF-189 | 5'TGT GAA TGC AGA CCA AAG AAA GA3' | 5'CGT TTT TGC CCC TTT CCC3' | 5'AGA GCA AGA CAA GAA AAA AAA TCA GTT GGA A3' |
| PCNA | 5'TTA AAT TGT CAC AGA CAA GTA ATG TCG3' | 5'TGG CTT TTG TAA AGA AGT TCA GGT AC3' | 5'TGG TTC ATT CAT CTC TAT GGT AAC AGC TTG CTC CT3' |
These oligonucleotides were designed using Primer Express software (PE-Applied Biosystems, Warrington, UK) and gene sequences obtained from the GenBank database. Primers and probes were intron spanning to prevent amplification of genomic DNA. The hTERT primers and probes are in exons 3 and 4 of the hTERT gene (accession number AF128893.1) and define a 73 base pair amplicon. The PCNA primers and probe define a 117 base pair amplicon in the fourth and fifth exons of the gene (accession number XM_056035). The isoforms of VEGF that were measured were VEGF-189 and VEGF-165. In the case of VEGF-189, the primers and probe bridge exons 4, 5 and 6 of the VEGF gene (accession number XM_166457) to amplify a 77 base pair amplicon. For the VEGF-165 isoform exons 4,5 and 7 are bridged (accession number AF486837.1) and the amplicon is 73 base pairs long.
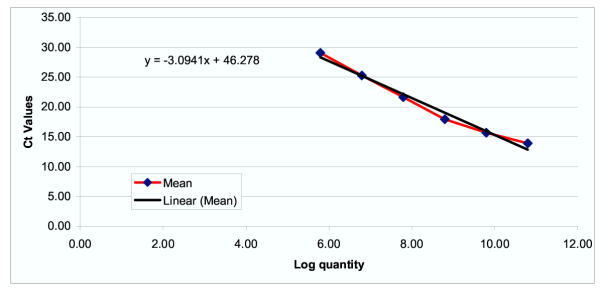
Conditions for the reverse transcription step were 50°C for 2 minutes, 60°C for 10 minutes, and 92°C for 5 minutes. The polymerase chain reaction was carried out for 50 cycles of 20 seconds at 92°C and 25 seconds at 62°C. For the negative control RNase free water was used in the RT-PCR mix instead of RNA. The positive control was the amplicon created for the standard curve. The standard curves for each mRNA were constructed using serial dilutions of a single stranded sense oligonucleotide specifying the amplicon as previously described by Bustin et al .26 and this together with the known RNA concentration was used to quantify the mRNA copy number per microgram of RNA for each gene. An example of a standard curve (that for hTERT) is shown in Figure 1.
Figure 1.
Standard curve for hTERT(Ct value = threshold cycle of RT-PCR at which fluorescence detectable)
Seventy-four specimens of breast RNA (36 from ANCT and 38 from tumour) were analysed using primers and probe for the VEGF-189 isoform, in 70 of these specimens hTERT mRNA levels were quantified. The VEGF-165 isoform was quantified in 39 of the specimens that had been analysed for VEGF-189, of which 37 had undergone hTERT mRNA measurement. Quantitative RT-PCR was carried out for PCNA mRNA in 78 breast specimens (40 ANCT, 38 tumour) that had had hTERT mRNA measured.
Statistical analysis was carried out using WinStat for Excel (R. Fitch Software, Staufen, Germany) and Stata 5.0 (Stata Corp, College Station, Texas).
Results and Statistical Analysis
All specimens had detectable mRNA for PCNA and both isoforms of VEGF. For PCNA expression ranged from 2.62 × 105 to 1.43 × 107 (mean 2.59 × 106) in ANCT and from 4.65 × 105 to 4.2 × 107 (mean 7.65 × 106) in tumour tissue. The ratio of PCNA mRNA expression in tumour to adjacent tissue ranged from 0.09 to 22.93 (mean 4.97) and PCNA was present in greater amounts in 25 out of 31 (80%) matched tissue specimens. Using the paired t test, the increased expression of PCNA in tumours compared to adjacent tissue was statistically significant (p = 0.02).
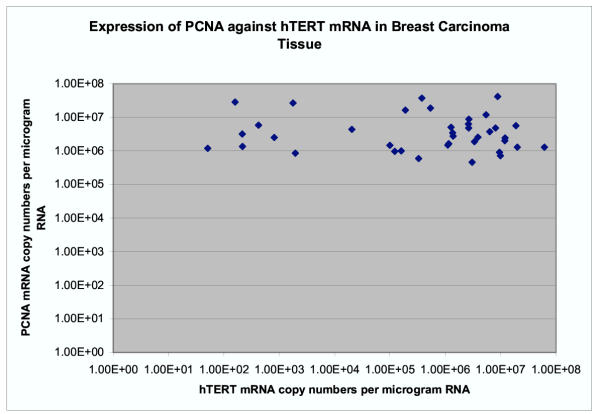
In 16 (51%) specimens, PCNA was higher by more than a factor of 2, in 12 (39%) by more than a factor of 3 and in 7 (23%) by more than a factor of 10. We were unable to detect any relationship between over-expression of hTERT in tumours (ratio of mRNA in tumour to ANCT > 1 × 104) and PCNA over-expression (ratio of mRNA in tumour to ANCT >2 or >3) using the χ2 test (p > 0.1). Using the product moment correlation coefficient on the log-transformed data no significant relationship was found between PCNA and hTERT mRNA levels in tumour (Figure 2) (correlationcoefficient-0.324, p = 0.3), or in adjacent non-cancerous tissue (correlation coefficient -0.199, p = 0.1).
Figure 2.
Shows no significant relationship between PCNA and hTERT mRNA levels in tumour samples.
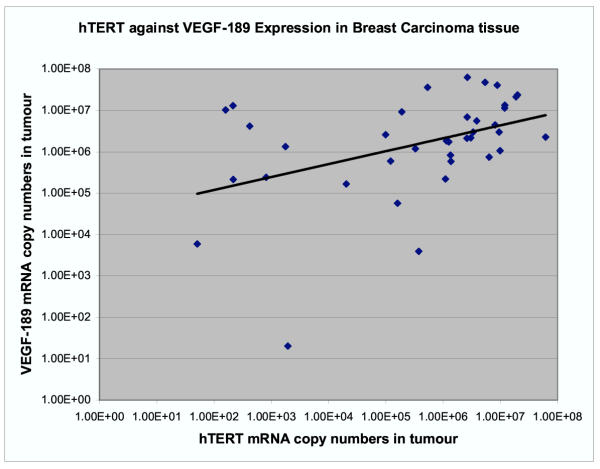
The expression of VEGF-189 ranged between 7.79 × 103 and 2.9 × 107 (mean 4.55 × 106) in ANCT and between 2.02 × 101 and 6.63 × 107 (mean 8.85 × 106) in tumour tissue. In the matched specimens, (n = 31) the VEGF-189 expression in tumour compared to ANCT was higher in 16 (52%) by a factor of between 1.5 and 63 although this was not significantly different using the paired t test (p = 0.12). We used the product moment correlation coefficient on the log-transformed data and found a significant positive correlation between hTERT and VEGF-189 mRNA levels in tumour tissue (coefficient 0.423, p = 0.008) but not in ANCT (Figure 3).
Figure 3.
There is a significant positive correlation between hTERT and VEGF-189 mRNA levels in tumour tissue (coefficient 0.423, p = 0.008).
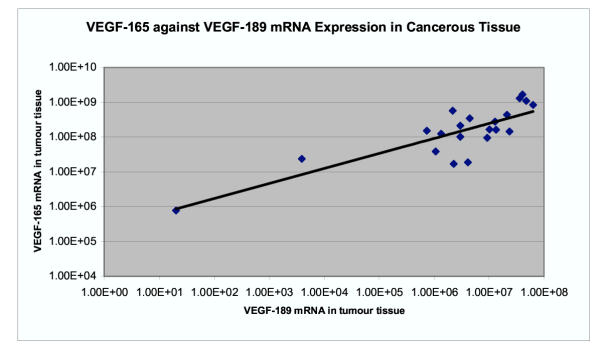
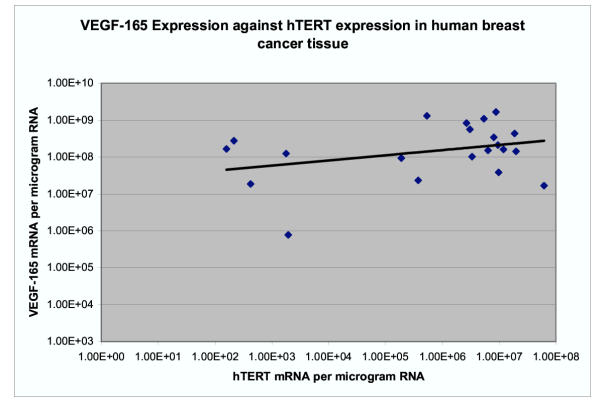
The expression of VEGF-165 mRNA in ANCT was between 1.68 × 107 and 2.60 × 108 copy numbers per μg of RNA (mean 7.2 × 107) and in tumour was between 7.7 × 105 and 1.67 × 109 (mean 3.57 × 108). The expression of VEGF-165 was significantly higher in tumour tissue than in adjacent tissue using the paired t test (p = 0.02). There was a significant correlation between the level of 165 isoform and the 189 isoform both in ANCT (coefficient 0.67, p = 0.001) and in tumour tissue (coefficient 0.83, p = 1.98 × 10-6) (Figure 4). The expression of VEGF165 mRNA positively correlated with that of hTERT in tumours but this just failed to reach statistical significance (coefficient 0.33, p = 0.07) (Figure 5). There was no correlation in ANCT (coefficient 0.02, p = 0.46). In fifteen specimens where both isoforms of VEGF were analysed in both cancerous and adjacent tissue, the ratio of VEGF165 to 189 mRNA expressions was examined. The ratio of isoform 165/189 expressions ranged between 4.00 and 46.59 (mean 15) in ANCT and between 7.44 and 38,421 (mean 3002) in cancerous tissue. In 80% (n = 12), the ratio of 165/189 isoform expressions was higher in tumours than in the adjacent tissue by a factor of between 1.3 and 9596 (mean 738). As the results were highly skewed, these data were log transformed before analysis with the paired t test and this showed a significant difference between the ratio of 165/189 isoform expression in tumours and adjacent tissue (p = 0.025).
Figure 4.
There is a significant correlation between the level of 165 isoform and the189 isoform both in tumour tissue (coefficient 0.83, p = 1.98 × 10-6).
Figure 5.
The expression of VEGF165 mRNA positively correlated with that of hTERT in tumours but this just failed to reach statistical significance (coefficient 0.33, p = 0.07).
Discussion
We found no statistically significant relationship between PCNA and hTERT over-expression in breast tumour tissue, or between absolute levels of these two mRNAs in tumour or adjacent non-cancerous tissue. To our knowledge, only one previous study has investigated hTERT and PCNA expression in malignancy, that of Nakanishi et al who also found no relationship between the two in urothelial cancers using in situ hybridisation and immunohistochemistry[27]. The correlation between PCNA and telomerase activity has been investigated in gastric carcinoma [28] and cervical intraepithelial neoplasia [29], and again no relationship found. These findings suggest that the association between hTERT expression and worsened prognosis that has been reported is not due to increased PCNA expression.
We have however found a significant correlation between the VEGF-189 isoform and hTERT mRNA expressions (p = 0.008) and a trend towards significant correlation between VEGF165 and hTERT mRNA expressions (p = 0.07) in breast cancer tissue, suggesting a possible pathway through which high expression of hTERT could be linked to worsened disease prognosis. Since VEGF expression is known to increase the likelihood of tumour metastasis .17, it might be expected that high levels of hTERT would therefore also be correlated with increased lymph node positivity. Although we did not detect such an association in our sample [14] and neither did Bieche et al [12], such a connection has been reported with telomerase and nodal status in breast cancer [30,31]. It may be that any relationship that does exist is masked in studies measuring mRNA by the presence of splice variants with different physiological activity, however it is surprising therefore that the correlation between VEGF-189 and hTERT mRNA is still apparent. The activities of the splice variants of VEGF-A, like those of hTERT, are currently poorly understood. The 121,165 and 189 isoforms are all reported to be present in breast tumour tissue with the VEGF165 isoform being the most important angiogenic factor [32,33]. The 165 variant has been found to be an independent negative prognostic indicator in breast cancer patients [32-35]. The VEGF189 isoform has been shown to be more strongly bound to the cell membrane than other isoforms and therefore has a high potential in local angigenesis and tumour growth in the cancer inductive microenvironment [36]. Furthermore, the VEGF189 mRNA has been reported to show a greater correlation with tumour angiogenesis and clinical outcome than other isoforms (121,165 and 206) in some epithelial cancers [37].
We have found both the overall expression of the 165 isoform and the ratio of VEGF-165 to 189 mRNA to be significantly increased in tumour tissue compared to adjacent tissue. This suggests that the 165 isoform may be important in breast carcinogenesis, although previously it has been reported that the proportions of each of the VEGF-A isoforms remains the same in breast carcinoma tissue as in benign tissue [15].
We have also detected high levels of both isoforms of VEGF in adjacent non-cancerous tissue in our study. This is in keeping with other studies where VEGF 189,121 and 165 isoforms have been found in normal tissues .38 and increased levels of VEGF have been reported in benign breast tissue from pre-menopausal women compared to post-menopausal women, suggesting hormonal regulation [15].
Further work in this area is required to corroborate this apparent relationship between VEGF-189 and hTERT, ideally using primers designed to amplify only the functional variants of hTERT mRNA. In addition, studies aimed at illustrating the physiological significance of the VEGF isoforms will be invaluable in clarifying the relevance of altered isoform ratios between benign and malignant tissues.
Contributor Information
Katharine L Kirkpatrick, Email: KAFFDABE@aol.com.
Robert F Newbold, Email: robert.newbold@brunel.ac.uk.
Kefah Mokbel, Email: kefahmokbel@hotmail.com.
References
- Mokbel K. The Role of Telomerase in Breast Cancer. European Journal of Surgical Oncology. 2000;26:509–14. doi: 10.1053/ejso.1999.0932. [DOI] [PubMed] [Google Scholar]
- Zhu J, Wang H, Bishop J, Blackburn E. Telomerase Extends the Lifespan of Virus-Transformed Human Cells Without Net Telomere Lengthening. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3723–8. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick K, Mokbel K. The Significance of Human Telomerase Reverse Transcriptase in Human Cancer. European Journal of Surgical Oncology. 2001;27:754–60. doi: 10.1053/ejso.2001.1151. [DOI] [PubMed] [Google Scholar]
- Liu K, Schoonmaker MM, Levine BL, June CH, Hodes RJ, Weng NP. Constitutive and Regulated Expression of Telomerase Reverse Transcriptase (HTERT) in Human Lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5147–52. doi: 10.1073/pnas.96.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Ichihashi M, Ueda M. Correlation of the Expression of Human Telomerase Subunits With Telomerase Activity in Normal Skin and Skin Tumors. Cancer. 1999;86:2038–44. doi: 10.1002/(SICI)1097-0142(19991115)86:10<2038::AID-CNCR22>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Kanaya T, Kyo S, Takakura M, Ito H, Namiki M, Inoue M. HTERT Is a Critical Determinant of Telomerase Activity in Renal-Cell Carcinoma. International Journal of Cancer. 1998;78:539–43. doi: 10.1002/(SICI)1097-0215(19981123)78:5<539::AID-IJC2>3.3.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bodnar A, Ouellette M, Frolkis M, Holt S, Chiu C, Morin G, Harley C, Shay J, Lichtsteiner S, Wright W. Extension of Life-Span by Introduction of Telomerase into Normal Human Cells. Science. 1998;279:349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick KL, Clark G, Ghilchick M, Newbold RF, Mokbel K. hTERT mRNA expression correlates with telomerose activity in human breast cancer. Eur J Surg Oncol. 2003;29:321–326. doi: 10.1053/ejso.2002.1374. [DOI] [PubMed] [Google Scholar]
- Dome JS, Chung S, Bergemann T, Umbricht CB, Saji M, Carey LA, Grundy PE, Perlman EJ, Breslow NE, Sukumar S. High Telomerase Reverse Transcriptase (HTERT) Messenger RNA Level Correlates With Tumor Recurrence in Patients With Favorable Histology Wilms' Tumor. Cancer Research. 1999;59:4301–7. [PubMed] [Google Scholar]
- Xu D, Gruber A, Peterson C, Pisa P. Telomerase Activity and the Expression of Telomerase Components in Acute Myelogenous Leukaemia. British Journal of Haematology. 1998;102:1367–75. doi: 10.1046/j.1365-2141.1998.00969.x. [DOI] [PubMed] [Google Scholar]
- Komiya T, Kawase I, Nitta T, Yasumitsu T, Kikui M, Fukuoka M, Nakagawa K, Hirashima T. Prognostic Significance of HTERT Expression in Non-Small Cell Lung Cancer. International Journal of Oncology. 2000;16:1173–7. doi: 10.3892/ijo.16.6.1173. [DOI] [PubMed] [Google Scholar]
- Bieche I, Nogues C, Paradis V, Olivi M, Bedossa P, Lidereau R, Vidaud M. Quantitation of HTERT Gene Expression in Sporadic Breast Tumors With a Real-Time Reverse Transcription-Polymerase Chain Reaction Assay. Clinical Cancer Research. 2000;6:452–9. [PubMed] [Google Scholar]
- Kirkpatrick KL. The Significantly of Human Telomerase Reverse Transcuptase (hTERT) in Cancer. European Journal of Surgical Oncology. 2001;27:776–83. doi: 10.1053/ejso.2001.1151. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick K, Ongunkulade W, Elkak A, Bustin SA, Jenkins P, Ghilchik M, Newbold RF, Mokbel K. HTERT Expression in Human Breast Cancer and Adjacent Non-Cancerous Tissue: Correlation With Tumour Stage and C-Myc Expression. Breast Cancer Res Treat. 2003;77:277–84. doi: 10.1023/A:1021849217054. [DOI] [PubMed] [Google Scholar]
- Greb R, Wallweiner D, Kiesel L. Vascular Endothelial Growth Factor A (VEGF-A) MRNA Expression Levels Decrease After Menopause in Normal Breast Tissue but Not in Breast Cancer Lesions. British Journal of Cancer. 1999;81:225–31. doi: 10.1038/sj.bjc.6690681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola J, Valve E, Karkkainen M, Joukov V, Alitalo K, Harkonen P. Vascular Endothelial Growth Factors Are Differentially Regulated by Steroid Hormones and Antiestrogens in Breast Cancer Cells. Molecular and Cellular Endocrinology. 1999;149:29–40. doi: 10.1016/S0303-7207(99)00003-9. [DOI] [PubMed] [Google Scholar]
- Scott PA, Smith K, Poulsom R, De Benedetti A, Bicknell R, Harris AL. Differential Expression of Vascular Endothelial Growth Factor MRNA Vs Protein Isoform Expression in Human Breast Cancer and Relationship to EIF-4E. British Journal of Cancer. 1998;77:2120–8. doi: 10.1038/bjc.1998.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Berse B, Jackman R, Tognazzi K, Guidi A, Dvorak H, Senger D, Connolly J, Schnitt S. Expression of Vascular Permeability Factor (Vascular Endothelial Growth Factor) and Its Receptors in Breast Cancer. Human Pathology. 1995;26:86–91. doi: 10.1016/0046-8177(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Toi M, Hoshina T, Takayanagi T, Tominaga T. Association of Vascular Endothelial Growth Factor Expression With Tumor Angiogenesis and With Early Relapse in Primary Breast Cancer. Japanese Journal of Cancer Research. 1994;85:1045–9. doi: 10.1111/j.1349-7006.1994.tb02904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini G. Prognostic Value of Vascular Endothelial Growth Factor in Breast Cancer. Oncologist. 2000;5:Suppl-44. doi: 10.1634/theoncologist.5-suppl_1-37. [DOI] [PubMed] [Google Scholar]
- Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, Abraham JA. The Human Gene for Vascular Endothelial Growth Factor. Multiple Protein Forms Are Encoded Through Alternative Exon Splicing. Journal of Biological Chemistry. 1991;266:11947–54. [PubMed] [Google Scholar]
- Robinson C, Stringer S. The Splice Variants of Vascular Endothelial Growth Factor (VEGF) and Their Receptors. Journal of Cell Science. 2000;114:853–65. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- Cardillo MR, Stamp GW, Pignatelli M. Proliferating Cell Nuclear Antigen in Benign Breast Diseases. European Journal of Gynaecological Oncology. 1995;16:476–81. [PubMed] [Google Scholar]
- Sheen-Chen S-M, Eng H-L, Chou F-F, Chen W-J. The Prognostic Significance of Proliferating Cell Nuclear Antigen in Patients With Lymph Node-Positive Breast Cancer. Archives of Surgery. 1997;132:264–7. doi: 10.1001/archsurg.1997.01430270050010. [DOI] [PubMed] [Google Scholar]
- Vidal H, Auboeuf D, DeVos P, Staels B, Riou J, Auwerx J, Laville M. Quantitation of Leptin MRNA in Human Adipose Tissue by RT-Competitive PCR. Qiagen News. 1997;2 [Google Scholar]
- Bustin SA, Gyselman VG, Williams NS, Dorudi S. Detection of Cytokeratins 19/20 and Guanylyl Cyclase C in Peripheral Blood of Colorectal Cancer Patients. British Journal of Cancer. 1999;79:1813–20. doi: 10.1038/sj.bjc.6690289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Hiroi S, Kawai T, Aida S, Kasamatsu H, Aurues T, Ikeda T. Expression of Telomerase Catalytic Subunit (HTERT) MRNA Does Not Predict Survival in Patients With Transitional Cell Carcinoma of the Upper Urinary Tract. Modern Pathology. 2001;14:1073–8. doi: 10.1038/modpathol.3880439. [DOI] [PubMed] [Google Scholar]
- Okusa Y, Shinomiya N, Ichikura T, Mochizuki H. Correlation Between Telomerase Activity and DNA Ploidy in Gastric Cancer. Oncology. 1998;55:258–64. doi: 10.1159/000011860. [DOI] [PubMed] [Google Scholar]
- Herbsleb M, Knudsen U, Orntoft T, Bichel P, Norrild B, Knudsen A, Mogensen O. Telomerase Activity, MIB-1, PCNA, HPV 16 and P53 As Diagnostic Markers for Cervical Intraepithelial Neoplasia. APMIS. 2001;109:607–17. doi: 10.1034/j.1600-0463.2001.d01-182.x. [DOI] [PubMed] [Google Scholar]
- Mokbel K, Parris CN, Ghilchik M, Williams G, Newbold RF. The Association Between Telomerase, Histopathological Parameters, and KI-67 Expression in Breast Cancer. American Journal of Surgery. 1999;178:69–72. doi: 10.1016/S0002-9610(99)00128-2. [DOI] [PubMed] [Google Scholar]
- Hiyama E, Gollahon L, Kataoka T, Kuroi K, Yokoyama T, Gazdar AF, Hiyama K, Piatyszek MA, Shay JW. Telomerase Activity in Human Breast Tumors. Journal of the National Cancer Institute. 1996;88:116–22. doi: 10.1093/jnci/88.2.116. [DOI] [PubMed] [Google Scholar]
- Linderholm B, Lindahl T, Holmberg L, Klaar S, Lennerstrand J, Henriksson R, Bergh J. The Expression of Vascular Endothelial Growth Factor Correlates With Mutant P53 and Poor Prognosis in Human Breast Cancer. Cancer Research. 2001;61:2256–60. [PubMed] [Google Scholar]
- Linderholm B, Tavelin B, Grankvist K, Henriksson R. Does Vascular Endothelial Growth Factor (VEGF) Predict Local Relapse and Survival in Radiotherapy-Treated Node-Negative Breast Cancer? British Journal of Cancer. 1999;81:727–32. doi: 10.1038/sj.bjc.6690755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderholm B, Lindh B, Tavelin B, Grankvist K, Henriksson R. P53 and Vascular Endothelial Growth Factor (VEGF) Expression Predicts Outcome in 833 Patients With Primary Breast Carcinoma. International Journal of Cancer. 2000;89:51–62. doi: 10.1002/(SICI)1097-0215(20000120)89:1<51::AID-IJC9>3.3.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Linderholm B, Tavelin B, Grankvist K, Henriksson R. Vascular Endothelial Growth Factor Is of High Prognostic Value in Node-Negative Breast Carcinoma. Journal of Clinical Oncology. 1998;16:3121–8. doi: 10.1200/JCO.1998.16.9.3121. [DOI] [PubMed] [Google Scholar]
- Tomii Y, Yamazaki H, Sawa N. Unique Properties of 189 Amino Acid Isoform of Vascular Endothelial Growth Factor in Tumourigenesis. International Journal of Oncology. 2002;21:1251–7. [PubMed] [Google Scholar]
- Yuan A, Yu C, Kuo S-H, Chen W-J, Lin F-Y, Luh K-T, Yang P-C, Lee Y-C. Vascular Endothelial Growth Factor 189 MRNA Isoform Expression Specifically Correlates With Tumor Angiogenesis, Patient Survival and Postoperative Relapse in Non-Small Cell Lung Cancer. Journal of Clinical Oncology. 2001;19:432–41. doi: 10.1200/JCO.2001.19.2.432. [DOI] [PubMed] [Google Scholar]
- Zhang H-T, Scott PA, Morbidelli L, Peak S, Moore J, Turley H, Harris AL, Ziche M, Bicknell R. The 121 Amino Acid Isoform of Vascular Endothelial Growth Factor Is More Strongly Tumorigenic Than Other Splice Variants in Vivo. British Journal of Cancer. 2000;83:63–8. doi: 10.1054/bjoc.2000.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]