Abstract
The N-terminus of Huntingtin, the protein encoded by the Huntington’s disease gene, contains a stretch of polyglutamine residues that is expanded in Huntington’s disease. The polyglutamine stretch is flanked by two conserved protein domains in vertebrates: an N1-17 domain, and a proline-rich region (PRR). The PRR can modulate the structure of the adjacent polyglutamine stretch, and is a binding site for several interacting proteins. To determine the role of the PRR in Huntingtin function, we have generated a knock-in allele of the mouse Huntington’s disease gene homolog that expresses full-length normal huntingtin lacking the PRR. Mice that are homozygous for the huntingtin PRR deletion are born at the normal Mendelian frequency, suggesting that the PRR is not required for essential huntingtin functions during embryonic development. Moreover, adult homozygous mutants did not exhibit any significant differences from wild-type controls in general motor function and motor learning. However, 18 month-old male, but not female, homozygous PRR deletion mutants exhibited deficits in the Morris water task, suggesting that age-dependent spatial learning and memory may be affected in a sex-specific fashion by the huntingtin PRR deletion.
Keywords: Huntington’s disease, huntingtin, proline-rich region, mouse model
Introduction
Huntingtin (htt), the protein product of the HTT gene, is a large (~350 kD), predominantly cytoplasmic protein with limited homology to other proteins. The htt polyglutamine (polyQ) stretch, which when expanded to >39Q causes Huntington’s disease (HD), is located near the N-terminus, and is flanked by two protein motifs that are conserved in vertebrates [1–5]: N1-17, an amino terminal domain that is a target for a number of post-translational modifications and is involved in htt’s association with membranes [6–10], and a proline-rich region (PRR) that is a potential binding site for many htt-interacting proteins [11]. In human htt, the 38 amino acid PRR consists of a stretch of 11 prolines that is separated from a stretch of 10 prolines by a 17 amino acid region containing 7 scattered proline residues [12]. The mouse htt PRR consists of 25 prolines in a 32 amino acid domain with stretches of 3, 10, 1, and 7 prolines interrupted by 1-3 amino acid stretches of glutamine [13, 14].
The polyQ stretch has been the focus of intense research, and is an obvious therapeutic target. However, a better understanding of the role of the polyQ flanking sequences in htt function could provide valuable information on how these sequences modulate normal and pathogenic htt function. PRRs in many proteins are generally exposed and located at either the N- or C-terminus, where they have the potential to form extended structures and flexible regions [15, 16]. They have been described as “sticky arms” that can rapidly and reversibly bind to other proteins. Typically, PRRs participate in processes that require the rapid recruitment or interchange of groups of interacting proteins, such as in transcription initiation, cytoskeletal rearrangements, and in signaling. PRRs can also function as protease cleavage sites, and as structural elements that separate one functional domain from another.
In vitro experiments and structural analysis of the htt N-terminus have suggested that the PRR might also have arisen during evolution as a defense against mutant htt aggregation and toxicity, either directly by affecting the structure of the N-terminus [17–20], or indirectly by its ability to bind interacting proteins [11, 21]. The htt PRR, for example, binds to WW domain- and Src homology 3 (SH3)-containing proteins [22–28]. WW domains are present in diverse signaling and structural proteins involved in non-receptor signaling, channel function, protein processing, and pre-mRNA splicing [29–31]. SH3 motifs are associated with catalytic domains in enzymes, structural proteins, and small adaptor proteins [30, 31]. Proteins with SH3 motifs can also function in signal transduction, and participate in vacuole sorting and receptor mediated endocytosis. Examples of proteins that can associate with htt through its PRR include: GAPDH, Grb2, HYP-A, HYP-C, IKKγ, MLK2, p53, PACSIN1, PSD-95, RasGAP, and SH3GL3 [11]. For many of these interacting proteins, the size of htt’s polyQ stretch can influence the strength of their interaction with the PRR domain. It has been hypothesized that the association of several of these proteins with mutant htt’s PRR could be responsible for mediating the resistance of many HD mouse models to excitotoxicity [21]. The IκB kinase complex (IKK) can interact with htt through its IKKγ regulatory subunit and can mediate phosphorylation of htt at S13 and S16, two critical posttranslational modifications within htt’s N1-17 domain that modulate pathogenesis and turnover of mutant htt [10, 32, 33]. Additional observations suggest that the htt PRR may serve as an aggresome-targeting signal, promoting the transport of small aggregates of mutant htt to the centrosomally located aggresome in mammalian and yeast cells [34]. Although a number of studies have contributed to our understanding of the role of the PRR in htt’s interaction with protein partners, and in modulating the toxicity of mutant htt N-terminal fragments, little is known about the contribution of the PRR to normal full-length htt function in vivo.
To determine the in vivo role of the mouse htt PRR in normal htt function, we have generated a knock-in mouse allele that expresses a version of the mouse homolog of the HD gene (Hdh) containing a deletion of the PRR encoded within exon-1 (HdhΔPRR). Mice homozygous for the PRR deletion (HdhΔPRR/ΔPRR) were obtained at the predicted Mendelian frequency, suggesting that the PRR deletion does not affect critical htt functions during development. Moreover, no significant differences were found between HdhΔPRR/ΔPRR mice and wild-type controls in general motor function, motor coordination and balance, and motor learning. However, 18 month-old male, but not female, HdhΔPRR/ΔPRR mice exhibited deficits in the Morris water task, suggesting that age-dependent spatial learning and memory may be affected by the htt PRR deletion in a sex-specific fashion.
Materials and Methods
All experiments with mice were carried out in accordance with the ethical guidelines described in the “Guide for the Care and Use of Laboratory Animals”, Institute of Laboratory Animal Resources, National Research Council, 1996 edition. All procedures were reviewed and approved by the Animal Care and Use Committee of the University of Virginia.
Generation of HdhΔPRR mice
A synthetic Hdh exon-1/intron-1 XmnI-KpnI restriction fragment containing the exon-1 7Q stretch and PRR deletion, 3´terminus of exon-1, and a portion of intron-1 containing a 100bp deletion was generated by annealing two HdhΔPRR exon-1 oligonucleotides (ΔPRR-1: 5´-CTGATGAAGGCTTTCGAGTCGCTCAAGTCGTTTCAGCAGCAACAGCAGCAGCAGCTGCCAGGTCCGGCAGAGGAA-3´ and ΔPRR-2: 5´-AAGGGAGGTACCGGACTCACGGTCGGTGCAGCGGTTCCTCTGCCGGACCTGGCAGCT-3´), that have 23 complementary nucleotides at their 3´ends, and generating a complete duplex fragment by extension with DNA Polymerase I (Klenow fragment). After digestion with the restriction enzymes XmnI and KpnI, the synthetic HdhΔPRR fragment was used to replace the wild-type Hdh exon-1 XmnI-KpnI restriction fragment.
A gene-targeting vector was assembled as described in [35]. ES cell transfection, selection, and genotyping were performed using standard procedures. The presence of the PRR deletion was confirmed by PCR (5´GCGTAGTGCCAGTAGGCTCCAAG-3´; 3´CCTCGTCTTGCGGGGTCT -5´), and five independent ES cell clones were injected into C57BL/6J host blastocysts to generate chimeric mice. Germline-transmitting male chimeras were obtained from ES clones 41 and 58, and DNA from tail biopsies was used to genotype progeny by PCR (5´GCTGCACCGACCGTGAGTCC-3´; 5´GGCCTGACCCGGCTCTGTCTC-3´). Once germline transmission was confirmed, sequencing of the DNA from tail biopsies was also performed to ensure that the desired sequence was present.
Motor and behavioral analyses
All motor and behavioral analyses were performed blind to genotype. Testing was conducted at the same time each day during the light phase of the diurnal cycle. Mice were habituated to the testing room for 30 minutes prior to behavioral analyses. The following testing sequence was used on separate cohorts of HdhΔPRR/ΔPRR mice at 6, 12 and 18 months of age: elevated plus-maze, open field, accelerating rotarod, observational and neurological reflex screen, and Morris water maze. n=7–12 mice per genotype and sex were used (with the exception of the 12 month-old male mice, where n=5), and each group contained mice derived from three to five litters.
Observational and neurological reflex screening
Body weight, general appearance, gait, limb clasping, and reflexes were assessed.
Forelimb grip strength
Forelimb grip strength was measured using a digital force gauge (San Diego Instruments) that measures the peak amount of force the mouse applies when grasping a pull bar assembly. Three trials per mouse were performed, and then averaged.
Visual acuity
The visual placing test was used to assess visual acuity. This test involved suspending the mouse by the base of its tail, approximately 15 cm above a wire grid, and noting the point of forelimb extension as it was lowered towards the grid. A mouse with good vision will extend its forepaws towards the approaching surface before its whiskers or nose touch the grid.
Neurological reflexes
Each mouse was evaluated for the presence or absence of neurological reflexes. The eye blink reflex was tested by lightly touching the eye with the tip of a clean cotton swab. The ear twitch reflex was tested by touching the ear with the tip of a clean cotton swap. The whisker twitch reflex was tested by lightly brushing the whiskers with a small brush. The flexion reflex was tested by lightly pinching the toes with forceps and noting the rapidity of foot withdrawal. The righting reflex was evaluated by turning the mouse onto its back and noting its ability to right itself onto all fours. The postural reflex involved the extension of all four limbs, and the mouse’s ability to maintain an upright, balanced position while in an empty cage that was being shaken from side to side.
Elevated plus-maze
The elevated plus-maze (Med Associates) has two open arms (35 × 6 cm2) and two closed arms (35 × 6 × 20 cm3) that extended from a common center square (6 × 6 cm2), and is elevated 72 cm above the floor. A small raised lip (0.6 cm) around the edges of the open arms helps to prevent the mice from falling off the maze. Each mouse was initially placed on the center square facing an open arm, and allowed to freely explore the maze for 5 minutes. The test session was recorded by a digital video camera mounted approximately 20 cm above the center square. Each 5 minute test session was scored in a blinded fashion following the conclusion of the experiment. Time spent in the open arms, number of open arm entries, and number of closed arm entries were scored. An open or closed arm entry was defined as when all four paws completely crossed the line demarking the center of the plus. Open and closed arm entries were combined to give a measure of general exploratory activity.
Open-field testing
The VersaMax Animal Activity Monitoring System (AccuScan Instruments) was used for open-field testing. Each mouse was initially placed in the bottom left corner of the arena, and monitored for 5 minutes. Animal activity levels, including horizontal activity, vertical activity, total distance traveled, and time spent in the center were analyzed using the VersaMax software.
Accelerating rotarod
Motor coordination and balance were assessed on an Economex accelerating rotarod (Columbus Instruments) that has the capacity to test four mice simultaneously. The testing procedure consisted of two training phases and a testing phase: stationary training on a non-rotating rod, constant-speed training on a rod rotating at a speed of 2.0 rpm, and three consecutive days of testing on an accelerating rotarod (acceleration = 0.1 rpm/sec). Latency to fall from the rotarod was recorded.
Morris water maze
The test was conducted over eight consecutive days, and included four consecutive training days on the hidden platform task, followed by a probe trial on day five, the reversal task on days six and seven, and the visible platform task on day eight. A standard circular pool, with a diameter of 123 cm was used. The water (23 ± 1°C) was whitened with non-toxic white tempera paint, and highly visible room cues were placed on the walls around the pool. The maze was digitally divided into four equal quadrants with a target area corresponding to the size of the platform, and swim paths were video tracked and analyzed using EthoVision XT 5.0 software (Noldus Information Technology). The water was changed following the probe trial.
Hidden platform task
The hidden platform, 10 cm in diameter and composed of clear Plexiglas, was submerged 1 cm below the surface of the water in the middle of one of the four quadrants. For each trial, the start position of the mouse was randomly assigned to one of four start positions in the quadrant opposite the target quadrant. Mice were placed into the maze at the edge of the pool, facing the wall. During the training trials, each mouse was given 60s to reach the hidden platform, climb up out of the water and remain on the platform. After 30s, the mouse was taken off the platform, dried with a paper towel and placed back into its cage. The trial was terminated after 60s. A value of 60s was recorded as escape latency for any mouse that did not reach the platform within the trial period. Mice that failed to reach the platform were either guided to or placed on the platform, and then removed after 30s. Mice were trained for four trials per day, with an average inter-trial interval of 5 minutes, on four consecutive days.
Probe trial
On day five, following training on the hidden platform task, the platform was removed from the pool. Mice were placed into the maze as usual, and allowed to swim for 60s. Time spent in each quadrant, number of entries into each quadrant, number of target crossings, latency to first target entry, path length to first target entry, average distance from the center of the platform location, total swim distance and swim speed were acquired using EthoVision XT 5.0 software (Noldus Information Technology).
Reversal task
On day five and six, the platform was placed back into the pool in the opposite quadrant, and the same protocol was followed as for the hidden platform task.
Visible platform task
A glow stick was mounted onto the platform such that it extended above the surface of the water for easy visibility. The platform was then placed in the middle of a third quadrant. Mice were placed into the maze as usual, and removed as soon as they climbed up out of the water and onto the platform. The escape latencies were recorded over a total of four trials.
Immunohistochemistry
Mice were deeply anaesthetized using isoflurane and euthanized by cervical dislocation. The brains were harvested and flash frozen in 2-methylbutane on dry ice. Frozen mouse brains were sectioned at 20µm using a cryostat (Bright Instruments), and stored at −80°C until use. Sections were thawed from storage at −80°C, washed briefly in 1X PBS, fixed for 15 minutes in 4% paraformaldehyde in 0.1M phosphate buffer pH 7.4, and washed three times for 5 minutes in 1X PBS. Sections were blocked in blocking solution (5% donkey serum, 0.1% Triton X100, 1X PBS) for 30 minutes, and incubated overnight at 4°C with primary antibody diluted in blocking solution (anti-htt, 1:250, BML-PW0595, Enzo Life Sciences). Following the primary antibody incubation, sections were washed in 1% donkey serum, 0.1% Triton X100, 1X PBS three times for 10 minutes, then incubated in secondary antibody (donkey anti-rabbit FITC, 711-095-152, Jackson ImmunoResearch Laboratories) and To-Pro-3 iodide (1:10000, T3605, Invitrogen) in the same washing solution for one hour. Sections were washed three times for 5 minutes in 1X PBS before adding the Autofluorescence Eliminator Reagent according to the manufacturer’s instructions (2160, Millipore). Finally, sections were mounted with Vectashield mounting medium (H-1000, Vector Laboratories), and examined using an Olympus BX51 microscope equipped with a MagnaFire CCD camera. (n = 3 for each genotype).
Immunocytochemistry
Primary mouse embryonic fibroblasts (PMEFS) were prepared from embryonic day 13.5 (E13.5) embryos, and cultured in vitro using standard procedures. PMEFs (at passage 3) were seeded at a concentration of 1 × 105 cells/well onto 24-well culture plates containing 12mm gelatinized coverslips. The following day, cells were incubated with MitoTracker Red CMXRos (100µm/mL, M-7512, Invitrogen) for 30 minutes at 37°C, washed twice in 1X PBS, fixed for 15 minutes in 4% paraformaldehyde in 0.1M phosphate buffer pH 7.4, and washed three times for 5 minutes in 1X PBS. Cells were then permeabilized in 0.25% Triton X100, 1X PBS for 10 minutes, washed three times for 5 minutes in 1X PBS, blocked in 5% donkey serum, 0.1% Triton X100, 1X PBS for 30 minutes, then incubated overnight at 4°C with primary antibody diluted in the same blocking solution. Secondary antibodies used were donkey anti-mouse or rabbit –Cy3 or –FITC (Jackson ImmunoResearch Laboratories), and nuclei were stained with To-Pro-3 iodide (1:10000, T3605, Invitrogen). Cells were mounted onto microscope slides using Vectashield mounting medium (H-1000, Vector Laboratories), and examined using a Nikon C-1 confocal microscope. Three htt primary antibodies were used: BML-PW0595 (1:100, Enzo Life Sciences), MAB2166 (1:100, Millipore), and RabMAb (1:100, Epitomics). Three independent experiments for each antibody were performed.
Western blotting
Mouse brains were homogenized in lysis buffer: 50mM Tris pH 8.8, 100mM NaCl, 5mM MgCl2, 1mM EDTA pH 8, 0.5% NP-40, 1mM NaF, and Halt™ Protease Inhibitor Cocktail (78430, Thermo Fisher Scientific), and centrifuged for 10 minutes at 4°C (16,100×g). Western blotting was performed as described in [35]. For densitometry, films in the linear exposure range were scanned on a flatbed scanner, and analyzed using the ImageJ program (Rasband, W.S., ImageJ, U.S. National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2005). Levels of protein in each sample were normalized to β-actin. Primary antibodies used were: anti-htt (1:3000, MAB2166, Millipore), PSD95 (1:1000, #2507, Cell Signaling), CREB (1:1000, #9197, Cell Signaling), anti-htt S13-P (1:1000, gift from L. Thompson), tubulin (1:1000, T6793, Sigma), and β-actin (1:1000, A4700, Sigma). In some cases, the blots were stripped (Restore™ Western Blot Stripping Buffer, 21059, Thermo Fisher Scientific) for 30 minutes, and re-probed with a new primary antibody. (n = 3 for each genotype).
Subcellular fractionation
PMEFs (P3) were plated onto 10 cm culture dishes, and grown until confluent. Nuclear and cytoplasmic protein fractions were obtained using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, 78833), and analyzed by western blotting, as described above.
Statistical analyses
Data were analyzed using the Student’s t-test and two way repeated measures ANOVA with the Holm-Sidak posthoc test. All statistical analyses used SigmaStat 3.5 software (Systat Software). Significance was accepted at P < 0.05.
Results
To generate the PRR deletion in the Hdh locus, we assembled a gene targeting construct by replacing an endogenous Hdh exon-1 XmnI–KpnI restriction fragment encoding the htt 7Q stretch and PRR with an oligonucleotide-generated synthetic fragment containing the 7Q stretch without the PRR (Fig. 1 and Methods). Germline transmission was obtained from two independent targeted ES cell clones, and mice were backcrossed to the C57BL6/J strain for at least six generations prior to characterization.
Figure 1. Generation of the HdhΔPRR allele.
(A) Schematic of the wild-type Hdh allele surrounding the first exon is shown (Hdh) along with the gene targeting construct (T) lacking the PRR, and the targeted locus following recombination (HdhΔP). The “~” in the targeting vector represents plasmid sequence, and the “II” indicates the restriction site used to linearize the targeting vector prior to ES cell electroporation. The location of the neomycin phosphotransferase cassette (pgkneo, box) flanked by loxP sites (black arrowheads) that was used for positive selection of the transfected ES cells is shown. The transcriptional orientations of the Hdh and pgkneo genes are indicated with arrows. The small gray arrows indicate the location of the forward and reverse oligonucleotide primers used for PCR genotyping. The sizes of the wild-type and targeted NcoI-digested genomic DNA fragments recognized by the 3´flanking probe (small black rectangle) are shown above (Hdh) and below (HdhΔP), respectively. Restriction enzyme sites are NotI (Not), NcoI (N), HindIII (H), XmnI (X) and KpnI (K). The schematic is not drawn to scale. (B) Exon-1 of the wild-type (Hdh) mouse htt gene was modified by gene targeting to generate a deletion of the sequence encoding the mouse PRR (HdhΔP). The XmnI and KpnI restriction sites used for the modifications are shown. (C) DNA from tail biopsies was used to genotype progeny by PCR with primers that were designed to discriminate between the wild-type (Hdh) and modified (HdhΔP) alleles: Lane 1- Hdh+/+; Lane 2 - HdhΔPRR/ΔPRR; Lane 3 - HdhΔPRR/+; M – 100bp DNA molecular weight marker. (D) Nucleotide and encoded amino acid sequence of the Hdh+ (top) and HdhΔPRR (bottom) exon-1 XmnI–KpnI restriction fragment. Intron-1 sequence is presented in lower case, and the first and last codons of the PRR are indicated in larger font
The PRR is not required for normal htt function during development
To determine if the PRR deletion affected htt’s essential functions during embryonic development, the genotypes of progeny from HdhΔPRR/+ heterozygous intercrosses were evaluated for any deviation from the expected Mendelian frequency (Table 1). The predicted number of HdhΔPRR/ΔPRR pups were obtained, and the HdhΔPRR/ΔPRR mice were indistinguishable from HdhΔPRR/+ and wild-type littermates at birth. HdhΔPRR/ΔPRR adults are fertile, and the mutation can be maintained in homozygosity. Hematoxylin and eosin stained sections of brains obtained from 20 month-old HdhΔPRR/ΔPRR and wild-type littermates did not exhibit any gross anatomical abnormalities compared to their wild-type littermates (data not shown). Taken together, these observations suggest that the deletion of the htt PRR does not affect substantially normal htt function during embryogenesis and neurogenesis.
Table 1.
Mendelian analysis of progeny.
| Cross | No. of litters | No. observeda (No. of expectedb) progeny | ||
|---|---|---|---|---|
| Hdh+/+ | HdhΔPRR/+ | HdhΔPRR/ΔPRR | ||
| HdhΔPRR/+ × HdhΔPRR/+ | 36 | 64 (72) | 149 (144) | 64 (72) |
All progeny genotyped at postnatal day 10
Calculation based on the predicted Mendelian ratio. Genotypes were obtained at the predicted Mendelian frequencies (χ2 P >0.05 for all genotypes).
Deletion of the PRR does not affect htt steady-state levels or subcellular localization, but does affect htt phosphorylation at S13
The htt PRR, in addition to providing a protein interaction domain, may affect htt’s N-terminal structure, as well as its subcellular localization and post-translational modifications. Although htt is predominantly a cytoplasmic protein, it has the capability to shuttle between the nucleus and cytoplasm [36–38], and mutations within the N1-17 domain have been shown to alter its subcellular localization [39]. To determine if the htt PRR deletion can affect the overall stability and steady-state localization of ΔPRR-htt, we performed western analyses of whole brain extracts from HdhΔPRR/ΔPRR, HdhΔPRR/+, and wild-type littermates using an anti-htt antibody (MAB2166) that recognizes both wild-type and ΔPRR-htt. Quantification of htt levels by densitometry revealed that the PRR deletion did not affect significantly steady-state levels of ΔPRR-htt in comparison to wild-type htt (Fig. 2A). However, the PRR deletion does affect htt’s migration on SDS-PAGE, as full-length ΔPRR-htt migrates slightly faster than wild-type htt (Fig. 2B). This difference in migration is greater than what would be expected for a 32 amino acid deletion, and likely reflects an effect of the PRR deletion on htt’s overall conformation.
Figure 2. Deletion of the PRR does not affect htt expression levels.
(A) Densitometry analysis of wild-type and ΔPRR-htt levels calculated from western blots of whole brain protein extracts obtained from wild-type (+/+) and HdhΔPRR/ΔPRR (ΔP/ΔP) mice. (B) Whole brain protein extracts (100µg) from HdhΔPRR/ΔPRR (ΔP/ΔP), HdhΔPRR/+ (ΔP/+), and Hdh+/+ (+/+) mice were fractionated on 5% SDS-PAGE, and analyzed by western blotting using an antibody that recognizes both wild-type and ΔPRR-htt (MAB2166). The position of a 250 kD protein standard is indicated on the left. (C) Immunohistochemical analyses of coronal brain sections obtained from 20 month-old wild-type (+/+) and HdhΔPRR/ΔPRR (ΔP/ΔP) mice using an anti-htt antibody (BML-PW0595, epitope: htt N2-17) recognizing both wild-type and ΔPRR-htt. Nuclei were stained with To-Pro-3 iodide. Scale bars = 100µm.
To determine if the htt PRR deletion affects the expression of ΔPRR-htt in the brain, we performed immunohistochemical analyses of htt expression in fresh frozen coronal brain sections from 20-month old wild-type and HdhΔPRR/ΔPRR mice (Fig. 2C). The pattern of htt immunostaining was indistinguishable in the wild-type and homozygous mutant cortex, striatum, and hippocampus, with both wild-type htt and ΔPRR-htt localized predominantly in the cell bodies and neuropil.
To confirm that the htt PRR deletion did not affect its subcellular localization, primary mouse embryonic fibroblasts (PMEFs) were obtained from E13.5 wild-type and HdhΔPRR/ΔPRR embryos, and immunocytochemical analyses were performed using three anti-htt antibodies: BML-PW0595 (recognizing htt aa 2-17), MAB2166 (recognizing htt aa 414-503), and RabMAb (recognizing htt between aa 585–606). Immunocytochemical staining for htt in the HdhΔPRR/ΔPRR PMEFs was indistinguishable from that observed in the wild-type PMEFs with all three antibodies (MAB2166 and RabMAb staining not shown) (Fig. 3A). In addition, the wild-type and HdhΔPRR/ΔPRR PMEFs were stained with MitoTracker Red CMXRos, a fluorescent mitochondrial dye. Htt can both associate with and modulate the trafficking of mitochondria in cells [40, 41]; therefore, a potential deficit in ΔPRR-htt’s function in organelle trafficking may impact the localization of mitochondria in the cell. We could not detect any obvious differences between the predominantly perinuclear mitochondrial staining we observed in wild-type and HdhΔPRR/ΔPRR PMEFs. Similarly, no obvious differences in the subcellular fractionation of wild-type and ΔPRR-htt in HdhΔPRR/+ PMEFs were detected (Fig. 3B). Both wild-type and ΔPRR-htt were enriched in the cytoplasmic fraction, with a lower amount present in the nuclear fraction.
Figure 3. Htt subcellular localization and fractionation are not affected by the PRR deletion.
(A) Immunocytochemical analyses of PMEFs obtained from wild-type (+/+) and HdhΔPRR/ΔPRR (ΔP/ΔP) embryos using an anti-htt antibody (BML-PW0595), as well as Mitotracker Red CMXRos, a red-fluorescent dye that stains mitochondria. Nuclei were stained with To-Pro-3 iodide. Scale bars = 50µm. (B) Nuclear (Nuc) and cytoplasmic (Cyto) protein fractions (40µg) isolated from HdhΔPRR/+ (ΔP/+) P3 PMEFs were fractionated on 9% SDS-PAGE, and analyzed by western blotting using antibodies recognizing htt (MAB2166), PSD95 and tubulin (proteins preferentially enriched in the cytoplasmic fraction), and CREB (a protein that preferentially associates with the nuclear fraction). (C) Whole brain protein extracts from 5 month-old wild-type, HdhΔPRR/+ and HdhΔPRR/ΔPRR mice were fractionated on 5% SDS-PAGE, and analyzed by western blotting using an anti-htt phospho-S13 antibody (S13-P), and MAB2166. A non-specific protein recognized by the phospho-S13 antibody in both wild-type and HdhΔPRR/ΔPRR protein samples is indicated with an “*”. The sizes (in kD) of protein standards are indicated on the left.
To determine if deletion of the PRR can influence post-translational modification of the htt N1-17 domain, whole brain extracts prepared from 5 month-old wild-type, HdhΔPRR/+, and HdhΔPRR/ΔPRR mice were analyzed by western blotting using an anti-phospho-S13 antibody (Fig. 3C). A protein species co-migrating with soluble full-length wild-type htt was detected in both the wild-type and HdhΔPRR/+ extracts, but not in the HdhΔPRR/ΔPRR extracts. The amount of this species was also reduced in the HdhΔPRR/+ brain in comparison to wild-type brain, suggesting that its level correlated best with the level of wild-type htt in the extracts. This species was also absent from extracts prepared from an Hdh conditional knock-out brain (data not shown). Taken altogether, these data suggest that the htt PRR can modulate phosphorylation of htt at its N-terminus.
Male HdhΔPRR/ΔPRR mice exhibit a subtle behavioral phenotype
To determine if deletion of the htt PRR affects any behavioral phenotypes, separate cohorts of male wild-type and HdhΔPRR/ΔPRR mice at 6, 12, and 18 months of age, as well as female wild-type and HdhΔPRR/ΔPRR mice at 6 and 18 months of age were subjected to a series of behavioral tests. Each genotype group contained mice from several litters to average out any home cage factors that could potentially influence behavior (such as parental care and social dominance status). Grip strength and neurological reflexes, including eye blink, ear twitch, whisker twitch, toe pinch, postural reflex, and the righting reflex were assessed in each mouse. In all of these preliminary tests, no significant differences were found between wild-type and HdhΔPRR/ΔPRR mice (Table 2 and Table S1).
Table 2.
Assessment of general health and neurological reflexes in male mice.
| Age: | 6 months | 12 months | 18 months | ||||
|---|---|---|---|---|---|---|---|
| +/+ (n=8) |
ΔP/ΔPa (n=8) |
+/+ (n=5) |
ΔP/ΔP (n=5) |
+/+ (n=9) |
ΔP/ΔP (n=12) |
||
| General Health | |||||||
| Body weight (g) | 30.8±1.1b | 29.2±1.2 | 33.9±1.5 | 34.7±2.3 | 34.3±1.2 | 34.4±1.0 | |
| Forelimb grip strength (g) | 89.4±2.5b | 91.6±2.2 | 100.7±3.5 | 101.8±3.5 | 92.1±2.3 | 97.0±2.0 | |
| Visual forepaw reach (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Limb clasping (%) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Neurological Reflexes | |||||||
| Eye blink (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Ear twitch (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Whisker twitch (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Toe pinch (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Righting reflex (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Postural reflex (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
HdhΔPRR/ΔPRR
Expressed as mean ± SEM
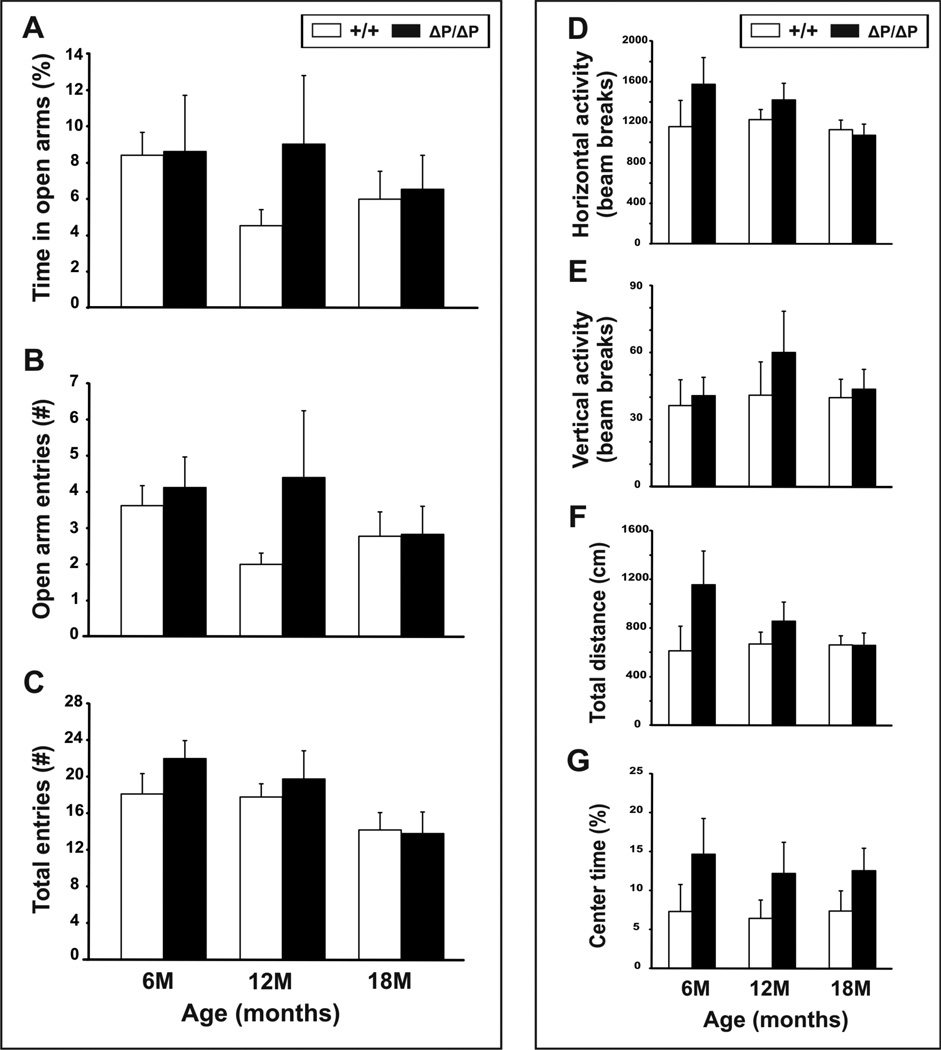
The elevated plus-maze was used to evaluate anxiety-like traits. Open and closed arm entries were combined to give a measurement of general exploratory activity. Reduced anxiety in the elevated plus-maze is indicated by an increase in the number of open arm entries, and the proportion of time spent in the open arms. No significant differences were observed between male wild-type and HdhΔPRR/ΔPRR mice in time spent in the open arms, number of open arm entries, and total number of open and closed-arm entries at any of the ages tested (Fig. 4A–C). A trend towards a significant difference was observed between wild-type and HdhΔPRR/ΔPRR females at 6 months of age in the proportion of time spent in the open arms of the maze (P=0.05, Student’s t-test) (Fig. S1A), but no differences were observed on any other parameters (Fig.S1A–C).
Figure 4. Deletion of the htt PRR does not affect anxiety and overall activity levels in HdhΔPRR/ΔPRR mice.
(A–C) The elevated plus maze was used to evaluate anxiety-like traits. Time spent in the open arms, number of open arm entries, and total number of entries were recorded for male mice at 6, 12, and 18 months of age. (D–G) General activity levels were collected using an automated activity cage. Horizontal activity, vertical activity, total distance, and percent time spent in the center were measured in male mice at 6, 12, and 18 months of age. All data are expressed as mean ± SEM.
To evaluate locomotion and exploratory behavior, open-field testing was performed. No significant differences in horizontal activity, vertical activity, total distance traveled, and time spent in the center of the cage were observed between wild-type and HdhΔPRR/ΔPRR mice (Fig. 4DG, Fig. S1D–G).
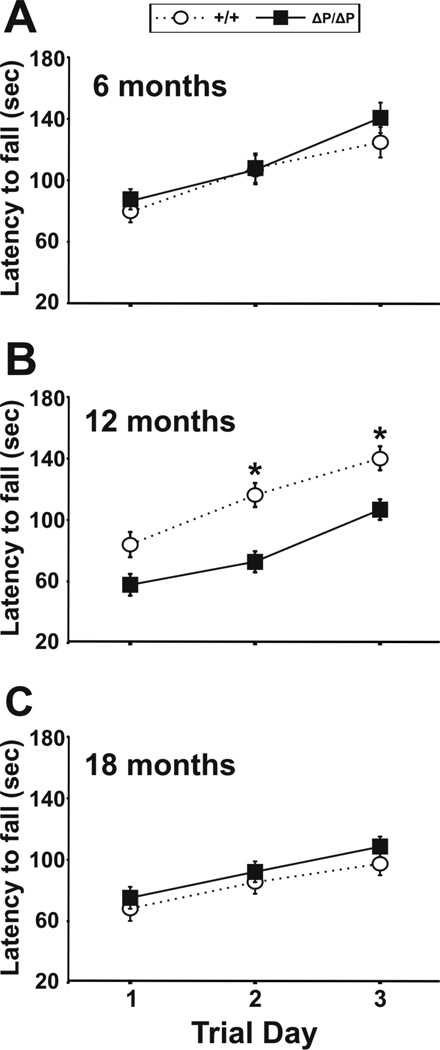
Performance on an accelerating rotarod was used to assess motor coordination and balance. A reduction in the latency to fall over repeated testing sessions on consecutive days provides a measure of motor learning. A trend towards a significant difference between male wild-type and HdhΔPRR/ΔPRR mice was observed on trial day 2 (P=0.05, ANOVA), and a significant difference was observed on trial day 3 (P=0.04, ANOVA) at 12 months of age (Fig. 5B). However, no significant differences were observed between genotypes at any other age among both males and females (Fig. 5A, 5C, Fig. S2A, S2B). We reason that the lower number of mice analyzed at 12 months of age may have contributed to the differences observed at this time point.
Figure 5. Rotarod testing of motor coordination and balance in HdhΔPRR/ΔPRR and control mice.
(A–C) Motor coordination and balance, as well as motor learning were tested on an accelerating rotarod. Male mice were tested at 6, 12 and 18 months of age. All data are expressed as mean ± SEM.
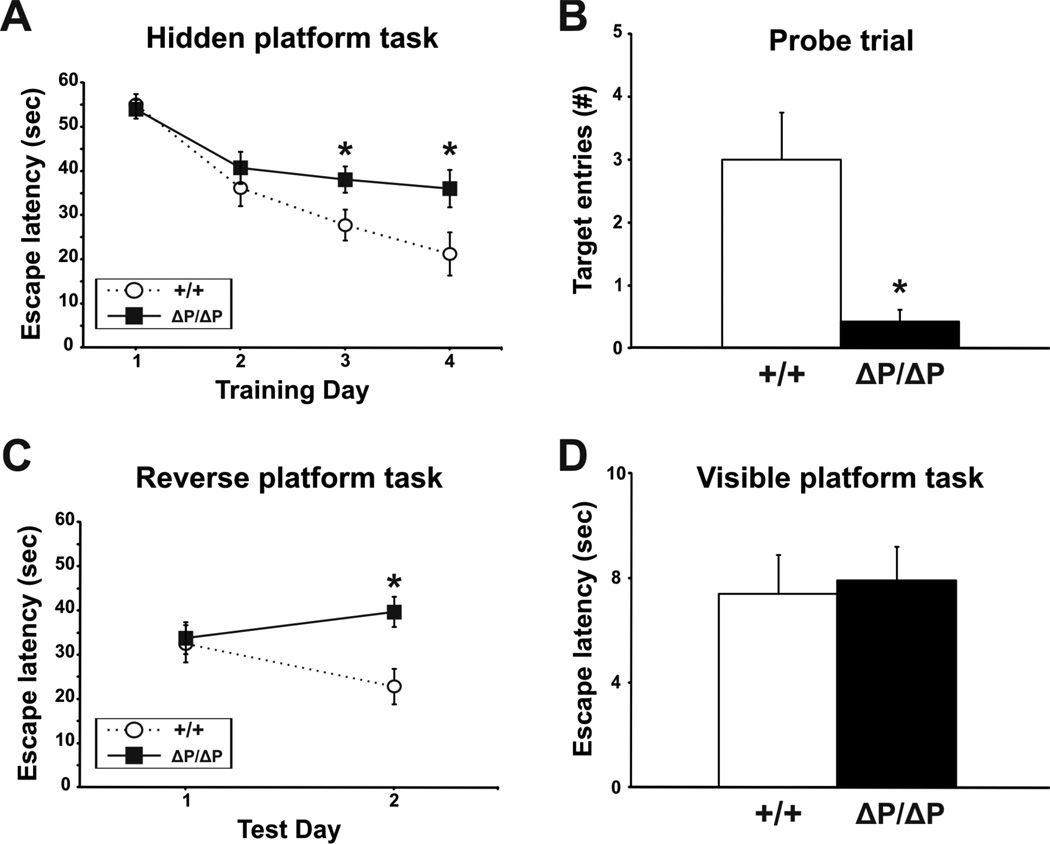
The Morris water maze task was conducted over eight consecutive days to assess spatial learning and memory. Wild-type and HdhΔPRR/ΔPRR mice were first tested on a hidden platform task for four days (four trials/day). A reduction in escape latency over repeated testing sessions provides a measure of the mouse’s ability to learn the task. A probe trial, in which the hidden platform was removed from the pool, was used on the fifth day to confirm that learning based on external environmental cues had occurred. If the mouse spent a significantly greater amount of time swimming near the former location of the platform, it was considered to have learned the location of the hidden platform. A reversal task, in which the hidden platform was moved to the opposite quadrant of the pool, was then used on the sixth and seventh day to measure the mouse’s ability to learn a new location for the platform. Finally, on the eighth day, a visible platform task was conducted to confirm that the mouse had the procedural ability to perform the water maze task. No significant differences were detected in escape latencies for the hidden platform, reversal and visible platform tasks, and in the number of target entries during the probe trial for both male and female wild-type and HdhΔPRR/ΔPRR mice at 6 months of age (data not shown). A significant difference between genotypes in male mice at 12 months of age was observed on training day 3 of the hidden platform task (P=0.009, ANOVA) (data not shown). In addition, significant differences were detected between genotypes in 18 month-old male mice on training days 3 (P=0.04, ANOVA) and 4 (P=0.03, ANOVA) in the hidden platform task (Fig. 6A), in the number of target entries during the probe trial (P=0.003, Student’s t-test) (Fig. 6B), and on test day 2 (P=0.005, ANOVA) in the reverse platform task (Fig. 6C). In contrast, no significant differences were found between wild-type and HdhΔPRR/ΔPRR female mice at 18 months of age (Fig. S3A–C). We also observed no significant differences between genotypes in the visible platform task or in the mean swim speeds of either male or female mice (Fig. 6D, Fig. S3D), suggesting that the 18 month-old HdhΔPRR/ΔPRR mice had the visual and motor ability to perform the tasks. Overall, these results suggest that spatial memory and navigation may be impaired in older HdhΔPRR/ΔPRR males.
Figure 6. Spatial learning and memory in the Morris water maze task is affected in older male HdhΔPRR/ΔPRR mice.
Escape latencies for the hidden platform (A), reversal (C) and visible platform tasks (D), and number of target entries during the probe trial (B) were recorded for male mice at 18 months of age. All data are expressed as mean ± SEM.
Discussion
Our results suggest that deletion of the PRR has a relatively subtle impact on normal htt function. Although a number of proteins have been proposed to interact with the htt PRR, many of these studies utilized truncated htt species as bait, and it is not clear whether the PRR is exposed or accessible to all potential protein binding partners in full-length htt. If we assume that most of these protein interactions also occur in the context of full-length htt, deletion of their binding site does not seem to affect appreciably htt’s core functions during development, as HdhΔPRR/ΔPRR mice are viable and fertile. However, we did observe that deletion of htt’s PRR reduced S13 phosphorylation within the N1-17 domain. Our inability to detect S13 phosphorylation in soluble full-length ΔPRR-htt is consistent with previous work suggesting that an interaction of IKK with the htt PRR and its adjacent polyQ stretch can modulate phosphorylation of the htt N-terminus and htt turnover [10, 32]. However, our western blotting protocol was not designed to detect the higher molecular weight phosphorylated htt species that are observed when using higher concentrations of SDS and reducing agent in the SDS-PAGE sample buffer together with modified transfer conditions [42]. Further experiments are needed to determine if such high molecular weight species are also absent in the HdhΔPRR/ΔPRR brain, and if ΔPRR-htt turnover in the proteosome or lysosome is affected by the PRR deletion.
The mild behavioral phenotypes observed in older male HdhΔPRR/ΔPRR mice contrasts with the progressive degenerative phenotype observed in Hdh conditional knock-out mice [43], suggesting that the PRR is not an essential htt domain, and more likely represents a region of htt that may modulate its conformation and function. This view is consistent with studies using synthetic peptides that suggest that the htt PRR can act as a structural element that protects against polyQ toxicity [17, 18]. Interestingly, the PRR in the context of an expanded polyQ stretch can also act as an aggresome targeting signal [34]. However, both of these PRR functions might not apply to the normal htt protein, as its short polyQ stretch does not form a stable β-sheet structure that is prone to aggregation. We note that in yeast and mammalian cell culture experiments expressing truncated normal htt N-terminal fragments containing a PRR deletion (as controls for the expression of mutant htt N-terminal fragments with a PRR deletion), their solubility and subcellular localization are similar to wild-type htt, and they do not produce a noticeable phenotype [44–47].
The phenotypic consequences of the htt PRR deletion are similar, in some respects, to the mild impact of deleting the normal htt polyQ stretch [35]. In both cases, homozygous mice are viable and fertile, and they exhibit relatively subtle behavioral deficits. However, the htt polyQ deletion in homozygosity can extend lifespan, and affects HD mouse model phenotypes in trans by stimulating neuronal autophagy [48]. With the limited numbers of older HdhΔPRR/ΔPRR mice that are currently available, we do not yet know if their lifespan is affected, and future experiments are needed to determine if the PRR deletion in trans can affect HD mouse model pathogenesis.
X-ray crystallographic analysis of the htt N-terminus has revealed that the N1-17 and PRR domains exhibit less conformational flexibility than the polyQ stretch [49]. Based on both X-ray crystallographic and circular dichroism (CD) analyses, the PRR forms a classic proline helix that can either be straight or kinked. Deletion of the PRR in the context of full-length htt alters its migration on SDS-PAGE, suggesting that htt’s overall structure is affected by the loss of its PRR. Nevertheless, this potential structural change does not significantly affect normal htt function, as HdhΔPRR/ΔPRR mice do not exhibit marked phenotypes. We note that full-length HTT that is expressed in insect cells and then purified by immunoaffinity chromatography can adopt multiple conformations [50], which is an observation that is consistent with the hypothesis that htt can act as a flexible scaffolding protein for diverse cellular processes. Thus, it is not surprising that deletion of the PRR may affect htt structure but not significantly affect its core functions.
Male HdhΔPRR/ΔPRR mice exhibit a behavioral phenotype in the Morris water maze task at 18 months of age. Significant differences were detected between mutants and controls in the hidden platform task, reverse platform task, and the probe trial, suggesting that spatial memory and navigation are affected. The HdhΔPRR/ΔPRR mice appear to have the necessary visual and motor skills that are required to successfully perform this task, because no significant differences were observed between the HdhΔPRR/ΔPRR and wild-type mice in the visible platform task, or in their mean swim speeds. It was difficult for us to assess visual ability with respect to the visual cues used to help the mice navigate the maze because these cues were placed outside of the maze, and the visible platform task uses a cue placed within the maze. Stress coping strategies, such as floating and attempts to escape along the wall of the maze could also produce confounding results, but these behaviors were not observed in our tests. Nevertheless, additional behavioral tests, such as the novel object recognition task, could be performed to confirm the presence of a learning and memory deficit in these mice.
In contrast to the males, female HdhΔPRR/ΔPRR mice did not show any significant differences from wild-type controls in the Morris water maze task. Sex differences have been observed in other models for HD, but they have not been studied extensively. In the CAG140 knock-in mouse model, Hdh140Q/140Q females spend more time grooming in the open-field, and on a running wheel during the diurnal dark phase, in comparison to Hdh140Q/140Q males and wild-type mice of both sexes [51]. Female R6/1 transgenic mice at 8 weeks of age also exhibit more depression-like phenotypes than males [52]. In contrast, both male N171-82Q transgenic mice and male HD transgenic rats exhibit a greater deficit in rotarod performance than females [53, 54]. In addition, a male deficit was observed in response to environmental enrichment with the R6/2 transgenic mouse model, with environmental enrichment having a positive effect on the cognitive performance of R6/2 female mice in the Morris water maze task, and a detrimental effect on male performance [55]. Male YAC128 mice also have a shorter lifespan than YAC128 females [56]. Moreover, age-at-onset is increased in female HD patients, and the progression of the disease in females is slower in comparison to males [57–59]. Taken together, these data suggest that further investigation into the role of sex differences in HD pathogenesis, and the contribution of the PRR to potential sex-specific functions for htt is warranted.
Supplementary Material
Acknowledgements
We thank Leslie Thompson for the gift of the htt S13-P antibody, Jeremy Tuttle for the use of his elevated plus-maze, Jonathan Kipnis for the use of his water maze, Paul Bonthuis for advice with behavioral and statistical analyses, Mark Fitzgerald for assistance with general troubleshooting, Noel Derecki for assistance with the Morris water maze, and Jeh-Ping Liu for critical reading of the manuscript. This work was supported by NIH NS43466 (S.O. Zeitlin), NS072453 (J.S. Steffan), and NS052789 (L.M. Thompson), with additional funding from the Hereditary Disease Foundation and CHDI Foundation, Inc. (J.S. Steffan).
Footnotes
Conflict of interest statement
The authors have no conflicts of interest arising from the publication of this paper.
References
- 1.Pecheux C, Gall AL, Kaplan JC, Dode C. Sequence analysis of the CAG triplet repeats region in the Huntington disease gene (IT15) in several mammalian species. Ann Genet. 1996;39(2):81–86. [PubMed] [Google Scholar]
- 2.Karlovich CA, John RM, Ramirez L, Stainier DY, Myers RM. Characterization of the Huntington's disease (HD) gene homologue in the zebrafish Danio rerio. Gene. 1998 Sep 14;217(1–2):117–125. doi: 10.1016/s0378-1119(98)00342-4. [DOI] [PubMed] [Google Scholar]
- 3.Baxendale S, Abdulla S, Elgar G, Buck D, Berks M, Micklem G, Durbin R, Bates G, Brenner S, Beck S. Comparative sequence analysis of the human and pufferfish Huntington's disease genes. Nat Genet. 1995 May;10(1):67–76. doi: 10.1038/ng0595-67. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Karlovich CA, Fish MP, Scott MP, Myers RM. A putative Drosophila homolog of the Huntington's disease gene. Hum Mol Genet. 1999 Sep;8(9):1807–1815. doi: 10.1093/hmg/8.9.1807. [DOI] [PubMed] [Google Scholar]
- 5.Gissi C, Pesole G, Cattaneo E, Tartari M. Huntingtin gene evolution in Chordata and its peculiar features in the ascidian Ciona genus. BMC Genomics. 2006;7:288. doi: 10.1186/1471-2164-7-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007 Nov 1;16(21):2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 7.Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007 Jan 1;16(1):61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 8.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004 Apr 2;304(5667):100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 9.Aiken CT, Steffan JS, Guerrero CM, Khashwji H, Lukacsovich T, Simmons D, Purcell JM, Menhaji K, Zhu YZ, Green K, Laferla F, Huang L, Thompson LM, Marsh JL. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009 Oct 23;284(43):29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O'Rourke JG, Khashwji H, Lukacsovich T, Zhu YZ, Lau AL, Massey A, Hayden MR, Zeitlin SO, Finkbeiner S, Green KN, LaFerla FM, Bates G, Huang L, Patterson PH, Lo DC, Cuervo AM, Marsh JL, Steffan JS. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009 Dec 28;187(7):1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrell-Pages M, Zala D, Humbert S, Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006 Nov;63(22):2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993 Mar 26;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 13.Lin B, Nasir J, MacDonald H, Hutchinson G, Graham RK, Rommens JM, Hayden MR. Sequence of the murine Huntington disease gene: evidence for conservation, alternate splicing and polymorphism in a triplet (CCG) repeat [corrected] Hum Mol Genet. 1994 Jan;3(1):85–92. doi: 10.1093/hmg/3.1.85. [DOI] [PubMed] [Google Scholar]
- 14.Barnes GT, Duyao MP, Ambrose CM, McNeil S, Persichetti F, Srinidhi J, Gusella JF, MacDonald ME. Mouse Huntington's disease gene homolog (Hdh) Somat Cell Mol Genet. 1994 Mar;20(2):87–97. doi: 10.1007/BF02290678. [DOI] [PubMed] [Google Scholar]
- 15.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000 Feb;14(2):231–241. [PubMed] [Google Scholar]
- 16.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994 Jan 15;297(Pt 2):249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006 Jan 20;355(3):524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 18.Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. 2007 Nov 30;374(3):688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Lakhani VV, Ding F, Dokholyan NV. Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLoS Comput Biol. 2010 Apr;6(4):e1000772. doi: 10.1371/journal.pcbi.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F. Protein sequences encode safeguards against aggregation. Hum Mutat. 2009 Mar;30(3):431–437. doi: 10.1002/humu.20905. [DOI] [PubMed] [Google Scholar]
- 21.Zuchner T, Brundin P. Mutant huntingtin can paradoxically protect neurons from death. Cell Death Differ. 2008 Mar;15(3):435–442. doi: 10.1038/sj.cdd.4402261. [DOI] [PubMed] [Google Scholar]
- 22.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998 Sep;7(9):1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 23.Gao YG, Yan XZ, Song AX, Chang YG, Gao XC, Jiang N, Zhang Q, Hu HY. Structural Insights into the specific binding of huntingtin proline-rich region with the SH3 and WW domains. Structure. 2006 Dec;14(12):1755–1765. doi: 10.1016/j.str.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Liu YF, Deth RC, Devys D. SH3 domain-dependent association of huntingtin with epidermal growth factor receptor signaling complexes. J Biol Chem. 1997 Mar 28;272(13):8121–8124. doi: 10.1074/jbc.272.13.8121. [DOI] [PubMed] [Google Scholar]
- 25.Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington's disease brains. Hum Mol Genet. 2002 Oct 1;11(21):2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 26.Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel JP, MacDonald ME. Huntingtin's WW domain partners in Huntington's disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum Mol Genet. 2000 Sep 1;9(14):2175–2182. doi: 10.1093/hmg/9.14.2175. [DOI] [PubMed] [Google Scholar]
- 27.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates GP, Lehrach H, Wanker EE. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol Cell. 1998 Oct;2(4):427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 28.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ. A map of WW domain family interactions. Proteomics. 2004 Mar;4(3):643–655. doi: 10.1002/pmic.200300632. [DOI] [PubMed] [Google Scholar]
- 30.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005 Sep 15;390(Pt 3):641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002 Feb 20;513(1):30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 32.Khoshnan A, Ko J, Watkin EE, Paige LA, Reinhart PH, Patterson PH. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J Neurosci. 2004 Sep 15;24(37):7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009 Dec 24;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem. 2008 Oct 10;283(41):27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 35.Clabough EB, Zeitlin SO. Deletion of the triplet repeat encoding polyglutamine within the mouse Huntington's disease gene results in subtle behavioral/motor phenotypes in vivo and elevated levels of ATP with cellular senescence in vitro. Hum Mol Genet. 2006 Feb 15;15(4):607–623. doi: 10.1093/hmg/ddi477. [DOI] [PubMed] [Google Scholar]
- 36.Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, Li XJ. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005 Feb;37(2):198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- 37.Tao T, Tartakoff AM. Nuclear relocation of normal huntingtin. Traffic. 2001 Jun;2(6):385–394. doi: 10.1034/j.1600-0854.2001.002006385.x. [DOI] [PubMed] [Google Scholar]
- 38.Truant R, Atwal RS, Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog Neurobiol. 2007 Nov;83(4):211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Atwal RS, Truant R. A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy. 2008 Jan-Feb;4(1):91–93. doi: 10.4161/auto.5201. [DOI] [PubMed] [Google Scholar]
- 40.Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008 Mar 12;28(11):2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004 Sep;24(18):8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Pardo A, Maglione V, Alpaugh M, Horkey M, Atwal RS, Sassone J, Ciammola A, Steffan JS, Fouad K, Truant R, Sipione S. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci U S A. 2012 Feb 28;109(9):3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000 Nov;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 44.Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, Patterson PH. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 2008 Sep 3;28(36):9013–9020. doi: 10.1523/JNEUROSCI.2747-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006 Nov 24;281(47):35608–35615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- 46.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006 Jul 18;103(29):11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin ZH, Wang Y, Sapp E, Cuiffo B, Wanker E, Hayden MR, Kegel KB, Aronin N, DiFiglia M. Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004 Jan 7;24(1):269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010 Feb;6(2):e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009 Sep 9;17(9):1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seong IS, Woda JM, Song JJ, Lloret A, Abeyrathne PD, Woo CJ, Gregory G, Lee JM, Wheeler VC, Walz T, Kingston RE, Gusella JF, Conlon RA, MacDonald ME. Huntingtin facilitates polycomb repressive complex 2. Hum Mol Genet. 2010 Feb 15;19(4):573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorner JL, Miller BR, Barton SJ, Brock TJ, Rebec GV. Sex differences in behavior and striatal ascorbate release in the 140 CAG knock-in mouse model of Huntington's disease. Behav Brain Res. 2007 Mar 12;178(1):90–97. doi: 10.1016/j.bbr.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renoir T, Zajac MS, Du X, Pang TY, Leang L, Chevarin C, Lanfumey L, Hannan AJ. Sexually dimorphic serotonergic dysfunction in a mouse model of Huntington's disease and depression. PLoS One. 2011;6(7):e22133. doi: 10.1371/journal.pone.0022133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orr AL, Huang S, Roberts MA, Reed JC, Li S, Li XJ. Sex-dependent effect of BAG1 in ameliorating motor deficits of Huntington disease transgenic mice. J Biol Chem. 2008 Jun 6;283(23):16027–16036. doi: 10.1074/jbc.M710606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bode FJ, Stephan M, Suhling H, Pabst R, Straub RH, Raber KA, Bonin M, Nguyen HP, Riess O, Bauer A, Sjoberg C, Petersen A, von Horsten S. Sex differences in a transgenic rat model of Huntington's disease: decreased 17beta-estradiol levels correlate with reduced numbers of DARPP32+ neurons in males. Hum Mol Genet. 2008 Sep 1;17(17):2595–2609. doi: 10.1093/hmg/ddn159. [DOI] [PubMed] [Google Scholar]
- 55.Wood NI, Carta V, Milde S, Skillings EA, McAllister CJ, Ang YL, Duguid A, Wijesuriya N, Afzal SM, Fernandes JX, Leong TW, Morton AJ. Responses to environmental enrichment differ with sex and genotype in a transgenic mouse model of Huntington's disease. PLoS One. 2010;5(2):e9077. doi: 10.1371/journal.pone.0009077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Raamsdonk JM, Pearson J, Rogers DA, Bissada N, Vogl AW, Hayden MR, Leavitt BR. Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum Mol Genet. 2005 May 15;14(10):1379–1392. doi: 10.1093/hmg/ddi147. [DOI] [PubMed] [Google Scholar]
- 57.Roos RA, Vegter-van der Vlis M, Hermans J, Elshove HM, Moll AC, van de Kamp JJ, Bruyn GW. Age at onset in Huntington's disease: effect of line of inheritance and patient's sex. J Med Genet. 1991 Aug;28(8):515–519. doi: 10.1136/jmg.28.8.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foroud T, Gray J, Ivashina J, Conneally PM. Differences in duration of Huntington's disease based on age at onset. J Neurol Neurosurg Psychiatry. 1999 Jan;66(1):52–56. doi: 10.1136/jnnp.66.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pekmezovic T, Svetel M, Maric J, Dujmovic-Basuroski I, Dragasevic N, Keckarevic M, Romac S, Kostic VS. Survival of Huntington's disease patients in Serbia: longer survival in female patients. Eur J Epidemiol. 2007;22(8):523–526. doi: 10.1007/s10654-007-9157-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.