Abstract
Melanomas and other cancers that do not express argininosuccinate synthetase (AS), the rate-limiting enzyme for arginine biosynthesis, are sensitive to arginine depletion with pegylated arginine deiminase (ADI-PEG20). However, ADI resistance eventually develops in tumors due to AS upregulation. Although it has been shown that AS upregulation involves c-Myc, the underlying mechanisms remain unknown. Here we show that ADI-PEG20 activates Ras signaling and the effector ERK and PI3K/AKT/GSK-3β kinase cascades, resulting in phosphorylation and stabilization of c-Myc by attenuation of its ubiquitin-mediated protein degradation mechanism. Inhibition of the induced cell signaling pathways using PI3K/AKT inhibitors suppressed c-Myc induction and enhanced ADI-mediated cell killing. Notably, in an animal model of AS-negative melanoma, combination therapy using a PI3K inhibitor plus ADI-PEG20 yielded additive anti-tumor effects as compared with either agent alone. Taken together, our findings offer mechanistic insight into arginine deprivation metabolism and ADI resistance, and they illustrate how combining inhibitors of the Ras/ERK and PI3K/AKT signaling pathways may improve ADI-PEG20 anti-cancer responses.
Keywords: ADI-PEG20, PI3K/AKT/ERK, c-Myc Stabilization, Drug-Resistance, Melanoma
Introduction
Malignant melanoma is the most aggressive type of skin cancer that accounts for most deaths from skin cancer and the cure rates remains <10% (1, 2). Effective treatment modalities for malignant melanoma are urgently needed. It has been demonstrated that most of malignant melanomas have abnormal urea cycle metabolism that are unable to perform de novo synthesis of arginine (Arg) from citrulline in a two-step reactions catalyzed by argininosuccinate synthetase (AS) and argininosuccinate lyase (3). AS is the rate-limiting enzyme and malignant melanomas do not express AS and therefore require Arg from extracellular source for tumor growth. This Arg-auxotrophicity provides a novel approach for using Arg-degrading enzymes to deplete Arg in the circulation to treat melanoma and other human malignancies (4). Pegylated recombinant bacterial arginine deiminase (ADI-PEG20) which converts Arg to citrulline and ammonia resulting in Arg deprivation, has been under various stages of clinical evaluation for the treatment of malignant melanoma (5). This strategy has also been used in the treatments of hepatocellular carcinoma (5–8).
Although ADI-PEG20 treatments have shown promising outcomes in most studies, one important mechanism associated with treatment failure is the development of drug resistance due to re-expression of AS in the tumors. Using cultured melanoma cells, we previously demonstrated that ADI-PEG20 treatments induced AS expression in A2058 and SK-MEL-2 cells, but not in A375 cells (9). Induction of AS expression was associated with upregulation of c-Myc and downregulation of HIF-1α. HIF-1α functions as a negative regulator by binding to the E-box at the AS promoter and suppressing AS expression prior to the induction. Upon ADI-PEG20 treatment, binding of HIF-1α at the E-box is replaced by c-Myc which functions as a positive regulator for the upregulation of AS. However, the mechanisms underlying this c-Myc upregulation are unknown. In this study, we report that upregulation of c-Myc by ADI-PEG20 is due to enhanced c-Myc protein stability elicited by activation of the Ras signaling-mediated protein kinase cascades. Our findings provide a plausible strategy for improving the treatment efficacy of ADI-PEG20 by intervention in this signal transduction pathway.
Materials and Methods
Reagents, antibodies, and recombinant DNA
Reagents were obtained from the following sources: ADI-PEG20 (specific activity, 5~10 IU/mg) from Polaris Pharmacologies Inc. (San Diego, CA); sulforhodamine B (SRB), MG-132, sodium selenite, and Ly294002 from Sigma-Aldrich (St. Louis, MO); perifosine from Selleck (Houston, TX) and PtdIns-(4, 5)-P2 from Cayman Chemical (Ann Arbor, MI); cycloheximide (CHX) and AKT inhibitor VIII from Calbiochem (La Jolla, CA); PI-103 from Echelon Biosciences (Salt Lake City, UT); GSK-3 siRNA from Cell Signaling (Boston, MA). Mouse anti-β-actin, mouse anti-hemagglutinin (HA), mouse anti-Flag, and anti-α-tubulin antibodies from Sigma-Aldrich; mouse anti-AS from Polaris Pharmacologies Inc; rabbit anti-c-Myc (N262), rabbit anti-p-T58-c-Myc and rabbit anti-AKT (H-136) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA); mouse p-S62-c-Myc (33A12E10) antibody from abcam (Cambridge, MA); rabbit anti-phospho AKT threonine 308, mouse anti-phospho-tyrosine, anti-PTEN (A2B1), anti-USP28, anti-extracellular receptor kinase (anti-ERK), anti-phospho-ERK and anti-glycogen synthease kinase-3 (anti-GSK-3) antibodies from Cell Signaling Technology (Danvers, MA).
Recombinant plasmid DNA encoding dominant negative (DN) forms of HA-AKT expression was described previously (10). DN GSK-3β and DN RasN17 expression plasmids were obtained from Dr. Geoffrey M. Cooper (11, 12) and HA-USP-28 recombinant DNA was from Dr. Stephen J. Elledge (13).
Cell culture, siRNA transfection, and SRB cytotoxicity assay
A2058, SK-MEL-2, and A375 melanoma cells and MDA-MB-231 breast cancer cells were purchased from American Type Culture Collection Center (ATCC) and were not further tested or authenticated. All cell cultures were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) at 5% CO2 atmosphere. For Arg depletion, cells were either maintained in the regular medium containing 0.3μg/mL ADI-PEG20 or cultured in Arg-free DMEM medium containing 10% dialyzed FBS. All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
For the cytotoxicity assay, cells were seeded in 96-well plates (4×103 cells per well) and cultured with different concentrations of inhibitors with or without ADI-PEG20 for 72 hrs. Cells were fixed with 50% trichloroacetic acid followed by staining with 0.4% SRB in 1% acetic acid for 30 min at room temperature. Plates were washed five times with 1% acetic acid to remove unbound dye. Bound dye was dissolved by adding 10 mM unbuffered Tris base. Cell proliferation was calculated by measuring OD at 564 nm using a spectrophotometer.
Immunoprecipitation, immunoblotting, and northern blot
Procedures for cell extract preparations and immunoprecipitation were previously described (9). Briefly, protein samples were incubated with an antibody and 50 μL 50% protein A sepharose beads. Protein A beads were collected and the immunopricipitates were resolved by SDS-PAGE followed by immunoblotting and visualized with an enhanced chemiluminescence kit (Thermo Scientific, Rockford, IL). For Northern blotting, cells were harvested by Trizol reagent (Invitrogen, Carlsbad, CA) and total RNA was extracted according to the manufacturer's instructions. Equal amounts of total RNA samples were separated by 2% agarose-formaldehyde gel electrophoresis, transferred to a nylon membrane, and hybridized to [α-32P]dCTP-labeled (PerkinElmer, Boston, MA) c-Myc cDNA probe according to the standard procedures.
Mouse experiments
Female athymic NCR nu/nu-nude mice (aged 7 weeks, weight ~20 grams, from National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD) were housed in a pathogen-free environment. The animals were inoculated subcutaneously with 2 × 106 A2058 melanoma cells in 100 μL physiological buffered saline (PBS) into the right flank of mice. Ten days later, when the tumor volumes reached ~20 mm3, the animals were randomly divided into four groups with six animals per group and the treatments were initiated by i.p. injections according to the following protocol. The first group received 100 μL PBS, the second group received Ly294002 (25 mg/kg), the third group received ADI-PEG20 (4 IU or 0.625 mg/100 μL), and fourth group received Ly294002 (25 mg/kg) plus ADI-PEG20 (4 IU). Each group of animals were received the same doses of drugs twice per week thereafter. Tumor size was measured by caliper. Tumor volume was calculated using the formula: (length × width2)/2. Statistical analysis was performed by Student t-test using Microsoft Excel 2007 program. p < 0.05 was regarded as significant. Error bars represent standard error of the mean (SEM).
Other procedures
Enzymatic activity assays for phosphatidylinositol-3 phosphate (10) and PTEN (14), and DNA fragmentation assay (15) followed the procedures previously described.
Results
ADI-PEG20 induces c-Myc protein stabilization
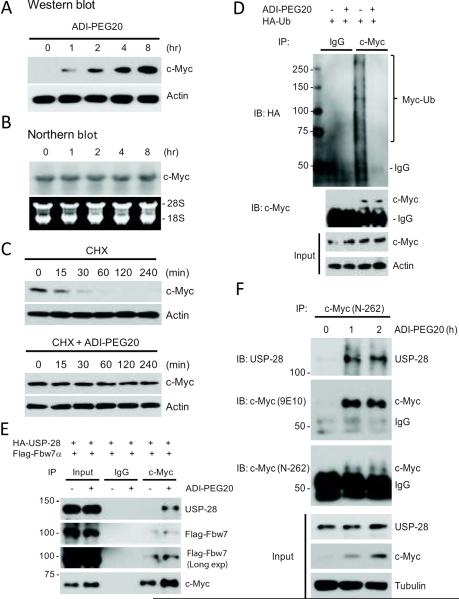
The enhancement of c-Myc expression by ADI-PEG20 could be regulated at the transcriptional level or at the post-transcriptional level. To distinguish between these two possibilities, we performed Northern blotting and Western blotting analyses to evaluate c-Myc mRNA and protein levels, respectively. Fig 1A shows that while induction of c-Myc protein was detectable within 1 hr of treatment and continued throughout the 8 hrs of treatment, no corresponding increases in c-Myc mRNA levels were seen (Fig. 1B). These results suggest that the induction mechanism is either by enhanced protein synthesis or by reduced protein degradation. To differentiate between these possibilities, we treated A2058 cells with the protein synthesis inhibitor CHX with or without ADIPEG20. In the absence of ADI-PEG20, c-Myc protein levels were reduced rapidly with a half-life (t½) of ~20 min (Fig. 1C, upper), consistent with the previous report that c-Myc is a very unstable protein with t½ between 20 ~ 30 min (16). In the presence of ADIPEG20, c-Myc degradation was attenuated, and > 70% of c-Myc remained even after 4 hrs of treatment (Fig. 1C, lower). In this experiment, we purposely overexposed the blot so that c-Myc expression level at the 0 time point could be visualized, as in contrast to those shown in Fig. 1A. These results demonstrate that ADI-PEG20 treatment induces c-Myc protein stabilization.
Fig. 1.
Induction of c-Myc by ADI-PEG20 is due to inhibition of c-Myc ubiquitination. (A) Western blot shows that c-Myc protein was increased in response to ADI-PEG20; whereas (B) northern blot shows that c-MYC mRNA levels were not affected by ADI-PEG20 treatment for up to 8-h in A2058 cells. (C) Effects of CHX on c-Myc expression in A2058 cells treated with ADI-PEG20. Cells were incubated with100 μM CHX in the presence or absence of ADI-PEG20. Protein extracts were obtained at the indicated time points. Expression levels of c-Myc, and actin were analyzed by Western blotting. (D) ADI-PEG20 prevented ubiquitination of c-Myc protein. A2058 cells transfected with HAUb-encoding plasmid were treated with 10 μM MG-132 in the absence or presence of ADI-PEG20 for 4-h. Cell lysates were immunoprecipitated with anti-c-Myc antibody, followed by western blotting with anti-HA and anti-c-Myc antibodies as indicated. (E) ADI-PEG20 induces association of USP-28, Fbw7α and c-Myc. A2058 cells were transfected with USP28 and Fbw7α-expressing plasmids for 16 hr. Cells were treated with 10 μM MG-132 in the absence or presence of ADI-PEG20 for 4 hours. Cells were lysed and immunoprecipitated with anti-Myc antibody. Precipitates were analyzed by immunoblotting with indicated antibodies. (F) ADI-PEG20 induces association of endogenous c-Myc and USP-28 in MDA-MB-231 cells. MDA-MB-231 cells were treated with ADI-PEG20 for 1 and 2-h. Cells were lysed and cell lysates were immunoprecipitated with anti-c-Myc antibody. The precipitates and input lysates were analyzed by immunoblotting with anti-c-Myc (N-262) and anti-c-Myc (9E10) and anti-USP-28 antibodies as indicated.
ADI-PEG20-induced c-Myc stabilization is due to inhibition of ubiquitin-mediated protein Degradation
It has been demonstrated that rapid c-Myc protein turnover can be mediated by the ubiquitin-dependent proteasome pathway (17, 18). To investigate whether the c-Myc stabilization induced by ADI-PEG20 was due to inhibition of ubiquitin-mediated protein degradation, we transfected A2058 cells with recombinant plasmid encoding HA-tagged ubiquitin followed by treating the cells with or without ADI-PEG20. Cell lysates were prepared and c-Myc was immunoprecipitated with anti-c-Myc antibody using normal rabbit IgG as a negative control. The immunoprecipitates were analyzed by Western blotting using anti-HA antibody. Fig. 1D shows that a significant amount of poly-ubiquitinated c-Myc was present in the ADI-PEG20-untreated cells; whereas almost no ubiquitinated c-Myc was detected in the ADI-PEG20-treated cells. These results demonstrate that ADI-PEG20-induced c-Myc accumulation was due to inhibition of c-Myc ubiquitination for protein degradation.
One of the important pathways for ubiquitin-dependent regulation of c-Myc turnover is controlled by the interactions with both USP-28 and Fbw7. Fbw7α is a subunit of the E3 ubiquitin ligase complex SCFFbw7 that recognizes c-Myc in response to specific stimuli leading to ubiquitination and subsequent proteasome degradation of c-Myc (19). In contrast, USP-28, which forms a complex with Fbw7α and counteracts the degradation of c-Myc by removing ubiquitins conjugated by Fbw7α (20). To investigate the possible roles of USP-28/Fbw7α in ADI-PED20-mediatd c-Myc stabilization, we transfected A2058 cells with recombinant plasmids encoding Frag-Fbw7α and HA-USP-28, in the presence of proteasome inhibitor MG132, followed by treatment with or without ADI-PEG20. c-Myc was subsequently immunoprecipitated and analyzed by Western blotting with anti-c-Myc, anti-USP-28, and anti-Flag antibodies. Our results show that ADI-PEG20 treatment resulted in a significant association between overexpresssed USP-28 and the c-Myc/Fbw7α complex (Fig. 1E). We next investigated the effects of ADI-PEG20 on the interactions between endogenous USP-28 and c-Myc. MDA-MB-231 cells were treated with ADI-PEG20 for either one or two hours. Cell lysates were prepared and c-Myc was immunoprecipitated with a polyclonal anti-c-Myc antibody (N-262). The precipitates were probed with anti-USP28 antibody, anti-c-Myc antibody (N-262), and anti-c-Myc monoclonal antibody (9E10). Fig. 1F shows that ADI-PEG20 treatment enhances the interaction between USP-28 and c-Myc. These results suggest the involvement of USP28/Fbw7α in the ADI-PEG20-mediated c-Myc accumulation.
ADI-PEG20-induced c-Myc stabilization is mediated by the ERK and PI3K/AKT-GSK3β signaling pathways
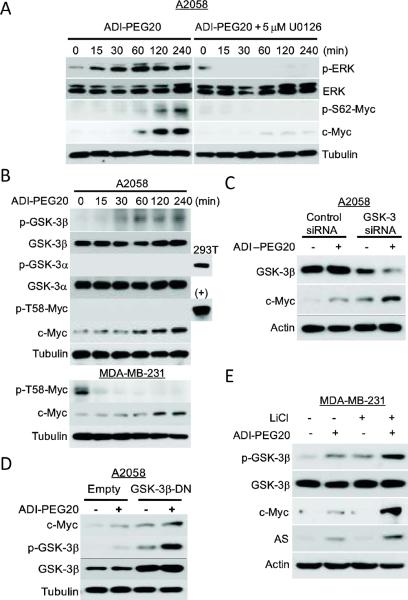
c-Myc protein is targeted for ubiquitin proteasomal degradation mechanism by phosphorylation at two specific amino acid residues at the N-terminus, serine 62 (S62) and threonine 58 (T58). S62 is a target of ERK and T-58 is targeted by GSK-3β (21, 22). ERK-mediated phosphorylation of S62 prevents c-Myc from degradation, whereas GSK-3β-phosphorylated T-58 promotes c-Myc degradation (21, 22). We investigated the possible involvement of these pathways and found that ADI-PEG20 treatment activated ERK and enhanced c-Myc phosphorylation at S62 in association with increased c-Myc levels (Fig. 2A). To test for the causal involvement of the ERK signaling, we used the chemical inhibitor U0126, which specifically inactivates MEK1/2 (ERK kinase). Western blot analysis revealed that U0126 was effective in completely inhibiting c-Myc phosphorylation and reducing c-Myc expression levels (Fig. 2A). These results suggest that ERK is involved in the posttranslational modification and stabilization of c-Myc in response to ADI-PEG20.
Fig. 2.
The roles of ERK and GSK-β in the regulation of c-Myc expression by ADIPEG20. (A) A2058 cells were treated with ADI-PEG20 alone (left) or co-treated with 5 μM U0126 (right) for the indicated times. Expression levels of c-Myc, p-S62-Myc, ERK, p-ERK and tubulin were analyzed by western blotting. (B) A2058 cells were treated with ADI-PEG20 for the given times. Expression levels of c-Myc, pGSK-3α(Ser21), GSK-3α, pGSK-3β(Ser9), GSK-3β, p-T58-Myc, and tubulin were analyzed by western blotting; (+) denotes lysates from A2058 trensfected with HA-c-Myc plasmid as a positive control. (C) Effects of GSK-3β siRNA on the expression of c-Myc by ADI-PEG20. A2058 cells were transfected GSK-3 (α and β) siRNA and its control for 48-h. Cells were treated with ADIPEG20 for another 4-h. Cell lysates were analyzed for indicated proteins by western blot. (D) A2058 cells were transfected with GSK3β-DN vector for 24 hr. Cells were maintained in normal medium, or medium containing 0.3 μg/mL ADI-PEG20 for 4-h. Expression levels of c-Myc, GSK-3β, p-GSK-3β and tubulin were analyzed by western blotting. (E) Inhibition of GSK-3 activity by LiCl increases the stability of c-Myc. MDA-MB-231 cells were treated with 10 mM LiCl in the absence or presence of ADI-PEG20 for 48-h. Expression levels of AS, c-Myc, GSK-3β, p-GSK-3β c-Myc, AS and actin were analyzed by Western blotting.
Phosphorylation of c-Myc at T58 by GSK-3β, a serine/threonine kinase, is recognized by Fbw7 in the proteasomal protein degradation signal (22). It has been demonstrated that GSK-3β itself is a target of PI3K/AKT-mediated phosphorylation at Ser9 position. GSK-3β phosphorylation inactivates its ability of phosphorylating c-Myc(T58), resulting in stabilization of c-Myc. PI3K/AKT also phosphorylates GSK-3β(Ser21). We asked whether this pathway regulates c-Myc stability in response to ADIPEG20 challenge in A2058 cells. Phosphorylation of GSK-3β(Ser9) and GSK-3α(Ser21) in the ADI-PEG20-treated cells were determined using anti-p-GSK-3α and anti-p-GSK-3β antibodies, respectively. We found that ADI-PEG-20 induces phosphorylation of GSK-3β but not GSK-3α (using lysate from 293T cells as a positive control) in a time-dependent manner (Fig. 2B). Because phosphorylated c-Myc(T58) levels were not detectable even in the un-stimulated A2058 cells (using lysate from the c-Myc-transfected A2058 cells as a positive control), we turned to MD-231 cells and observed that ADI-PEG20 treatment indeed reduced p-c-Myc(T58) expression in these cells (Fig. 2B, lower). These results demonstrate that ADI-PEG20 treatment induces GSK-3β phosphorylation in association with reduced c-Myc(T58) phosphorylation.
We also demonstrated that knockdown of GSK-3β using siRNA led to increased c-Myc levels even without ADI-PEG20 treatment, whereas ADI-PEG20 plus GSK-3β siRNA treatments further increased c-Myc levels (Fig. 2C). Likewise, expression of GSK-3β-DN alone enhanced the expression of c-Myc compared with empty vector-transfected cells, and together with ADI-PEG20 treatment further enhanced the expression of c-Myc in the GSK-3β-DN-transfected cells (Fig. 2D). Additionally, we used a potent mammalian GSK-3 inhibitor, LiCl, which competes against the cofactor Mg2+ for GSK-3 activity (23). We found that LiCl treatment enhanced the expression of c-Myc and its downstream target AS in ADI-PEG20-treated MDA-MB-231 cells (Fig. 2E). Taken together, these results demonstrate that GSK-3β plays an inhibitory role in ADI-PEG20-mediated c-Myc stabilization.
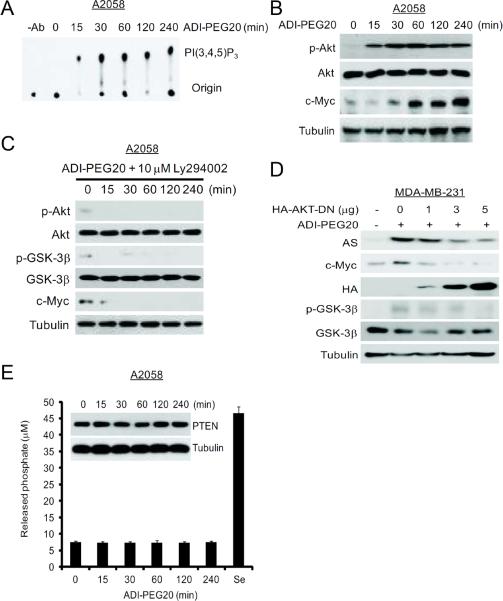
ADI-PEG20-induced phosphorylation of GSK-3β could be due to activation of PI3K/AKT signaling. To investigate the role of PI3K/Akt in ADI-PEG20-induced GSK-3β phosphorylation, we measured PI3K activity in extracts from ADI-PEG20-treated cells using phosphotidylinositol as a substrate. Fig. 3A shows that induction of PI3K activity occurred as early as 15 min and persisted throughout the 4 hrs of ADI-PEG20 treatment. Activation of PI3K by ADI-PEG20 was also demonstrated by the induction of Akt(T308) phosphorylation, a downstream effector of PI3K (Fig. 3B). We further demonstrated that inhibition of PI3K activity with LY294002 suppressed the ADI-PEG20-induced phosphorylation of Akt(T308) and GSK-3β(Ser9), and expression levels of c-Myc (Fig. 3C). Likewise, a dominant negative mutant form of AKT suppressed the induction of p-GSK-3β(Ser9), and the expression of c-Myc and AS in a dose-dependent manner in MDA-MB-231 cells (Fig. 3D). Activation of PI3K by ADI-PEG20 is apparently not due to downregulation of PTEN which is a phosphatase that negatively regulates PI3K signaling (24, 25), because we failed to detect reduction of PTEN expression levels by Western blotting within the first 4 hrs of the treatment (Fig. 3B). Nor did we see reduction of PTEN phosphatase activities using sodium selenite treatment, which is known to induce PTEN activity, as a positive control (26) (Fig. 3E). Overall, these results indicate that upregulation of c-Myc is through the inhibition of GSK-3β activity which is mediated by the activation of PI3K/AKT in response to ADI-PEG20.
Fig. 3.
Roles of the PI3K/AKT signal cascade in the regulation of c-Myc expression by ADI-PEG20. (A) Time-course induction of PI3K activity in A2058 cells treated with ADI PEG20. A2058 cells were treated with 0.3 μg/mL ADI-PEG20 for various time intervals. Cells were harvested. PI3K was immunoprecipitated with anti-phospho-tyrosine antibody/protein G agarose. The kinase activity was determined. –Ab, no anti-phosphotyrosine antibody. (B) Protein extracts were obtained from (A). The expression levels of pT308-AKT, AKT, c-Myc, PTEN, and Tubulin were analyzed by Western blotting.(C) Effects of the PI3K inhibitor Ly294002 on ADI-PEG20-induced c-Myc expression. A2058 Cells were incubated ADI-PEG20 plus 10 μM Ly294002. Protein extracts were obtained at the indicated time points. AKT phosphorylation was determined with an antibody specific to AKT phosphor-Thr308. GSK-3β phosphorylation was determined with an antibody specific to GSK-3β phosphor-Ser9. AKT, GSK-3β or tubulin proteins were probed with their respective antibodies. (D) Effects of DN AKT on ADI-PEG20-induced c-Myc expression. MDA-MB-231 cells were transfected with different amounts of HA-AKTDN vector for 24-h. Cells were maintained in normal medium or in medium containing 0.3 μg/mL ADI for 48-h. The expression levels of AS, c-Myc, pGSK-3β(Ser9), GSK-3β, HA, and actin were analyzed by western blotting. (E) Effects of ADI-PEG20 on PTEN expression and activity. A2058 cells were treated with 0.3 μg/mL ADI-PEG20 for various time intervals. Enzymatic activities of PTEN were measured using an 1-h treatment of sodium selenite lysate as a positive control for phosphatase activity assay. Insert shows Western blot measurement of PTEN protein levels in the treated cells.
Involvement of Ras signaling in ADI-PEG20-induced c-Myc stabilization and cell line specificity
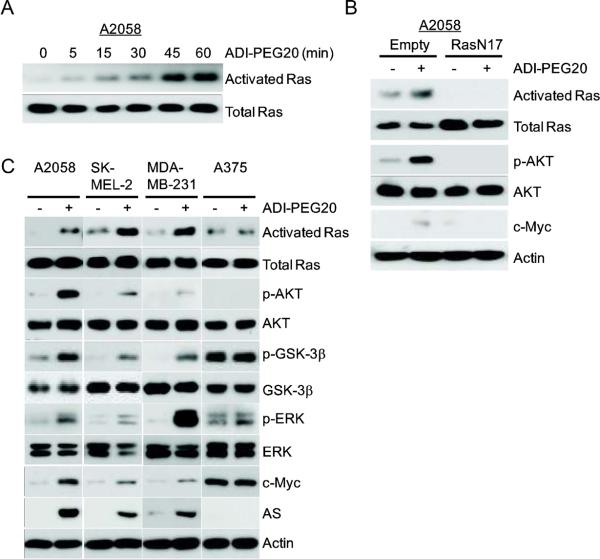
One important upstream activator for PI3K/Akt pathway is Ras. To investigate whether Ras is involved in ADI-induced PI3K/AKT activation that leads to c-Myc accumulation, we measured Ras activity in ADI-treated A2058 cells. Fig. 4A shows induction of Ras activity by ADI-PEG20 occurred as early as 5 min after A2058 cells were exposed to ADI-PEG20. Moreover, a dominant negative mutant form of Ras (RasN17) could reduce the signal of pT308-AKT and c-Myc protein levels in response to ADI-PEG20 in A2058 cells (Fig. 4B). These results, collectively, demonstrate that induction of c-Myc expression by ADI-PEG20 is due to activation of Ras/PI3K/Akt/ERK pathways in A2058 cells.
Fig. 4.
Activation of Ras by ADI-PEG20 treatment and cell line-specificity of the induction. (A) A2058 cells were treated with ADI-PEG20 for the indicated times. Whole-cell lysates were prepared. Activated Ras were eluted after pull-down using the Ras-binding domain (RBD) of RAF-1 from the cell lysates. Levels of activated Ras and total Ras from cell lysates are determined by immunoblotting. (B) Effects of DN Ras on the induction of c-Myc by ADI-PEG20. A2058 cells were transfected with empty vector or RasN17 vector for 24-h. Cells were maintained in normal medium or medium containing 0.3 μg/mL ADI-PEG20 for 1-h. Levels of activated Ras, total Ras, p-T308-AKT, AKT, c-Myc and actin were analyzed by western blotting. (C) Cell line specificity of activation of Ras and its down-stream signals by ADI-PEG20 treatment. A2058, SK-MEL-2, MDAMB-231, and A375 cells were treated with ADI-PEG20 for 1-h. Expression levels of activated Ras, total Ras, pT308-AKT, AKT, pGSK-3β(Ser9), GSK-3β, ERK, p-ERK, c-Myc, AS and actin were analyzed by western blotting.
Our results described thus far were mainly carried out in the A2058 cell line. To assess the generality of these findings, we examined the effects of ADI-PEG20 in three additional human cancer cell lines, SK-MEL-2, MDA-MB-231, and A375. Expression of AS and c-Myc in SK-MEL-2 (9) and MDA-MB-231 cells, like A2058 cells, can be induced by ADI-PEG20 treatment; whereas expression of c-Myc/AS is not inducible in A375 cells (9). Fig. 4C shows that ADI-PEG20 treatment induced activation of Ras, phosphorylation of AKT, phosphorylation of GSK-3β, and activation of ERK signals in A2058, SK-MEL-2 and MDA-MB231 cells, but not in A375 cells. These effects were correlated with induced expression of c-Myc and AS proteins in these cells (Fig. 4C). This correlation suggests an important role of Ras/PI3K/Akt/GSK-3β/ERK signaling in the upregulation of c-Myc under Arg-deprivation conditions in multiple cell lines.
Inhibitors of PI3K/AKT signaling can enhance ADI-PEG20-mediated cell killing through apoptosis
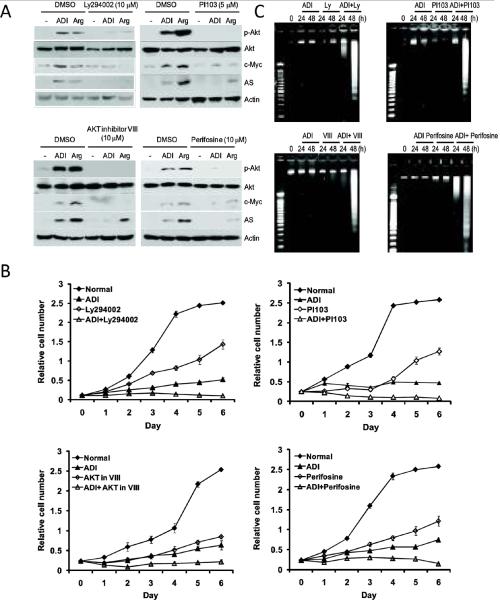
The results above suggest that targeting the Ras/PI3K/AKT pathway may enhance ADI-PEG20's cell killing capacity by preventing AS expression. As a proof of principle, we tested two PI3K inhibitors, Ly294002 and PI103, both are pan-PI3K inhibitors for class I (PI3Kα/β/γδ) (27). We also included two AKT inhibitors, VIII which is an AKT1 and AKT2 isozyme-selective inhibitor (28) and perifosine which targets the pleckstrin homology (PH) domain of Akt, preventing its translocation to the plasma membrane (28, 29). These inhibitors may have various off-targets when high concentrations (50 μM) were used. A2058 cells were maintained in normal medium, ADI-containing medium, or Arg-deficient medium, each with or without these inhibitors (10 or 5 μM). As anticipated, cells cultured in Arg-deficient medium, like those grown in the ADI-PEG20-containing medium, upregulated AS and c-Myc expression (Fig. 5A). All four inhibitors suppressed the induction of pT308-AKT, c-Myc, and AS expression by either ADI-PEG20 or Arg-negative culture conditions (Fig. 5A). While each of these inhibitors shows dose-dependent anti-cell proliferative activities; when in combination with ADI-PEG20, each of these inhibitors showed additive effects on cell growth as compared with ADI-PEG20 or individual inhibitors alone (Fig. 5B). Furthermore, while we found that A2058 cells treated with each of these four inhibitors or arginine-deprivation for up to 48 hrs only produced minimally detectable DNA fragmentation; combination treatment with ADI-PEG20 produced significant effects on genomic DNA fragmentation (Fig. 5C). These results demonstrate that inhibition of PI3K/AKT pathway enhances the cell killing capacity of ADI-PEG20.
Fig. 5.
Enhanced ADI-PEG20-mediated cell killing by PI3K and AKT inhibitors in A2058 cells. (A) Cells were maintained in normal medium, ADI-PEG20-containing, or Arg-free medium without (DMSO as vehicle) or without PI3K inhibitors (Ly294002, PI103) or AKT inhibitors (VIII and perifosine) for 48-h. Expression levels of pT308-AKT, c-Myc, and AS were analyzed by western blotting. (B) Effects of various PI3K and AKT inhibitors on ADI-PEG20 sensitivity. A2058 cells maintained in normal medium or medium containing 0.3 μg/mL ADI were treated with or without 5 μM LY294002, 1 μM PI103, 5 μM AKT inhibitor VIII, and 5 μM perifosine for 72-h. The antiproliferative effects of the combination of PI3K/AKT inhibitors and ADI-PEG20 were determined by the SRB assay. Error bars represent standard deviation from 6 independent experiments. (C) DNA fragmentation assay of ADI-PEG20-induced apoptosis. DNA extracted from A2058 cells treated with 5 μM LY294002, 1 μM PI103, 5 μM AKT inhibitor VIII, or 5 μM perifosine in the absence or presence ADI-PEG20 for 24 or 48 hours or ADI-PEG20 alone were subjected to the DNA fragmentation assay. DNA samples and molecular size markers (M) were analyzed by electrophoresis on 2% agarose gels. DNA was stained with ethidium bromide and photographed.
PI3K inhibitor can enhance ADI-PEG20's antitumor activity in animal tumor model
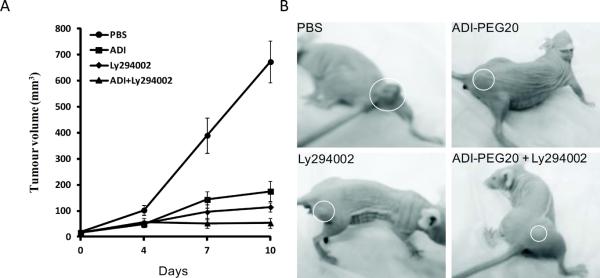
To test whether PI3K inhibitor would enhance antitumor activity of ADI-PEG20 in vivo, we treated A2058 xenografts in animals with vehicle, ADI-PEG20, Ly294002, or combination of both drugs. Ten days after treatments, we found that the average tumor volumes were 672.6±80.9, 174.6±38.4, 114.2±17.3, and 51.6±19.5 mm3, in the PBS-, and ADI-PEG20-, Ly294002-, and combination of ADI-PEG20 and ADI-PEG20)-treated groups, respectively (Fig. 6A). These corresponded to 74% (p <0.001),83% (p <0.001),and 92% (p <0.0005) reduction of tumor volumes in the ADI-PEG20-, Ly294002-, and ADI-PEG20+Ly294002-treated groups, respectively, as compared with the PBS-treated group. Representative tumor volumes from each group are shown in Fig. 6B. These results demonstrate that Ly294002 can enhance ADI-PEG20's antitumor activity in vivo.
Fig. 6.
Enhancement of ADI-PEG20's antitumor activity by Ly294002 in the tumor xenografts. (A) Inhibition of tumor growth in the ADI-PEG20-, Ly294002-, and combination of ADI-PEG20+Ly294002-treated group as compared with the PBS-treated group; and (B) representative tumor sizes in animals from the four different treated groups.
Discussion
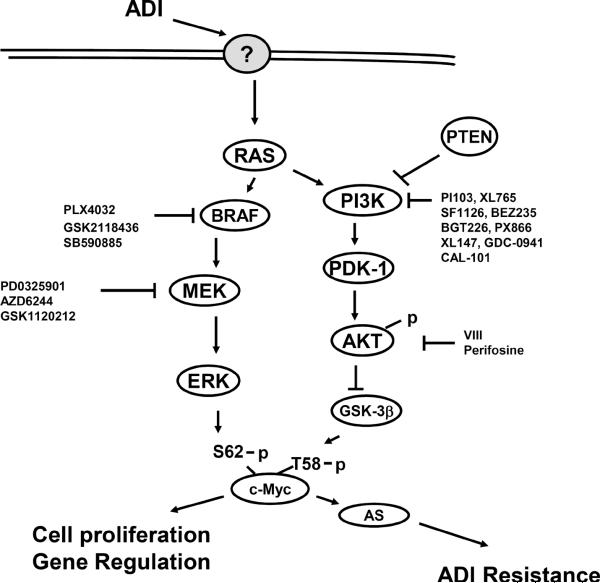
In this study, we elucidated the underlying mechanism associated with c-Myc induction by ADI-PEG20. Our results, as schematically summarized in Fig. 7, show that ADI-PEG20 activates Ras signal and its downstream kinase cascades, resulting in c-Myc stabilization. C-Myc has been recognized as one of important mammalian transcription regulator. Chromatin immunoprecipitation studies have revealed that about 10 ~ 20% of total cellular genes are regulated by c-Myc through E-box interactions (30, 31). These genes are involved in a vast variety of cellular functions including cell proliferation, differentiation, survival, apoptosis, metabolism, invasion and metastasis (32, 33). Multiple regulation mechanisms starting from transcriptional initiation to protein turnover are involved in c-Myc expression in response to various extracellular challenges (18). The molecular mechanism underlying how c-Myc stabilization is induced in response to Arg deprivation as elucidated in this study exemplifies the exquisite regulation mechanism of c-Myc expression in drug resistance research.
Fig. 7.
Signal transduction pathways involved in the induction of c-Myc stabilization by ADI-PEG20 treatment elucidated in this study. Some inhibitors of the components of this pathway are listed. ? denotes the hypothetical upstream target of the Ras signal as elucidated in this study.
At least 8 protein factors (including Fbw7, USP28, Fbx29, Skp2, β-TRCP, Truss, Hect-19, and Trim32) have been reported to control c-Myc stability through ubiquitination (for review, see ref (18)). Among these, the Fbw7/USP28 ubiquitination/deubiquitination system is the best characterized partners. Whereas Fbw7 binds to phosphorylated c-Myc, USP-28 does not bind to c-Myc directly but through interaction with Fbw7 and reverses the ubiquitination built by Fbw7 (20, 34), thereby protecting c-Myc from degradation. These observations suggest that cellular levels of USP-28 have a profound influence on Fbw7-mediated c-Myc stabilization. In the current study we found that ADI-PEG20 treatment drastically increased the association between USP-28 and c-Myc. These findings underscore the important role of USP28 in ADI-PEG20-induced c-Myc accumulation, although it remains possible that other protein factors could also be involved.
The importance of Fbw7/USP-28 in the ADI-PEG20-dependent c-Myc regulation is also supported by the elucidation of their upstream signal transduction pathways. The role of PI3K in regulation of c-Myc has been supported by several recent reports showing that inhibition of PI3K activity resulted in amplification or overexpression of c-Myc, and that c-Myc overexpression could confer resistance to PI3K inhibitors, although the underlying mechanism was not elucidated in these reports (35–37).
We could detect activation of Ras signaling in melanoma cells within 5 min after ADI-PEG20 treatment. Such rapid activation suggests an intricate sensing mechanism for Arg deficiency in melanoma cells. Whether the sensing mechanism involves activation of membrane-bound receptor(s) that transmit signal to Ras resulting in c-Myc accumulation remains to be investigated (Fig. 7). The Ras pathway is a key regulator of cancer cell survival and has been shown to be constitutively active in many cancer types, including the four cell lines used in this study, either by BRAF(V600E) mutations (A2058 and A375) or Ras mutations (SK-MEL-2), or both (MDA-MB-231) (38). Notably, we found correlations between Ras/PI3K/AKT/ERK activation and c-Myc/AS inducibility by ADI-PEG20 among four melanoma cell lines. While the steady-state level of c-Myc level in A375 cells is higher than those in other three cell lines, the inability of c-Myc/AS induction in A375 cells is due to defective Ras upstream signal, and elucidation of this defective mechanism is currently underway in our laboratories. We previously demonstrated that lack of c-Myc/AS induction by ADI-PEG20 in A375 cells is associated with the inability of developing resistance to ADI-PEG20 (9). While the number of cell lines used remains small and further in-depth studies are needed, our current results suggest that Ras pathway may offer not only as a potential target for therapeutic intervention but also as a useful biomarker for predicting ADI-PEG20's treatment outcomes in clinical setting.
Because of the importance of the Ras/PI3K/AKT/ERK pathway in many aspects of human malignancies, including tumor growth, proliferation, invasion and sensitivity to therapy, clinical studies have been performed using inhibitors of this pathway in human cancers (39–41). Examples of these drugs are listed in Fig. 7. Of particular relevance to malignant melanoma are those targeting mutant BRAF(V600E), a recurrent mutation occurs in 50% to 60% of malignant melanomas. Clinical trials of inhibitors targeting this mutation have shown remarkable results (42); however, resistance to BRAF(V600E) almost inevitably developed after prolonged treatment, through activation of alterative/compensatory survival pathways, such as PDGFRβ or N-Ras (43), MAPK (44) and IGF-1R/PI3K (45), and combined treatment with inhibitors of IGF-1F/PI3K induces cell death in BRAF-resistant variants (45). The present demonstration that inhibitors of the PI3K/AKT pathway can enhance the cell killing capacity by ADI-PEG20 suggests that intervention of this pathway may have added advantages for ADI-PEG20 cancer chemotherapy. We envision that inhibitors of BRAF(V600E), like the inhibitors of anti-PI3K and anti-AKT that have been demonstrated in this study, would also enhance ADI-PEG20's cell killing effects. These approaches of simultaneously targeting two prevalent genetic abnormalities in melanoma (BRAF mutation and Arg-auxotrophicity) may improve the treatment efficacy of malignant melanoma.
Finally, our present study on ADI-PEG20 in melanoma cells may provide important information for the use of other Arg-degrading enzymes in cancer chemotherapy, such as recombinant arginases which convert Arg into orinithine and urea. Recombinant arginases have also been in various stages of clinical development for human cancer chemotherapy (46–49).
Acknowledgement
We thank Dr. Bor-wen Wu (Polaris) for ADI-PEG20 and anti-AS antibody, Drs. Geoffrey Cooper (Boston Univ.) and Stephen Elledge (Harvard Med. School) for the recombinant DNA.
Grant Support This research was supported in part by ROI-CA152197 (to M. T. K. and L. F.) from the National Cancer Institute.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Blank CU, Hooijkaas AI, Haanen JB, Schumacher TN. Combination of targeted therapy and immunotherapy in melanoma. Cancer Immunol Immunother. 2011;60:1359–71. doi: 10.1007/s00262-011-1079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 3.Feun L, Savaraj N. Topoisomerase I inhibitors for the treatment of brain tumors. Expert Rev Anticancer Ther. 2008;8:707–16. doi: 10.1586/14737140.8.5.707. [DOI] [PubMed] [Google Scholar]
- 4.Kuo MT, Savaraj N, Feun LG. Targeted cellular metabolism for cancer chemotherapy with recombinant arginine-degrading enzymes. Oncotarget. 2010;1:246–51. doi: 10.18632/oncotarget.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ascierto PA, Scala S, Castello G, Daponte A, Simeone E, Ottaiano A, et al. Pegylated arginine deiminase treatment of patients with metastatic melanoma: results from phase I and II studies. J Clin Oncol. 2005;23:7660–8. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- 6.Ni Y, Schwaneberg U, Sun ZH. Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 2008;261:1–11. doi: 10.1016/j.canlet.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 7.Cheng PN, Lam TL, Lam WM, Tsui SM, Cheng AW, Lo WH, et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67:309–17. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 8.Izzo F, Marra P, Beneduce G, Castello G, Vallone P, De Rosa V, et al. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: results from phase I/II studies. J Clin Oncol. 2004;22:1815–22. doi: 10.1200/JCO.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 9.Tsai WB, Aiba I, Lee SY, Feun L, Savaraj N, Kuo MT. Resistance to arginine deiminase treatment in melanoma cells is associated with induced argininosuccinate synthetase expression involving c-Myc/HIF-1alpha/Sp4. Mol Cancer Ther. 2009;8:3223–33. doi: 10.1158/1535-7163.MCT-09-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo MT, Liu Z, Wei Y, Lin-Lee YC, Tatebe S, Mills GB, et al. Induction of human MDR1 gene expression by 2-acetylaminofluorene is mediated by effectors of the phosphoinositide 3-kinase pathway that activate NF-kappaB signaling. Oncogene. 2002;21:1945–54. doi: 10.1038/sj.onc.1205117. [DOI] [PubMed] [Google Scholar]
- 11.Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–43. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–32. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–42. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 14.Cai Z, Semenza GL. PTEN activity is modulated during ischemia and reperfusion: involvement in the induction and decay of preconditioning. Circ Res. 2005;97:1351–9. doi: 10.1161/01.RES.0000195656.52760.30. [DOI] [PubMed] [Google Scholar]
- 15.Tsai WB, Chung YM, Zou Y, Park SH, Xu Z, Nakayama K, et al. Inhibition of FOXO3 tumor suppressor function by betaTrCP1 through ubiquitin-mediated degradation in a tumor mouse model. PLoS One. 5:e11171. doi: 10.1371/journal.pone.0011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–97. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–26. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2010;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 19.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 21.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–7. [PubMed] [Google Scholar]
- 23.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–8. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 24.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 25.Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim Biophys Acta. 2000;1470:M21–35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 26.Berggren M, Sittadjody S, Song Z, Samira JL, Burd R, Meuillet EJ. Sodium selenite increases the activity of the tumor suppressor protein, PTEN, in DU-145 prostate cancer cells. Nutr Cancer. 2009;61:322–31. doi: 10.1080/01635580802521338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–57. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calleja V, Laguerre M, Parker PJ, Larijani B. Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. doi: 10.1371/journal.pbio.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gills JJ, Dennis PA. Perifosine: update on a novel Akt inhibitor. Curr Oncol Rep. 2009;11:102–10. doi: 10.1007/s11912-009-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res. 2011;71:2034–7. doi: 10.1158/0008-5472.CAN-10-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popov N, Herold S, Llamazares M, Schulein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–31. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 35.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7:787–93. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowles DW, Jimeno A. New phosphatidylinositol 3-kinase inhibitors for cancer. Expert Opin Investig Drugs. 2011;20:507–18. doi: 10.1517/13543784.2011.562192. [DOI] [PubMed] [Google Scholar]
- 40.Pal SK, Reckamp K, Yu H, Figlin RA. Akt inhibitors in clinical development for the treatment of cancer. Expert Opin Investig Drugs. 2010;19:1355–66. doi: 10.1517/13543784.2010.520701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 42.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsui SM, Lam WM, Lam TL, Chong HC, So PK, Kwok SY, et al. Pegylated derivatives of recombinant human arginase (rhArg1) for sustained in vivo activity in cancer therapy: preparation, characterization and analysis of their pharmacodynamics in vivo and in vitro and action upon hepatocellular carcinoma cell (HCC) Cancer Cell Int. 2009;9:9. doi: 10.1186/1475-2867-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal V, Woo JH, Mauldin JP, Jo C, Stone EM, Georgiou G, et al. Cytotoxicity of human recombinant arginase I (Co)-PEG5000 in the presence of supplemental L-citrulline is dependent on decreased argininosuccinate synthetase expression in human cells. Anticancer Drugs. 2011;23:51–64. doi: 10.1097/CAD.0b013e32834ae42b. [DOI] [PubMed] [Google Scholar]
- 48.Lam TL, Wong GK, Chow HY, Chong HC, Chow TL, Kwok SY, et al. Recombinant human arginase inhibits the in vitro and in vivo proliferation of human melanoma by inducing cell cycle arrest and apoptosis. Pigment Cell Melanoma Res. 2010;24:366–76. doi: 10.1111/j.1755-148X.2010.00798.x. [DOI] [PubMed] [Google Scholar]
- 49.Stone EM, Glazer ES, Chantranupong L, Cherukuri P, Breece RM, Tierney DL, et al. Replacing Mn(2+) with Co(2+) in human arginase i enhances cytotoxicity toward l-arginine auxotrophic cancer cell lines. ACS Chem Biol. 2010;5:333–42. doi: 10.1021/cb900267j. [DOI] [PMC free article] [PubMed] [Google Scholar]