Abstract
Over the last 15 years, double stranded DNA (dsDNA) has been used as a model polymeric system for nearly all single polymer dynamics studies. However, dsDNA is a semiflexible polymer with markedly different molecular properties compared to flexible chains, including synthetic organic polymers. In this work, we report a new system for single polymer studies of flexible chains based on single stranded DNA (ssDNA). We developed a method to synthesize ssDNA for fluorescence microscopy based on rolling circle replication, which generates long strands (>65 kb) of ssDNA containing “designer” sequences, thereby preventing intramolecular base pair interactions. Polymers are synthesized to contain amine-modified bases randomly distributed along the backbone, which enables uniform labelling of polymer chains with a fluorescent dye to facilitate fluorescence microscopy and imaging. Using this approach, we synthesized ssDNA chains with long contour lengths (>30 μm) and relatively low dye loading ratios (~1 dye per 100 bases). In addition, we used epifluorescence microscopy to image single ssDNA polymer molecules stretching in flow in a microfluidic device. Overall, we anticipate that ssDNA will serve as a useful model system to probe the dynamics of polymeric materials at the molecular level.
Introduction
Polymeric materials exhibit complex behaviour upon deformation and exposure to non-equilibrium conditions. Polymer chains stretch and orient in solution-based fluid flows, which gives rise to flow-dependent material properties such as bulk stress and viscosity. Ultimately, the macroscopic material response is determined by the underlying microstructure and molecular properties of deformed polymer chains. Polymer dynamics have been studied using a wide array of experimental and theoretical tools.1,2 Single molecule techniques allow for the direct observation of chain dynamics at the molecular level, thereby enabling characterization of polymer backbone motion and chain stretching pathways. Over the last 15 years, double stranded DNA (dsDNA) has served as the model system to study single polymer dynamics using fluorescence microscopy.3 In particular, dsDNA has been extensively used in single polymer experiments to study chain relaxation,4 coil diffusion,5 stretching in uniform flow,6 dynamics in extensional,7–9 shear10 and mixed flow,11 polymer conformation hysteresis,12 and in bulk measurements of turbulent drag reduction.13,14 Recently, dsDNA has been used to directly observe polymer dynamics in confined geometries.15–18
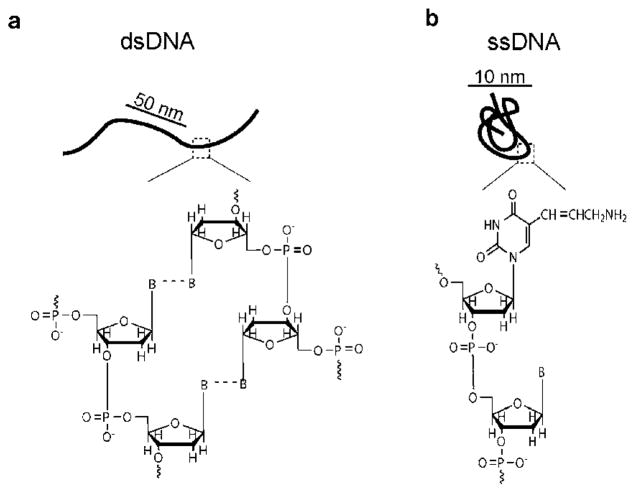
Single polymer experiments probing the dynamics of dsDNA have largely relied on lambda (λ)-DNA as a model system. Lambda DNA exhibits several advantages as “model” polymer, including facile sample preparation and fluorescent labelling. In addition, monodisperse samples of λ-DNA are readily available from commercial vendors. However, dsDNA is a semiflexible polymer with markedly different molecular properties compared to flexible polymer chains (Fig. 1). Under physiological salt concentrations, the persistence length of native, unlabeled dsDNA is ~53 nm, which is approximately two orders of magnitude larger than the persistence length of common flexible synthetic organic polymers. Using single polymer techniques based on fluorescence microscopy, dsDNA is commonly labelled with intercalating dyes,3 which alter the local double helix structure and increase both the polymer persistence length and contour length.5 Chain flexibility and local molecular properties play a key role in overall chain dynamics. Recently, the elasticity of single flexible polymer molecules was measured using ssDNA, and the force–extension relationship revealed pronounced differences in the low-force elastic behaviour for ssDNA compared to semiflexible polymers such as dsDNA.19,20 Overall, there is a general need for development of new experimental approaches to extend the techniques of single polymer visualization to broader classes of polymeric materials, including flexible polymer chains.
Fig. 1.
Polymer chain structures are shown for (a) double stranded DNA, a semiflexible polymer with a double helix backbone and (b) single stranded DNA, a flexible chain consisting of 5-carbon sugars linked by phosphodiester bonds. For ssDNA, a modified base with primary amine (aminoallyl-uracil) is shown, and ‘B’ represents a natural nucleobase.
In this work, we report a new model system for single polymer studies of flexible chains based on single stranded DNA (ssDNA). We developed a versatile platform to synthesize ssDNA containing “designer” sequences, which prevents intra-molecular base pair formation. Using a biochemical synthesis scheme for nucleic acids called rolling circle replication, we generated long chain polymers of ssDNA containing amine-modified bases, which serve as reactive chemical moieties for uniform labelling along the polymer backbone with fluorescent dyes. Using this technique, we synthesize a new class of molecules for single polymer experiments, and we explore the practical sequence space for producing ssDNA molecules using this replication method. Following synthesis and characterization, we directly imaged fluorescently labelled ssDNA molecules in a microfluidic device using epifluorescence microscopy. Images of single polymer chains stretching in fluid flow reveal pristine polymer backbones, thereby demonstrating the utility of the biochemical labelling strategy. In the following discussion, we present the synthesis and characterization scheme for ssDNA polymers. Overall, this method will enable investigation of a new class of polymeric materials at the molecular-level.
Experimental
Rolling circle replication
Oligonucleotide templates and primers for rolling circle replication (RCR) were designed using VectorNTI software and synthesized by Integrated DNA Technologies (Coralville, IA). Table 1 shows the sequences of template oligonucleotides used in these experiments. Template oligonucleotides were designed to generate ssDNA products rich in either purine or pyrimidine nucleotides. In some cases, product ssDNA was designed to be nearly homopolymeric. All template oligos contained a 5′ phosphorylated terminus to enable formation of the minicircle template by ligation.
Table 1.
RCR template sequences
| Sequence | Template sequencea | # nt | Longest 1 nt stretch | RCR success |
|---|---|---|---|---|
| 1 | 5′-ACTCTTC(T)3A(T)3C(T)4ACTTTCCAT | 28 | 4 | Yes |
| 2 | 5′-CTTTCTTACACT(TC)4ATTC(T)5AC(T)4ACTTCTTCCTACT | 48 | 5 | Yes |
| 3 | 5′-ACTCTTTCTTACTTCTCTCTACTTC(T)6CACTTCTCTCTAC(T)4ATTTCCTTACCTTCTTCT | 66 | 6 | Yes |
| 4 | 5′-AGGTGT(G)4A(G)4T(G)4ATTGGATG | 28 | 4 | Yes |
| 5 | 5′-GGTAGGTGGT((G)3AT)2(G)3T((G)3AT)2GGATGGTGAGTGGA | 48 | 3 | Yes |
| 6 | 5′-AGGTGAGTGGA(G)4TA(G)3TT(G)3T(G)3A(G)3TGA(G)3TGGTTGGA(G)4TA(G)3ATGGTGGATG | 66 | 4 | Yes |
| 7 | 5′-AGAGGG(G)16GGAGGA | 28 | 21 | No |
| 8 | 5′-ATATTT(T)7A(T)8TTATTA | 28 | 10 | Yes |
| 9 | 5′-ACACCCCCCCCCA(C)9A(C)9A(C)9A(C)9ACCCCCCCCCACCA | 66 | 9 | No |
| 10 | 5′-ATATTTTTTTTTA(T)9A(T)9A(T)9A(T)9ATTTTTTTTTATTA | 66 | 9 | Yes |
| Pyr 1 | 5′-GGAAGA(G)5A(G)4A(G)4AGAGAAG | 28 | 5 | No |
| Pyr 2 | 5′-AAGAGA(G)7A(G)7AGAAGGA | 28 | 7 | No |
| Pyr 3 | 5′-AGA(G)3GAGAGAGAGAGAGAGA(G)4AA | 28 | 4 | No |
| Pyr 4 | 5′-GAGGAAGAGAGAGAGAGAGAGAGGAAAG | 28 | 3 | No |
| Pyr 5 | 5′-GAGGAAGGGAGGAGAGAGGAGAGGAGGGAGAGAGAGAGGAGGAAA | 46 | 3 | No |
| Pyr 6 | 5′-GGGAGA(A)4G(A)7GAAAGGAGAA | 28 | 7 | Yes |
| Pyr 7 | 5′-GAGAATGAGGGATGAGAGGAAGAGGAAA | 28 | 3 | No |
| Pyr 8 | 5′-AGGAGA(A)4G(A)7G(A)3AGAAGG | 28 | 7 | Yes |
| Pyr 9 | 5′-GAG(A)7AAG(A)9G(A)9G(A)9G(A)9GAAA(A)6GAAG | 66 | 9 | Yes |
| Pyr 10 | 5′-AGGAGT(G)4A(G)4T(G)4AAAGGATG | 28 | 4 | Yes |
Bold values indicate nucleotides that participate in hybridization.
Linear templates were circularized by a ligation reaction using T4 DNA ligase (New England Biolabs).21 First, a reaction mixture (500 μL) consisting of 200 nM template and primer oligonucleotides in T4 DNA ligase buffer (10 mM Tris/Tris–HCl, 200 μM EDTA, 2 mM NaCl) was prepared. Template and primer oligonucleotides were hybridized by heating at 70 °C for 2.5 minutes, followed by slow cooling to room temperature (20 °C). Next, 600 units of T4 DNA ligase were added, and the ligation reaction proceeded for 5 hours at 16 °C. In the case of Exonuclease I treatment, 20 μL (4 pmol) of the ligated/primed minicircle mixture was incubated in Exonuclease I reaction buffer (67 mM glycine–KOH, 6.7 mM MgCl2, 10 mM 2-mercaptoethanol, pH 9.5) with 10 units of Exonuclease I at 37 °C for 30 minutes. The Exo I enzyme was deactivated by heating to 80 °C for 20 minutes. Ligated/primed minicircle templates were stored at 4 °C.
RCR reactions consisted of 50 nM ligated/primed minicircle, 200 μM dTTP/aa-dUTP, 500 μM custom-mix dNTPs (containing only nucleotides in the synthesized product sequence) and 200 μg mL−1 BSA in phi29 DNA polymerase reaction buffer (50 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM MgCl2, 4 mM dithiothreitol, pH 7.5) in a total reaction volume of 50 μL. The polymerization reaction was initiated by addition of 5 units of phi29 polymerase, and the reaction proceeded for 30 minutes at room temperature, unless otherwise noted. In negative control (NC) reactions, no phi29 polymerase was added. Reactions were terminated by addition of EDTA to a final concentration of 20 mM, and the products were stored at 4 °C.
Gel electrophoresis was used to characterize oligonucleotide products from ligation reactions and ssDNA generated by RCR. Polyacrylamide gels (8%) were run in 1× TAE buffer (Biorad) to assay the efficiency of ligation reactions in generating ligated/primed minicircles. Polyacrylamide gels were initially equilibrated in running buffer without DNA for 20 min at 50 V, and ligated/primed oligos were loaded and run for 45 minutes at 80 V. Low molecular weight DNA ladder (NEB) was used as a standard marker. Polyacrylamide gels were post-stained with SYBR Gold (1× in TAE buffer) for 15 minutes and imaged. To assay ssDNA products from RCR reactions, agarose gels (0.6%) were run in TAE buffer using 1 kb DNA ladder (NEB) supplemented with 50 ng of λ-DNA as a standard marker. Agarose gels were run for 30 minutes at 120 V and post-stained with SYBR Gold (1× in TAE buffer) for 30 minutes. All gels were imaged using a Gel Doc 2000 (Biorad), and fluorescent gel images are inverted for display. All enzymes, including T4 DNA ligase, phi29 DNA polymerase, Exonuclease I, and DNA markers for electrophoresis and dNTP stocks were purchased from New England Biolabs. Aminoallyl dUTP (aa-dUTP) and SYBR Gold were purchased from Invitrogen. All other chemicals were obtained from Sigma-Aldrich or Fischer Scientific and were molecular biology grade purity.
Fluorescent dye labelling and quantification
Single polymer imaging was performed using ssDNA products generated from template Sequence 1 (Table 1). RCR reaction times were increased to 90 minutes to generate long ssDNA products appropriate for single molecule fluorescence microscopy. Prior to fluorescent labelling, ssDNA from RCR reactions was purified using a HiTrap desalting column and fast protein liquid chromatography (AKTA FPLC, GE Biosciences). Before sample purification, the RCR reaction solution should be supplemented to ~8 M guanidine–HCl, thereby allowing removal of template minicircle oligos (complementary to ssDNA product) and dNTPs via FPLC/desalting. Approximately 800 μL of the RCR reaction was loaded onto the HiTrap column and eluted into Alexa Fluor 532 labelling buffer (100 mM sodium bicarbonate, pH 8.0).22 Purification and ssDNA concentration prior to labelling was performed using Vivacon2 spin columns, 100 kDa MWCO (Sartorius). Approximately 2 mL of eluted ssDNA sample was loaded onto the spin columns and centrifuged at 2500 × g for 45 minutes, which yielded a ~25 to 30× increase in DNA concentration, as determined by measuring absorbance at 260 nm using a Nanodrop 1000 (Thermo Scientific). Purified ssDNA (5 μg) was labelled with 2 μL of 30 μg μL−1 NHS-ester Alexa Fluor 532 (Invitrogen). Solutions were mixed thoroughly by pipet and the reaction proceeded for 1 hour at room temperature in the dark. In all labelling reactions, DNA and dye concentrations were maintained constant to standardize reaction conditions. Labelling reactions were quenched by addition of 1 M Tris/Tris–HCl (pH 8.0) to a final volume of 70 μL.
Following the dye labelling reaction, ssDNA products were purified from unreacted fluorescent dye to enable single molecule imaging and for quantification of dye labelling ratios. Immediately following quenching of the ssDNA labelling reactions, reaction mixtures were purified using two successive Biospin 6 columns (Biorad), which also served as a buffer exchange to elute fluorescently labelled ssDNA products in 10 mM Tris/Tris–HCl buffer (pH 8.0). Dye labelling ratios were determined using absorbance measurements. Fluorescently labelled ssDNA for single molecule experiments was supplemented with EDTA (10 mM) and diluted with 50% (v/v) glycerol for storage at −20 °C.
Bulk fluorescence measurements of enzymatically digested fluorescently labelled ssDNA were used to quantify the extent of putative dye interactions (if any) along the polymer backbone. Here, 1.5 μg of labelled ssDNA sample was split into two, equal concentration reactions. One reaction treated with an excess of DNase I for 30 minutes at 37 °C, and the second reaction was untreated. Bulk fluorescence measurements were performed by illuminating samples at excitation wavelengths between 400 and 540 nm, while monitoring emission at 555 nm using a Cary Eclipse Fluorescence Spectrophotometer (Varian).
Epifluorescence microscopy
We directly visualized single fluorescently labelled ssDNA molecules using epifluorescence microscopy. Single stranded DNA was imaged using an Olympus IX71 inverted microscope equipped with a 100× oil immersion objective lens (Olympus UPlanSApo) and an Andor Ixon EMCCD camera. A solid-state laser (CrystaLaser) was used as an illumination source at a wavelength of 532 nm. Polymers were imaged in viewing solution containing 50 mM Tris/Tris–HCl (pH 8.0), 1 mM EDTA, 5 mg mL−1 glucose, 20 mM NaCl, and ~95% glycerol by weight. Prior to microscopy, solutions were purified with a 0.45 μm membrane filter, followed by addition of β-mercaptoethanol (140 mM), glucose oxidase (~65 U mL−1) and catalase (1.1 kU mL−1) as oxygen scavenging reagents to minimize photobleaching of the Alexa Fluor dye. For viewing, ~50 ng of fluorescently labelled ssDNA was added to 1 mL of viewing solution (yielding ~1 to 10 pM ssDNA). Individual ssDNA molecules were visualized both in quiescent conditions on a microscope slide and in planar extensional flow generated in a PDMS-based microfluidic device, which effectively stretches ssDNA molecules for backbone visualization (ESI†). Images were processed using ImageJ software. All images were corrected for background, false colour was applied, and in some cases, levels were adjusted to filter approximately 15–20% of the lowest intensity pixels for noise reduction.
Results and discussion
Polymer backbone structures for double stranded DNA and single stranded DNA are shown in Fig. 1. Single stranded DNA is a flexible polymer containing monomers that are free to rotate around torsional bonds. In the following discussion, we describe a biochemical synthesis scheme to generate flexible ssDNA polymer chains for single polymer studies.
Rolling circle replication
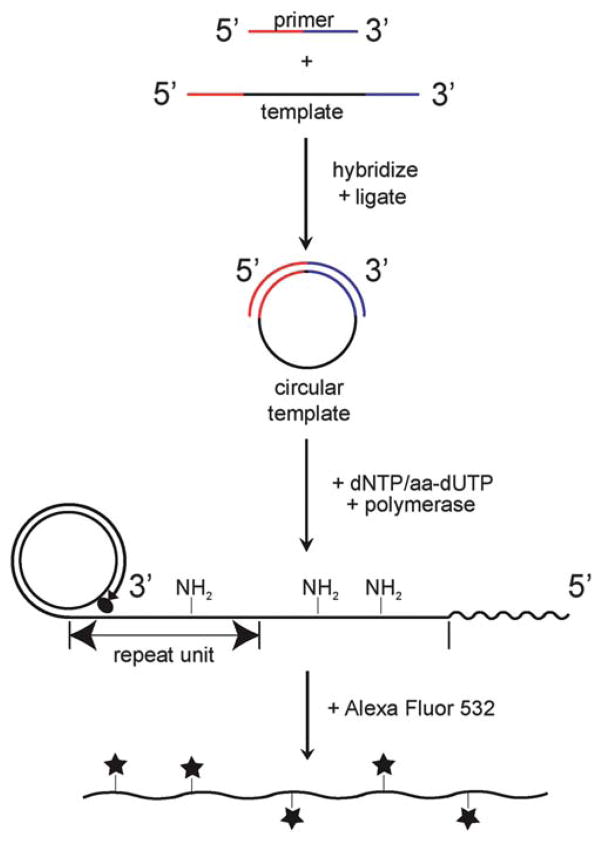
We synthesized ssDNA containing user-defined sequences using rolling circle replication (RCR). RCR is a useful method to generate long-chain molecules of ssDNA amenable for direct visualization by fluorescence microscopy. Previously, RCR has been used for multiple applications,23 including biodetection and DNA synthesis in microfluidic systems.24 Fig. 2 shows a schematic of the RCR reaction scheme. First, an oligonucleotide template is hybridized to a complementary primer strand, thereby circularizing the template strand. Next, the template strand is ligated to yield covalently closed circular ssDNA.25–27 During RCR, DNA polymerase binds to the free 3′ hydroxyl on the primer strand and initiates replication of the circular template. In rolling circle synthesis, DNA polymerase replicates around and traverses circular templates for hundreds to thousands of cycles, thereby generating long strands of ssDNA. In this work, we use phi29 DNA polymerase, which exhibits exceptional processivity and strand displacement activity28,29 and yields long ssDNA product (>65 kb). RCR reactions are supplemented with aminoallyl dUTP (aa-dUTP), which is a modified nucleotide containing a primary amine (Fig. 1b) as a reactive chemical moiety to facilitate subsequent dye-labelling with succinimidyl ester dyes. The aa-dUTP nucleotide contains only a small modification compared to natural dTTP and was readily incorporated by phi29 DNA polymerase, with no observable change to ssDNA product length distributions.
Fig. 2.
Schematic of rolling circle replication scheme for ssDNA synthesis. DNA polymerase is represented by an oval shape, and stars represent fluorescent dye molecules (Alexa Fluor 532).
Table 1 contains a summary of oligonucleotide template sequences and RCR reaction results. Template ssDNA sequences were designed to allow for dye labelling and to prevent intramolecular base pairing. To facilitate labelling, adenine monomers were designed to occur in template strands at semi-random locations, thereby yielding amine-modified nucleotides in ssDNA product strands at tuneable intervals. In addition, we designed purine-rich or pyrimidine-rich templates consisting of only 2 nucleotides (A/G or C/T), thereby preventing intrachain base pairing. In some cases, a third nucleotide (A) was present at low ratios to facilitate dye-labelling by incorporation of amine-modified nucleotides (aa-dUTP). Using this method, a wide-range of template sequences was successfully replicated by RCR, yielding long ssDNA products (>65 kb). In some cases, we observed limitations on template sequence design, as evidenced by low reaction yields or short products. In general, RCR reactions were inhibited for templates containing consecutive stretches of a single nucleotide, particularly cytosine (C) or guanine (G). Indeed, previous research has demonstrated that some polymerases are inhibited by homopolymeric sequences due to enzymatic ‘slippage’, and guanine-rich sequences can form quaternary structures, which may prevent ligation.30 For some sequences, purine-rich templates failed to replicate entirely, and many of the unsuccessful reactions contained templates with larger amounts of guanine compared to the successful reactions. Overall, RCR success or failure appeared to be independent of sequence length for circular DNA templates in the range of 28–66 bases, as observed for Sequences 1–3 and Pyr 4 and 5.
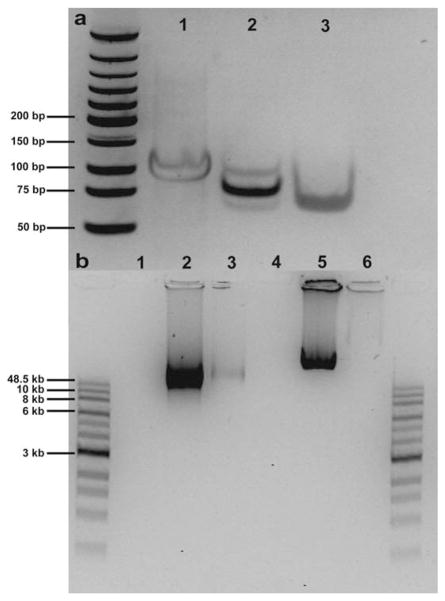
Polyacrylamide gel electrophoresis (PAGE) was used to assay ligation of circular template strands. Fig. 3a shows a gel image containing linear templates and ligated/primed mincircles for Sequence 10, where linear templates migrate slower than minicircles. In Lane 2, the bright band corresponds to primed minicircle templates, and the upper and lower weaker bands correspond to unreacted linear templates and unprimed mini-circles, respectively. For some ligation reactions, primed mini-circle templates were treated with exonuclease (Exo I), which specifically degrades linear DNA. Samples treated with Exo I show single bands on PAGE gels corresponding to unprimed closed circular ssDNA (Lane 3). Following ligation, primed minicircle templates were carried forward to 30 minute RCR reactions. Fig. 3b shows an image of ssDNA products from RCR reactions analysed using native, non-denaturing agarose gel electrophoresis. For minicircle templates treated with Exo I, ssDNA product bands are faint due to near complete digestion of the minicircle primer, which is required for replication. In addition to native gel electrophoresis, we also used denaturing alkaline gel electrophoresis to determine ssDNA product lengths (ESI†). In general, higher molecular weight products were obtained for ssDNA rich in purines (A/G) compared to pyrimidine rich products. For most ssDNA sequences, products migrated with similar mobility on both native and denaturing gels, suggesting the absence of base pairing in ssDNA products. Interestingly, ssDNA products rich in pyrimidine bases (generated by Sequences 4–6 and Pyr 6, 8–10) showed inhibited migration through native agarose. For these sequences, denaturing gels were used to quantify product length distributions, in addition to single molecule visualization.
Fig. 3.
Rolling circle replication for ssDNA. (a) PAGE gel showing minicircle template formation (Sequence 10). Lane 1: linear template, Lane 2: ligated/primed minicircles, Lane 3: Exo I treated minicircles. (b) Agarose gel showing ssDNA products from RCR for Sequence 1 (Lanes 1–3) and Sequence 10 (Lanes 4–6). Lanes 1 and 4 are negative controls (no polymerase), Lanes 2 and 5 show ssDNA products generated from primed minicircle templates, and Lanes 3 and 6 show ssDNA products from Exo I treated minicircle templates.
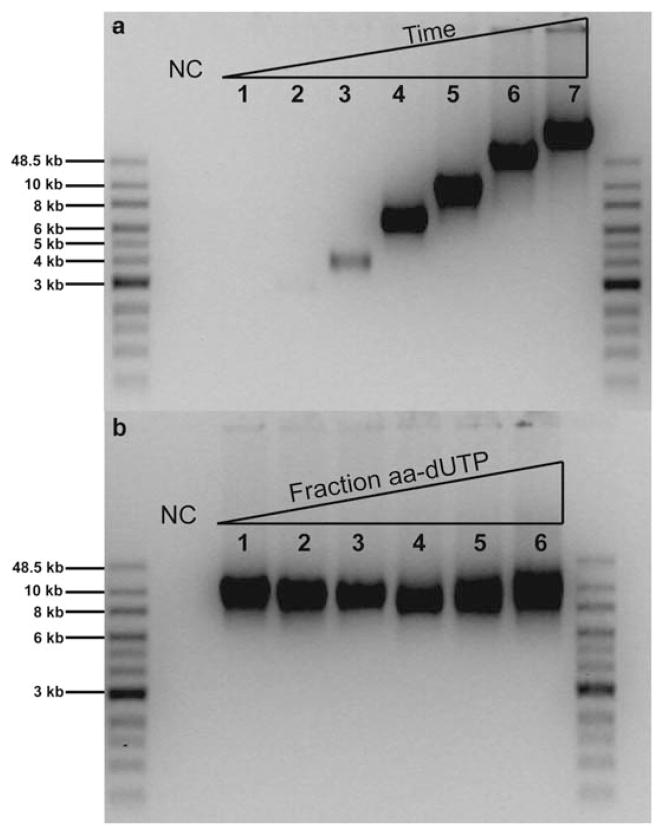
We characterized the effect of RCR reaction time on ssDNA product length distributions using Sequence 1. Fig. 4a shows an image of an agarose gel containing ssDNA products from timed RCR reactions with durations ranging from 0.5 to 180 minutes. As expected, ssDNA product length increases with increasing reaction time. In addition, product bands appear to broaden and intensify for long reaction times, indicating increases in both product yield and polydispersity. Typical yields for a single RCR reaction are on the order of 1–2 μg ssDNA product, using the reaction protocol described in this work. To increase yield for dye labelling, multiple reactions were carried out in parallel. We also varied the ratio of natural to modified nucleotide (dTTP : aa-dUTP) in the reaction mixture to determine the effect of unnatural bases on product lengths (Fig. 4b). Upon increasing the amount of aa-dUTP, product length, yield, and polydispersity remained nearly constant, which suggests that the base modification is non-perturbative for replication by phi29 polymerase. Finally, RCR reactions were successful using chemically modified oligonucleotide primers containing a 5′ biotin moiety, which could enable further ssDNA functionalization or surface immobilization for dynamic studies.
Fig. 4.
ssDNA product lengths for rolling circle replication. (a) RCR reaction time was varied between 0.5, 2, 5, 10, 30, 60, and 180 minutes in Lanes 1–7, respectively. (b) RCR products for varying ratios of natural to modified nucleotide, with dTTP : aa-dUTP ranging between 1 : 0, 4 : 1, 3 : 2, 2 : 3, 1 : 4, and 0 : 1 in Lanes 1–6, respectively. Agarose gels (0.6%) were run for 30 min at 120 V. NC: negative controls.
Fluorescent dye labelling and quantification
Following rolling circle replication, ssDNA backbones were labelled with fluorescent dye. First, we optimized RCR synthesis and labelling conditions to generate ssDNA molecules suitable for single molecule fluorescence microscopy. Overall, the goal was to label ssDNA with low amounts of fluorescent dye in order to allow for efficient visualization of single polymers, while minimizing putative modifications to the native ssDNA backbone. For determination of optimal backbone labelling, we synthesized three batches of ssDNA (Sequence 1) with varying amounts of aa-dUTP, specifically with aa-dUTP : dTTP ratios of 1 : 9, 3 : 7, and 3 : 2. All three samples were labelled with dye, purified and analysed using bulk and single molecule techniques.
Table 2 contains a summary of dye labelling reactions. The number of dye molecules per 100 bases was experimentally determined using UV and visible wavelength absorption measurements. The theoretical maximum dye labelling ratio was determined by assuming stoichiometric incorporation of aa-dUTP and quantitative completion of the labelling reaction. In general, each sample had a dye labelling ratio less than the theoretical maximum, though we determined similar labelling ratios for all samples (~1 dye : 100 bases). However, bulk fluorescence data and single molecule experiments suggest that ssDNA molecules become moderately (~2×) brighter upon increasing amounts of aa-dUTP. Experimentally measured dye-labelling ratios represent maximum labelling amounts for chain backbones and serve as an effective upper limit for dye incorporation. Overall, a labelling ratio of ~1 dye : 100 bases is low and significantly less than the typical labelling ratio for dsDNA (1 dye : 4 bp) using intercalating dyes such as YOYO-1.
Table 2.
Quantitative analysis of dye labelling reactions
| aa-dUTP : dTTP | Dyes per 100 bases | Theoretical dyes per 100 bases | Fluorescence (a.u.) |
|---|---|---|---|
| 1 : 9 | 1.60 ± 0.06 | 1.43 | 84.0 ± 5.6 |
| 3 : 7 | 1.03 ± 0.01 | 4.29 | 97.2 ± 6.8 |
| 6 : 4 | 1.40 ±0.17 | 8.57 | 159.6 ± 10.0 |
Finally, we used bulk-level fluorescence measurements in combination with a nuclease assay to probe for putative dye–dye interactions arising from π bond stacking for dye molecules along the polymer backbone. The nuclease assay showed no dye interactions, which is expected because the degree of labelling for ssDNA polymers is low (~1 dye : 100 bases) and far below labelling amounts for previously observed dye–dye interactions using Alexa fluorophores.31
Direct visualization of single polymer molecules
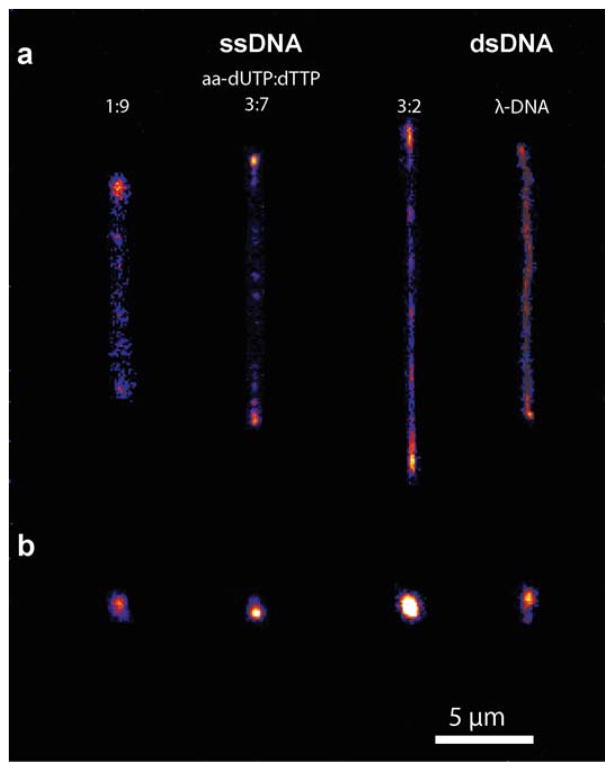
We used fluorescence microscopy to directly image single fluorescently labelled ssDNA molecules. Single ssDNA molecules generated by RCR synthesis are bright, photostable and suitable for single molecule fluorescence microscopy (Fig. 5). We directly compared images of fluorescently labelled ssDNA to images of λ-DNA labelled with an intercalating dye. Currently, λ-DNA is the standard molecule for single molecule polymer studies, and our comparison serves as a useful benchmark for fluorescent labelling studies. Single λ-DNA molecules were labelled with YOYO-1 as described elsewhere,8 and dsDNA was imaged using epifluorescence microscopy using a mercury lamp as the illumination source.
Fig. 5.
Direct visualization of fluorescently labelled ssDNA molecules using fluorescence microscopy. Single molecules of ssDNA and ds-λ-DNA are shown for (a) stretched and (b) coiled configurations. ssDNA (Sequence 1) with variable dye-labelling ratios are imaged.
Fig. 5 shows images of stretched and coiled ssDNA molecules (Sequence 1) for ssDNA samples with variable dye loadings, with a side-by-side comparison to fluorescently labelled λ-DNA. Polymer molecules are stretched using a microfluidic flow device, as described in the ESI†. Stretched ssDNA molecules were chosen to be of similar length to stretched λ-DNA (~20 μm contour length). Clearly, fluorescently labelled ssDNA molecules appear as bright polymer chains suitable for single molecule visualization. Overall, the ssDNA sample synthesized with a 3 : 2 ratio of aa-dUTP : dTTP showed comparable image quality to λ-DNA. Chain backbones appeared brighter, and the signal to noise levels for labelled ssDNA molecules improved as the aa-dUTP : dTTP ratio was increased, which is generally consistent with the fluorescence data in Table 2. Overall, these experiments serve as a useful guide to generate ssDNA for single molecule studies.
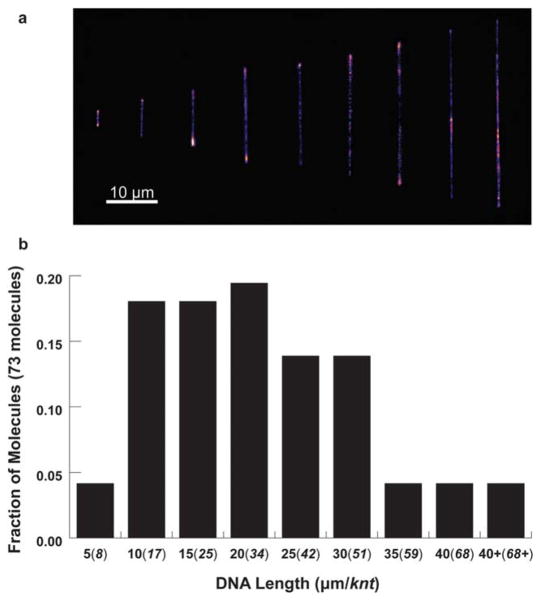
In this work, rolling circle replication is used for ssDNA synthesis, which generates polydisperse polymer samples. To estimate chain length distributions in replicated ssDNA samples, we used pulsed field gel electrophoresis (PFGE), which showed that ssDNA (Sequence 1) synthesized by RCR using 60 minute reactions produced chains with an average size of ~50 kb. To further quantify sample polydispersity, we directly visualized single ssDNA molecules stretched to near full extension using fluorescence microscopy (ESI†). We determined chain length distributions and constructed a histogram of ssDNA molecule size (Fig. 6). In particular, ssDNA polymers were stretched in a stagnation point flow generated in a PDMS-based microfluidic device,12 which is a useful method for stretching polymers to high degrees of extension (~90% contour length). Polymer length was determined by measuring chain end-to-end distance using image analysis.
Fig. 6.
Histogram of ssDNA size distribution using single molecule visualization. (a) Single ssDNA molecules are classified based on size32,33 and binned into (b) size distributions over a 73 molecule ensemble.
As shown in Fig. 6, the majority of ssDNA molecules were sized larger than ~20 μm (equal to the contour length of stained ds-λ-DNA), which agrees with PFGE analysis. In addition, ~10 to 15% of molecules exhibited stretched lengths in excess of 30 μm, which is ideal for single molecule polymer dynamics. Although the contour lengths of ssDNA molecules are similar to λ-DNA, ssDNA is extraordinarily flexible. Whereas λ-DNA contains ~150 Kuhn steps, ssDNA contains ~104 Kuhn steps for a chain of similar contour length, assuming a bare persistence length of ~0.62 nm for ssDNA with screened electrostatic interactions.19
For single molecule polymer studies conducted in dilute solution, polydisperse samples can be easily analysed, because the experimenter can select “targeted” polymer chains with specific contour lengths when acquiring data, which overcomes issues with chain length distributions. Therefore, monodisperse polymer samples are not essential for dilute solution single polymer studies. Of course, monodisperse samples of ssDNA can be obtained by the direct denaturation of monodisperse dsDNA molecules such as λ-DNA. However, this approach presents significant challenges, including fluorescent labelling of the polymer backbone without using modified nucleotides. Furthermore, the denaturation approach does not allow for control of ssDNA sequence, which opens the possibility for base pairing interactions and hairpin formation along the polymer backbone.
Base pairing and base stacking
In this work, ssDNA sequences are designed to prevent intra-chain base pairing and base stacking interactions. In nature, base stacking is an important phenomenon that aids in stabilization of the double helix in dsDNA. In ssDNA, base stacking can give rise to helical domains. Stacking interactions in ssDNA are dominant for poly(dA) and poly(dC) homopolymeric sequences, as evidenced by plateaus in force–extension elasticity data for homopolymeric ssDNA molecules.32,34–36 Poly(dT) shows minimal or no evidence of base stacking and exhibits force–extension curves similar to random ssDNA sequences (denatured λ-DNA).37
For the majority of our designer sequences (including Sequence 1), base stacking is not expected; stacking is primarily relevant for homopolymers with long poly(dA) domains,34 which do not occur in any of the designer sequences except Sequences 8 and 10, which were not used in labelling experiments. Furthermore, base stacking interactions are weak between chemically distinct bases34 and mainly dominate in homopolymeric sequences, which has been previously verified in molecular beacon experiments where a single base defect in a poly(dA) loop significantly impacted the enthalpic barrier to hairpin closing.38 Base stacking involves only interactions between neighbouring nucleobases and is either non-cooperative or weakly cooperative,34 which implies that short stretches of single base connected by a differing nucleotide are not impacted by base stacking.
Finally, we aimed to minimize or completely eliminate base pairing in ssDNA products. By design, hairpins are avoided in all sequences used in this work. Templates for RCR are designed to be either pyrimidine-rich (C/T) or purine-rich (A/G) sequences that never contained all four nucleotides. For some pyrimidine-rich templates, a third base (A) is included in minicircles in small amounts (~1 : 10 bases) to facilitate labelling with aa-dUTP (e.g., Sequence 1). We calculated base pairing energies for all sequences, and in all cases, the energy was low and on the order of thermal energy (kbT). A single isolated dATP–dTTP base pair in a long, non-interacting chain (e.g., Sequence 1) is a relatively weak interaction with energy ~1.3 kbT.39 Previous work based on Monte Carlo simulations was used to model force-extension data for random and hairpin sequences (poly-(dA-dT) and poly(dG-dC)) using standard base pairing rules, and random sequences did not alter the elastic chain behavior.40
Single molecule images of stretched ssDNA reveal clean and pristine linear polymer backbones, with no discernible evidence of base interactions. We do not anticipate that base pairing plays a role for ssDNA molecules containing user-defined tailored sequences used in this work, in particular Sequence 1. In future studies, strictly pyrimidine-only (C/T) ssDNA products can be synthesized in RCR reactions with alkyne-modified dCTP and labelled via click chemistry, thereby circumventing the requirement for a third nucleotide for fluorescent labelling.
Conclusions
In this work, we extend single molecule polymer dynamics studies to a new class of materials. Although λ-DNA has served as a model system for single polymer studies over the last several years, dsDNA is a semiflexible polymer chain with local molecular properties that differ from flexible polymers. Here, we present ssDNA as a new model system to study single flexible chains and polyelectrolytes. We developed a straightforward synthesis technique based on rolling circle replication to generate long strands of fluorescently labelled ssDNA. Dye labelling is achieved using covalent dye incorporation, which provides enhanced robustness to solvent conditions including low or high salt concentrations, thereby avoiding complications associated with using intercalating dyes in variable solvents. Precise control over ssDNA sequence will enable systematic investigation of the effect (if any) of DNA sequence on polymer chain dynamics. In addition, the RCR synthesis method described in this work generates reasonable product yields (~5 μg labelled ssDNA samples), which allows for the repeated use of the same sample for multiple single molecule polymer experiments (~100 to 200 experiments per synthesized batch). Furthermore, the ssDNA synthesis reaction can be scaled up to generate even larger amounts of material (~100 μg), which will facilitate dissemination of this new method for single polymer studies through collaborations with polymer science research groups.
The ssDNA synthesis platform described in this manuscript will enable a myriad of molecular-level studies of polymeric materials. In future work, template-based synthesis of ssDNA will enable generation of copolymers using chemical modification schemes for polymer backbones. End-functionalized oligo-nucleotide primers used in RCR synthesis will allow for surface tethering of ssDNA chains for force-extension measurements or additional surface-based assays. Using ssDNA, the dynamics of single flexible polymers can be studied, including relaxation and stretching dynamics of flexible polymer chains in flow. In addition to materials-based applications and polymer physics, fluorescently labelled ssDNA may also be useful for biological studies. For example, ssDNA “curtains”41,42 can be created in microfluidic devices, which would facilitate high-throughput measurements of nucleic acid enzymology and protein–DNA interactions using ssDNA.
Supplementary Material
Acknowledgments
We thank Prof. Paul Kenis for access to clean room facilities. This work was funded by an NIH Pathway to Independence (PI) Award, under Grant No. 4R00HG004183-04.
Footnotes
Electronic supplementary information (ESI) available: Schematic of PDMS microfluidic device for ssDNA stretching (Fig. S1), alkaline agarose gel electrophoresis for Sequence 1 with varying dTTP : dUTP ratios (Fig. S2), and native and alkaline agarose gel electrophoresis for pyrimidine rich sequences (Fig. S3). See DOI: 10.1039/c1sm05297g
Notes and references
- 1.Doiand M, Edwards SF. The Theory of Polymer Dynamics. Oxford University Press; 1986. [Google Scholar]
- 2.Larson R. The Structure and Rheology of Complex Fluids. Oxford University Press; 1999. [Google Scholar]
- 3.Shaqfeh ESG. J Non-Newtonian Fluid Mech. 2005;130:1–28. [Google Scholar]
- 4.Perkins TT, Quake SR, Smith DE, Chu S. Science. 1994;264:822–825. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 5.Smith DE, Perkins TT, Chu S. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 6.Perkins TT, Smith DE, Larson RG, Chu S. Science. 1995;268:83–87. doi: 10.1126/science.7701345. [DOI] [PubMed] [Google Scholar]
- 7.Perkins TT, Smith DE, Chu S. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 8.Smith DE, Chu S. Science. 1998;281:1335–1340. doi: 10.1126/science.281.5381.1335. [DOI] [PubMed] [Google Scholar]
- 9.Jendrejack RM, de Pablo JJ, Graham MD. J Chem Phys. 2002;116:7752–7759. [Google Scholar]
- 10.Smith DE, Babcock HP, Chu S. Science. 1999;283:1724–1727. doi: 10.1126/science.283.5408.1724. [DOI] [PubMed] [Google Scholar]
- 11.Hur JS, Shaqfeh ESG, Babcock HP, Chu S. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2002;66:011915. doi: 10.1103/PhysRevE.66.011915. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder CM, Babcock HP, Shaqfeh ESG, Chu S. Science. 2003;301:1515–1519. doi: 10.1126/science.1086070. [DOI] [PubMed] [Google Scholar]
- 13.Choi HJ, Lim ST, Lai P-Y, Chan CK. Phys Rev Lett. 2002;89:088302. doi: 10.1103/PhysRevLett.89.088302. [DOI] [PubMed] [Google Scholar]
- 14.Lim ST, Choi HJ, Lee SY, So JS, Chan CK. Macromolecules. 2003;36:5348–5354. [Google Scholar]
- 15.Jendrejack RM, Schwartz DC, Graham MD, de Pablo JJ. J Chem Phys. 2003;119:1165–1173. doi: 10.1063/1.1637331. [DOI] [PubMed] [Google Scholar]
- 16.Balducci A, Mao P, Han JY, Doyle PS. Macromolecules. 2006;39:6273–6281. [Google Scholar]
- 17.Balducci A, Hsieh CC, Doyle PS. Phys Rev Lett. 2007;99:238102. doi: 10.1103/PhysRevLett.99.238102. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, Trahan DW, Doyle PS. Macromolecules. 2010;43:3081–3089. doi: 10.1021/ma902689c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleh OA, McIntosh DB, Pincus P, Ribeck N. Phys Rev Lett. 2009;102:068301–068304. doi: 10.1103/PhysRevLett.102.068301. [DOI] [PubMed] [Google Scholar]
- 20.McIntosh DB, Ribeck N, Saleh OA. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2009;80:041803. doi: 10.1103/PhysRevE.80.041803. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson SV, Magnusson G. Nucleic Acids Res. 1982;10:1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molecular Probes HTML reference. Molecular Probes. 2004. Aminoallyl dUTP. [Google Scholar]
- 23.Zhao W, Monsur Ali M, Brook MA, Li Y. Angew Chem, Int Ed. 2008;47:6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 24.Reiß E, Holzel R, Bier FF. Small. 2009;5(20):2316–2322. doi: 10.1002/smll.200900319. [DOI] [PubMed] [Google Scholar]
- 25.Duabendiek SL, Ryan K, Kool ET. J Am Chem Soc. 1995;117:7818–7819. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fire A, Xu SQ. Proc Natl Acad Sci U S A. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu DY, Daubendiek SL, Zillman MA, Ryan K, Kool ET. J Am Chem Soc. 1996;118:1587–1594. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 29.Baner J, Nilsson M, Mendel-Hartvig M, Landegren U. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen D, Gilbert W. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 31.Cox WG, Beaudet MP, Agnew JY, Ruth JL. Anal Biochem. 2004;331:243–254. doi: 10.1016/j.ab.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Ke C, Humeniuk M, S-Gracz H, Marzalek PE. Phys Rev Lett. 2007;99:018302. doi: 10.1103/PhysRevLett.99.018302. [DOI] [PubMed] [Google Scholar]
- 33.Olson WK, Sussman JL. J Am Chem Soc. 1982;104:270–278. [Google Scholar]
- 34.Buhot A, Halperin A. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2004;70:020902. doi: 10.1103/PhysRevE.70.020902. [DOI] [PubMed] [Google Scholar]
- 35.Mishra G, Giri D, Kumar S. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2009;79:031930. doi: 10.1103/PhysRevE.79.031930. [DOI] [PubMed] [Google Scholar]
- 36.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Phys Rev Lett. 2007;98:031930. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 37.Smith SB, Cui YJ, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 38.Goddard NL, Bonnet G, Krichevsky O, Libchaber A. Phys Rev Lett. 2000;85:2400–2403. doi: 10.1103/PhysRevLett.85.2400. [DOI] [PubMed] [Google Scholar]
- 39.Rief M, Clausen-Schaumann H, Gaub HE. Nat Struct Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 40.Bommarito S, Peyret N, SantaLucia J. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granéli A, Yeykal CC, Prasad TK, Greene EC. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 42.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu M. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.