SUMMARY
Hepatitis C virus (HCV) is a major causative agent of chronic liver disease in humans. To gain insight into host factor requirements for HCV replication we performed a siRNA screen of the human kinome and identified 13 different kinases, including phosphatidylinositol-4 kinase III alpha (PI4KIIIα) as required for HCV replication. Consistent with elevated levels of the PI4KIIIα product phosphatidylinositol-4-phosphate (PI4P) detected in HCV infected cultured hepatocytes and liver tissue from chronic hepatitis C patients, the enzymatic activity of PI4KIIIα was critical for HCV replication. Viral nonstructural protein 5A (NS5A) was found to interact with PI4KIIIα and stimulate its kinase activity. The absence of PI4KIIIα activity induced a dramatic change in the ultrastructural morphology of the membranous HCV replication complex. Our analysis suggests that the direct activation of a lipid kinase by HCV NS5A contributes critically to the integrity of the membranous viral replication complex.
INTRODUCTION
Hepatitis C virus (HCV), the sole member of the genus Hepacivirus within the family Flaviviridae, poses a global health problem. Treatment of infected individuals with a combination of pegylated interferon and ribavirin leads to a sustained virological response in only about 50% of patients and provokes serious side effects. Neither a prophylactic nor a therapeutic vaccine is available and their development has been hampered by the high variability of the virus.
HCV has a single-stranded RNA genome of positive polarity encoding for a polyprotein that is cleaved by cellular and viral proteases into 10 different proteins: core, envelope glycoprotein 1 (E1), E2, p7, non-structural protein 2 (NS2), NS3, NS4A, NS4B, NS5A and NS5B. The structural proteins core, E1 and E2 are the main constituents of the virus particle whereas most of the non-structural proteins are required for RNA replication (Moradpour et al., 2007; Tang and Grise, 2009). These include the NS3/4A protein that has serine-type protease as well as NTPase and helicase activities, the NS5B RNA-dependent RNA polymerase and the NS5A replicase factor. The latter is composed of 3 domains (Marcotrigiano and Tellinghuisen, 2009): RNA-binding domain I required for RNA replication, domain that, to the most part, is dispensable for viral replication and domain III that is essential for virion assembly (Appel et al., 2008). NS4B is a highly hydrophobic protein triggering rearrangements of intracellular membranes, designated the membranous web (Egger et al., 2002). This web is composed of membranous vesicles of heterogeneous size and morphology and assumed to serve as site of HCV RNA replication (Gosert et al., 2003).
The possibility to propagate either subgenomic HCV replicase constructs (replicons) or infectious HCV in cultured human hepatoma cells opened new avenues to gain insight into the intimate interaction between HCV and its host cell (reviewed in Poenisch and Bartenschlager, 2010) By using various screening assays, an increasing number of host factors possibly promoting or restricting HCV replication has been described (Tai et al., 2009; Ng et al., 2007; Supekova et al., 2008; Vaillancourt et al., 2009; Borawski et al., 2009; Trotard et al., 2009; Randall et al., 2007; Berger et al., 2009; Li et al., 2009; Coller et al., 2009; Jones et al., 2010). However, the overlap of identified genes is minimal and mechanistic insights how they contribute to the HCV replication cycle is sparse.
Phosphoinositides (PIs) are membrane phospholipids regulating numerous cell processes by recruitment of distinct effector proteins (D'Angelo et al., 2008). Seven PIs are known that have been named according to the specific position in the inositol ring where phosphorylation occurs. They are generated by a set of kinases residing in different subcellular compartments, thus providing a molecular signature to the membrane where a given PI resides. A critical role of PI4kinase III (PI4KIII) for HCV replication has been suggested earlier (Tai et al., 2009; Vaillancourt et al., 2009; Borawski et al., 2009; Trotard et al., 2009; Berger et al., 2009; Li et al., 2009; Hsu et al., 2010). However, conflicting reports exist as to which specific isoform is required, and the molecular mechanism by which PI4KIII contributes to HCV RNA replication has not been explored in detail.
In this study, we undertook an unbiased large-scale human kinome screen and identified the PIP pathway as a central element of HCV replication. We report that NS5A binds to and activates PI4KIIIα. Local synthesis of PI4P at distinct membrane sites appears to provide a molecular signature required for structural and functional integrity of the membranous HCV replication complex.
RESULTS
Screening for human kinases involved in HCV entry or replication
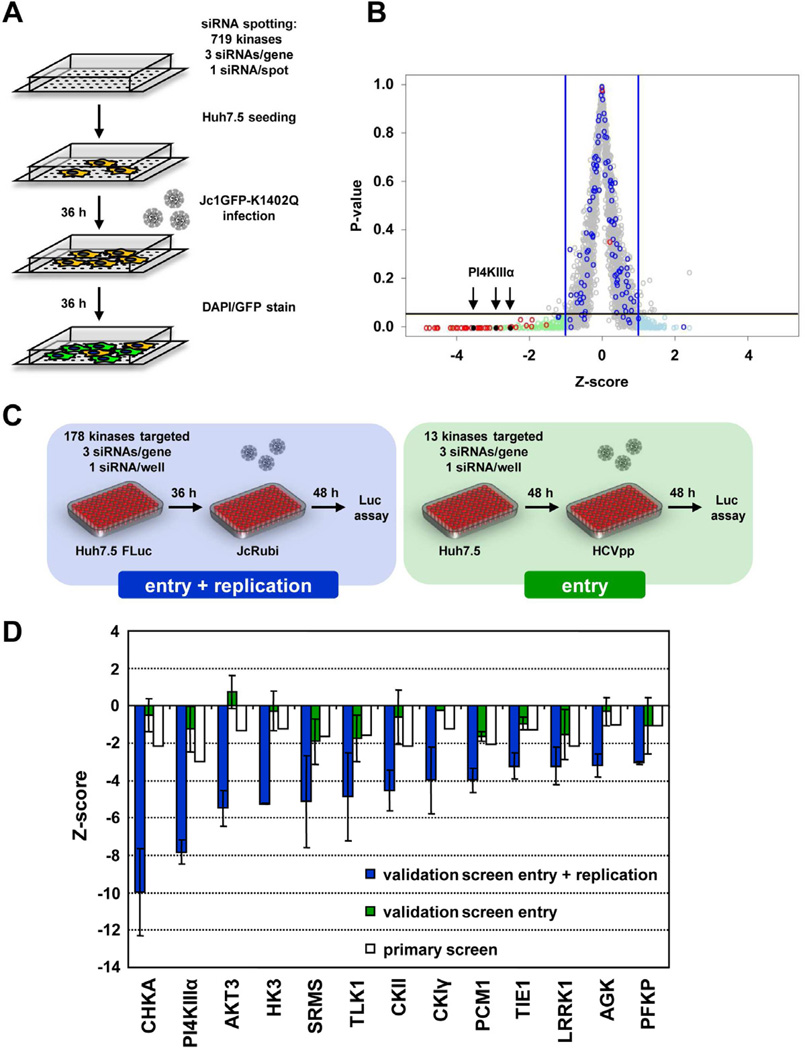
To screen a siRNA library targeting all known and predicted 719 human kinases we generated a suitable reporter virus system (Jc1GFP-K1402Q; described in detail in the Supplemental Data). Reverse transfection of siRNAs into Huh7.5 cells in a chamber slide format was optimized according to a previously described protocol (Erfle et al., 2007; and Supplemental Data). Overall, 2157 siRNAs targeting 719 human kinase genes plus positive controls targeting the entry receptor CD81 or the viral genome itself (HCV321 and HCV138) and four different negative controls were spotted in transfection mixture onto chamber slides (Fig. 1A). After seeding of Huh7.5 cells we allowed silencing for 36 h. Cells were then infected with Jc1GFP-K1402Q, 36 h later fixed and stained with a GFP-specific antibody. This staining was necessary, because after PFA-fixation required for biosafety reasons, GFP fluorescence was too weak for reliable HCV detection and signal to noise ratios were too small. Fluorescence intensity was quantified by automated image analysis (Matula et al., 2009). The screen was repeated 12 times in four independent experiments. Data were normalized as described in the Supplemental Experimental Procedures. Figure 1B depicts the mean z-scores over the 12 replicates for each siRNA (Table S1). With a hit criterion of −1 > z > 1 and p < 0.05, we defined 83 potential dependency and 95 potential restriction factors for HCV entry and replication, which were selected for further analysis.
Fig. 1. High throughput siRNA screen to identify kinases involved in HCV entry and replication.
(A) Assay setup of the primary siRNA screen. Each chamber slide was coated with 384 different siRNA spots. Huh7.5 cells were infected 36 h after seeding and another 36 h later, cells were fixed and stained. (B) Virus specific signal intensities obtained in the primary screen were quantified and values were normalized intra- and inter-slide wise as well as between the 12 repetitions. Mean z-scores for all siRNAs tested are depicted in grey. Z-scores of negative and positive controls are highlighted in blue and red, respectively. With a hit criterion of −1 > z > 1 and a p-value < 0.05 indicated by the blue and black bars, respectively, 83 potential dependency and 95 potential restriction factors were identified (light green and light blue, respectively). Positions of PI4KIIIα hits are indicated by arrowheads. (C) Assay setup of the entry and replication validation screen (left panel). For each candidate 3 unique siRNAs were tested in a 96-well plate format. Huh7.5 FLuc cells were infected with JcR-ubi to measure effects of the silencing on HCV entry and replication (4 repetitions). To measure effects of the silencing of validated hits on entry (right panel), Huh7.5 cells were infected with HCVpp containing a FLuc reporter gene and luciferase activity was determined 48 h later (7 repetitions). (D) Summary of mean z-scores of hits determined in the two independent validation screens. Z-scores determined in the primary screen (white bars) are shown for comparison.
For the validation of the 178 hit candidates, three unique siRNAs per gene were used to minimize the number of potential off-target hits. In addition, the format of the assay was changed to a statistically more robust 96-well plate format to increase the number of transfected cells per siRNA and thus statistical power (Fig. 1C). This assay format also allowed use of a renilla luciferase (RLuc) reporter virus (JcR-ubi) facilitating the analysis of the screen. This virus reaches infectivity titres up to 8 × 104 TCID50/ml (not shown) and after infection of Huh7.5 cells with a MOI of ~0.3 TCID50 per cell, relative light units (RLU) were about 100-fold above background.
To determine effects on entry and RNA replication, Huh7.5 FLuc cells constitutively expressing firefly luciferase (FLuc), were infected with JcR-ubi 36 h after seeding and replication was determined by RLuc assay 48 h later (Fig. 1C). Virus-specific RLuc counts were normalized to cell counts by determining FLuc activity in the same lysate (see Supplemental Data). The assay was repeated four times (Table S2). With a hit criterion of −2.5 > z > 2.5 for at least two siRNAs per gene, we identified 13 dependency factors (Fig. 1D) whereas none of the potential restriction factors could be confirmed.
To discriminate whether the identified kinases play a role in virus entry or replication, we screened the 13 validated kinase genes by using the HCV pseudoparticle (HCVpp) system. Z-scores for each tested siRNA were calculated as described in Materials and Methods (Fig. 1D; summary in Table S2). Silencing of the tested candidates did not reduce HCVpp entry to an extent corresponding to a z-score lower −2.5, arguing for a role of these kinases in a step after virus entry such as uncoating, establishment of the replication complex or HCV RNA replication itself.
Bioinformatics analysis of HCV dependency factors
To map those cellular pathways to which the 13 dependency factors identified in our study are linked, we integrated various molecular datasets, comprising information on protein-protein interactions (PPIs), protein complexes, molecular pathways, and functional annotations (see Supplemental Data). The inclusion of all dependency factors identified in previous siRNA-based HCV screens enabled a meta-analysis revealing numerous direct interactions between dependency factors with 8 of our 13 dependency factors being directly connected to at least one previously reported dependency factor (Table S3). Figure 2 depicts this network and a subset of those cellular pathways in which HCV dependency factors are significantly overrepresented (p-value < 0.05) (for detailed information see Table S3). These pathways include, amongst others, apoptosis, integrins in angiogenesis, endocytosis, focal adhesion, signaling in the immune system as well as the ErbB and the MAP kinase signaling pathways. The latter two have been described by Li and colleagues to be important for HCV and flaviviruses in general (Li et al., 2009). From our 13 dependency factors, the one having the most inhibitory siRNAs, choline kinase alpha (CHKA), and the only factor confirmed by all individual siRNAs, PI4KIIIα, are both associated with glycerophospholipid biosynthesis. CHKA is required for the biosynthesis of the membrane components phosphatidylcholine and ethanolamine. While this result suggests that lipid membrane biosynthesis plays a crucial role in HCV replication, a direct interaction of CHKA with another dependency factor in the network shown in Figure 2 is not known yet.
Fig. 2. Network of HCV dependency factors.
The depicted network highlights pathways in which dependency factors identified in our study (hexagonal nodes) and in previous HCV screens (ellipsoid nodes) are involved. Squares represent kinases that were found in our study and have already been known as dependency factors. The mean z-score of the two most inhibitory siRNAs per gene in the entry and replication validation screen is indicated by the color intensity of the nodes. Factors in white nodes were either not studied in this work or below the hit criterion in the primary screen; red-shaded nodes display candidates analyzed in the validation screen. Solid lines indicate proteins that are known to interact with each other or to participate in the same protein complex. Pathways for which we found a significant enrichment of HCV dependency factors are linked by colored areas and labeled at the top and bottom.
PI4KIIIα is necessary for HCV replication in a genotype- and cell line-independent manner
The most consistent hit of our screen was PI4KIIIα, also known as PIK4CA or PI4KA. In fact, it was the only gene for which all 6 siRNAs tested in the primary and the validation screen scored positive. In the initial set of experiments, we corroborated our results from the siRNA screen and consistently found that silencing of PI4KIIIα expression in two cell lines harboring FLuc reporter replicons of two different HCV genotypes (JFH-1, gt 2a and Con1ET, gt 1b) inhibited HCV RNA replication (Fig. 3A and B). In contrast, in spite of efficient knockdown of the beta isoform of PI4KIII replication of the JFH-1 replicon was not affected and only a minor, yet statistically significant effect on Con1 RNA replication could be observed.
Fig. 3. Effect of PI4KIIIα and PI4KIIIβ silencing on HCV replication and colocalization of these lipid kinases with NS5A.
(A, B) Cells containing a genotype (gt) 2a or 1b luciferase reporter replicon specified in the top of each panel were transfected with siRNAs targeting PI4KIIIα or PI4KIIIβ (three individual siRNAs each) or with the positive control siRNA (HCV321) or the negative control siRNAs (DV-3’NTR or NegC#1). SiRNAs were transfected twice and RNA replication was determined 48 h after the second transfection by luciferase assay. RLUs were normalized to the mean of the negative controls. Data (mean +/− SD; n = 3 in triplicates) were analyzed using a one-way t-test. (C) Lysates prepared from Con1ET replicon cells 48 h after the second transfection were analyzed by immunobloting for PI4KIIIα, PI4KIIIβ, Transferrin receptor (TfR) and NS5A. Levels of NS5A were quantified and normalized to the corresponding TfR levels. Values are given in % relative to the mean of the NS5A levels of the negative control treated cells. (D, E) Huh7-Lunet cells (mock) or cells containing a selectable JFH-1 replicon (neo-sgJFH-1) were stained with a NS5A- (red) and either a PI4KIIIα- (D) or a PI4KIIIβ-specific antibody (green) (E). Nuclear DNA was stained with DAPI (blue). An enlargement of the sections indicated by a white square in each of the merged images is shown in the corresponding crop panel in the bottom. Images were acquired with a confocal microscope.
The reduction of replication measured by luciferase assay was confirmed by immunoblotting for Con1 NS5A (Fig. 3C). The degree of silencing of PI4KIIIα expression correlated well with the reduction of NS5A amounts with the most effective PI4KIIIα-specific siRNA (#3) suppressing HCV replication as efficient as the siRNA targeting the viral genome itself (HCV321). The same was found for JFH-1 (data not shown). Likewise, knock-down efficiency of PI4KIIIβ correlated with NS5A amounts in Con1 replicon-containing cells (Fig. 3C) whereas no effect was found with JFH-1 replicon cells (not shown).
We next analyzed the localization of endogenous PI4KIIIα in cells with or without persistent subgenomic JFH-1 replicons (neo-sgJFH-1 and mock, respectively) (Fig. 3D). In both cell pools the kinase localized in punctuate structures throughout the cytoplasm, but mainly in a perinuclear region, as reported elsewhere (Kakuk et al., 2006). Staining for NS5A, a presumed marker for HCV replication sites revealed a striking colocalization with the kinase. In contrast, we did not observe colocalization of PI4KIIIβ with NS5A (Fig. 3E). In addition, also in cells with a persistent subgenomic Con1 replicon, we could observe a colocalization of NS5A with PI4KIIIα, but not with PI4KIIIβ (not shown).
To exclude that the dependency of HCV RNA replication on PI4KIIIα is a cell line specific phenomenon, we compared knock-down phenotypes in Huh7.5 and LH86 cells (Zhu et al., 2007). For this experiment, we used a more efficient RLuc reporter virus genome (JcR-2a) producing much higher infectivity titers as compared to JcR-ubi and thus an about 10-fold higher signal over background (not shown). Silencing of PI4KIIIα clearly decreased viral replication in both cell lines also in this infection-based assay (Fig. 4A), comparable to the results obtained with the replicon cell lines (Fig. 3A and B). Knock-down of PI4KIIIα expression did not affect replication of Dengue virus, arguing for a specific role of PI4KIIIα in HCV replication (Fig. 4B).
Fig. 4. Impact of PI4KIIIα silencing on HCV production and integrity of the membranous web.
(A) Expression of PI4KIIIα in Huh7.5 or LH86 cells was silenced by using individual siRNAs and cells were infected with JcR-2a. SiRNA HCV321 served as positive control, siRNAs DV-3’NTR and NegC#1 were used as negative controls. Virus replication was determined by luciferase assay. Data (mean +/− SD; n = 2 in duplicates) were analyzed using a one-way t-test. (B) Same as in (A) but measuring Dengue virus replication by using a Renilla reporter virus which will be described elsewhere. Data (mean +/− SD; n = 2 in duplicates) were analyzed using a one-way t-test. P-values below 0.05 or 0.001 are indicated by one or three asterisks, respectively. (C) Huh7.5 cells with stable knock-down of PI4KIIIα expression (sh-PI4KIIIα) or expressing a non-targeting shRNA (sh-NT) were stably transduced with shRNA-resistant PI4KIIIα wild type (wt) or D1957A mutant (inactive) expression constructs. For comparison, the PI4KIIIα knock-down cell line was transduced with the empty expression vector in parallel (empty vector). Cell lines were additionally transfected with siRNA #3 targeting PI4KIIIα (corresponding to the sequence of the shRNA used for stable silencing) prior to infection with JcR-2A with a MOI of 0.5 TCID50/cell. Virus replication was determined by luciferase assay 48 h post infection. As negative control, sh-NT cells were infected and analyzed in parallel. (D) Huh7-Lunet/T7 cells stably expressing a PI4KIIIα-specific shRNA (sh-PI4KIIIα) or a non-targeting shRNA (sh-NT) (as described in (C)) were transfected with a T7 promoter-based NS3 to NS5B expression construct. Cells were stained with a NS5A-specific antibody (red) and nuclear DNA was stained with DAPI (blue). Note the formation of profound NS5A “clusters” in PI4KIIIα knock-down cells, but not in sh-NT cells. (E) Cell lines described in panel C were transfected with a NS3 to NS5B polyprotein expression construct and the percentage of “wildtype” and “cluster” phenotype (D) was determined for 250 HCV-positive cells per condition. (F) Control (upper panels) and PI4KIIIα-silenced cells (lower panels) were transfected with a NS3 to NS5B expression construct containing a functional NS5A with a GFP insertion in domain III (Schaller et al., 2007) and prepared for EM analysis. The GFP insertion had no impact on the morphology of membrane alterations induced by the HCV NS-proteins in IF or EM (not shown) and does not interfere with RNA replication in a replicon context (Schaller et al., 2007). Consecutive enlargements of the boxed areas are shown from left to right. Note the very heterogeneous membranous web (MW) in control cells and the clusters of small double membrane vesicles (DMVs) (indicated by yellow arrows) in sh-PI4KIIIα cells. Size markers are given in the lower right of each panel. N, nucleus; LD, lipid droplet; MMVs, multi-membrane vesicles; rER, rough endoplasmic reticulum; m, mitochondrium.
Enzymatic activity of PI4KIIIα is required for HCV replication
For rigorous exclusion of off-target effects and to clarify whether PI4KIIIα enzymatic activity was needed for HCV replication, we generated Huh7.5-based cell lines with stable knock-down of PI4KIIIα or a non-targeting control (sh-NT) (for details see Supplemental Data). Rescue cell lines were established by stable expression of shRNA-resistant PI4KIIIα wild type (WT) or the D1957A mutant, which is devoid of enzymatic activity (Fig. S1). Upon infection of the PI4KIIIα-wt rescue cell line with JcR-2a, HCV replication was comparable to replication in the sh-NT control cells (Fig. 4C). However, no rescue was obtained in cells stably transduced with the inactive PI4KIIIα D1957A mutant or an empty vector, demonstrating that kinase activity of PI4KIIIα was required for HCV RNA replication.
PI4KIIIα is required for integrity of membranous web structures
Studies on the potential contribution of PI4KIIIα to the morphology of HCV replication sites were hampered by the fact that interference with lipid kinase activity directly impaired viral replication. We therefore generated stable PI4KIIIα-silenced Huh7-Lunet cells stably expressing the T7 RNA polymerase (Huh7-Lunet/T7), allowing the cytoplasmic expression of HCV proteins and thus induction of membranous web formation independent of viral RNA replication. Expression of the polyprotein fragment encoding the replicase (NS3 to NS5B) in sh-NT cells did not affect the punctuate staining pattern of NS5A that was well comparable to the one found in replicon cells (compare Fig. 4D, left panel with Fig. 3D and E). However, silencing of PI4KIIIα expression led to the formation of abnormal NS5A “clusters” (Fig. 4D, right panel) in approx. 40 % of the silenced cells (Fig. 4E). Importantly, this phenotype could be rescued by overexpression of the WT kinase, but not with the enzymatically inactive mutant. These results suggested that enzymatic activity of PI4KIIIα was necessary for integrity of the membranous replication complex.
Ultrastructural analyses of the same sh-NT cells expressing the NS3 to NS5B polyprotein revealed membrane rearrangements with accumulation of heterogeneous vesicles (Fig. 4F) resembling the membranous web detected in Jc1 infected cells (not shown). Prominent membrane rearrangements only detected in cells expressing the HCV polyprotein (Fig. S2) included double membrane vesicles (DMVs) with an average diameter of 211+/−71 nm and multi-membrane vesicles (MMVs) with an average diameter of 287+/−85 nm. In contrast, NS3 to NS5B expression in sh-PI4KIIIα cells did not trigger formation of MMVs. In addition, the DMVs were smaller in size, very homogeneous (134 +/−30 nm) and aggregated into huge clusters resembling the phenotype observed in immunofluorescence analysis.
Elevated PI4P levels in HCV containing cells in vitro and in vivo
As reported previously, PI4P, the product synthesized by PI4kinases was detected predominantly in the Golgi membrane (Fig. 5A) (colocalization with the Golgi marker TGN46 not shown) (reviewed in D'Angelo et al., 2008). Upon infection with HCV a profound and time-dependent altered subcellular distribution as well as an increase of PI4P was found resulting in a pattern that very much resembled HCV replication sites. In fact, a fraction of NS5A-positive dot like structures (19.7% +/− 4.4 SEM) colocalized with PI4P, arguing that the kinase generated this product at HCV replication sites. In contrast, Huh7 cells harboring a DENV replicon neither showed this relocalization of PI4P nor a colocalization of PI4P with assumed sites of viral replication (not shown). Quantitative analysis of PI4P immunofluorescence in HCV-infected cells revealed a ~3-fold induction in PI4P levels compared to mock treated cells (Fig. 5B). Consistently, we found that PI4P levels in neo-sgCon1, neo-sgJFH and Jc1-transfected cells were increased ~3-fold compared to mock cells when measuring PI4P levels in an immunoblot assay (Fig. 5B). Importantly, PI4KIIIα protein levels were not elevated by HCV RNA transfection or infection (Fig. 5C), arguing for activation of PI4KIIIα by HCV (Fig 5C).
Fig. 5. PI4P and PI4KIIIα levels in HCV containing cells.
(A) HCV permissive Lunet/CD81 cells were mock treated or infected with Jc1. Cells were fixed at given time points and stained with a NS5A- (red) and a PI4P-specific antibody (green). Nuclear DNA was stained with DAPI (blue). An enlargement of the section indicated by the white square is shown in the crop panel in the bottom. Images were acquired with a confocal microscope. (B) Quantification of PI4P levels by immunofluorescence analysis (green bars) and immunoblot (black bars). For immunofluorescence of mock- or Jc1-transfected cells mean values and standard error of the mean (SEM) of 80 cells per condition are shown. For immunoblot, cells transfected with Jc1 or subgenomic replicons specified in the bottom were lysed after 48 h. Lipids were extracted, spotted onto membranes and PI4P was quantified using the PI4P Mass strip kit (Echelon). Signals were quantified by using the Quantity One software (Bio-Rad). Error bars represent the SEM of three independent experiments. IF, immunofluorescence; IB, immunoblotting; n.t., not tested (C) Lysates used in (B) were prepared from mock- or Jc1-transfected cells at time points post transfection given in the top and analyzed by immunobloting for PI4KIIIα, NS5A and Calnexin (loading control). (D) Immunohistochemistry on snap-frozen liver tissues from a HCV negative person (10H6; Table S4, S5) or a patient with chronic HCV infection (3E6; Table S4, S5). Consecutive liver sections were stained for NS5A, PI4P and core. Note that cells staining positive for viral proteins and PI4P reside in the same region of each section.
We furthermore investigated PI4P levels in hepatocytes of patients persistently infected with HCV by using immunohistochemistry on consecutive sections of frozen liver samples (Fig. 5D and S3; Table S4 and S5). NS5A- or core-reactive cells were only found in infected tissues obtained from HCV positive patients, demonstrating specificity of the staining method. Importantly, staining for NS5A, core and PI4P revealed that HCV positive areas were always enriched in PI4P, thus corroborating our in vitro data and indicating that HCV infection indeed resulted in elevated PI4P levels.
Role of NS5A domain 1 in recruitment of PI4KIIIα and induction of PI4P
Given the colocalization of PI4KIIIα with NS5A and the accumulation of PI4P at sites of HCV replication, we assumed that the kinase might be recruited by interaction with one or several replicase proteins. Individual HCV proteins were therefore coexpressed with HA-tagged PI4KIIIα in Huh7-Lunet/T7 cells and coimmunoprecipitation experiments were performed using monospecific antisera (Fig. 6A). We found that NS5A and NS5B, but not the NS3/4A protease/helicase or NS4B interacted with the kinase. Moreover, expression of only NS5A, but not NS5B lead to an induction and altered staining pattern of PI4P (Fig. S4) and this pattern was well comparable to the one observed in infected cells (Fig. 5A). These results suggested that NS5A is the key player altering PI4P amounts and subcellular distribution most likely by modulating PI4KIIIα activity.
Fig. 6. Role of NS5A domain I in recruitment of PI4KIIIα and induction of PI4P.
(A) Huh7-Lunet/T7 cells were cotransfected with HA-tagged PI4KIIIα and expression constructs encoding individual viral proteins specified in the top. After 8 h cell proteins were radiolabeled for 16 h with [35S] methionine/cysteine-containing medium. Cells were lysed and immunoprecipitation was performed using antibodies indicated in the bottom. Filled arrowheads point to HCV proteins; the open arrowhead marks PI4KIIIα. (B) Huh7-Lunet/T7 cells were cotransfected with HA-tagged PI4KIIIα and pTM-based NS3 to NS5B polyprotein constructs containing given deletions of individual NS5A domains. Immunoprecipitation was performed as described in (A) using an NS5A-specific antibody. Filled arrowheads on the right refer to NS5A variants; the open arrowhead marks PI4KIIIα. (C) Huh7-Lunet/T7 cells were transfected with constructs encoding NS3 to NS5B harboring wildtype NS5A or NS5A deletions mutants specified in the top. Twenty four hours after transfection NS5A (red) and PI4P (green) were detected by immunofluorescence. Nuclear DNA was stained with DAPI (blue). Images were acquired with a confocal microscope. Numbers in the bottom refer to percentage and SEM of NS5A positive dot-like structures costaining with PI4P. Values are derived from analysis of 20 individual cells.
To identify the region of NS5A interacting with the kinase, we performed pull-down experiments by using NS3 to NS5B polyproteins containing different in frame deletions in NS5A (Fig. 6B) (Appel et al., 2008; Backes et al., 2010). Only by deleting domain I we observed a reduced interaction between NS5A and PI4KIIIα. In addition, the deletion of NS5A domain I also hampered the induction of PI4P whereas deletions of domain II or III had no effect (Fig. 6C). We thus concluded that NS5A domain I is required for interaction with PI4KIIIα.
Activation of PI4KIIIα kinase activity by NS5A
We next investigated whether HCV proteins directly affected PI4KIIIα activity by using in vitro kinase assays and recombinant purified NS3, NS5A or NS5B (Fig. 7A). Incubation of the kinase with a phosphatidylinositol-containing substrate and radiolabeled γ-[32P] ATP led to clearly detectable incorporation of radioactive phosphate into phosphatidylinositol (Fig 7B). Incorporation of radioactivity could be reduced by addition of Wortmannin (WM), a known inhibitor of the kinase (Balla et al., 1997), to levels obtained with an inactive mutant (Fig. S1), thus confirming specificity of the assay. Addition of NS3 or NS5B only slightly reduced or increased kinase activity, respectively (Fig. 7B). In contrast, NS5A enhanced PI4KIIIα activity dose-dependently up to 5-fold. This stimulation was abolished by Wortmannin excluding an irrelevant kinase activity copurified with NS5A.
Fig. 7. Activation of PI4KIIIα by NS5A.
(A) Analysis of recombinantly expressed and purified HCV nonstructural proteins and PI4KIIIα. NS3 and NS5A were expressed as full-length proteins with N-terminal hexahistidine tag, NS5B was C-terminally tagged and lacked the C-terminal membrane insertion sequence. One µg of each protein was loaded onto a SDS 7 % polyacrylamide gel and proteins were visualized by Coomassie blue staining. (B) Purified PI4KIIIα was incubated with a phosphatidylinositol containing substrate and radiolabeled γ-[32P] ATP in the presence or absence of the indicated purified viral proteins. Molar ratios of PI4KIIIα and a given viral protein are indicated in the bottom. PI4KIIIα activity was determined by measuring the incorporation of [32P] into the phosphatidylinositol substrate. As controls, the PI4KIIIα inhibitor Wortmannin (WM; 100 µM) was added to the reaction. Data (mean +/− SD; n = 3 in duplicates) were analyzed using a two-way t-test. P-values below 0.05 or 0.001 are indicated by one or three asterisks, respectively. n.d., not detectable (C) Naïve Huh7-Lunet/T7 cells or cells with stable knock-down of PI4KIIIα expression (sh-PI4KIIIα) or expressing a non-targeting shRNA (sh-NT) were transfected with a NS3 to NS5B polyprotein expression construct under control of the T7 RNA polymerase promoter (pTM-NS3-5B) and analyzed for NS5A (red) and PI4P (green) distribution. A fraction of the cells were treated with 30 µM PIK93 for 16 h. Mock transfected Huh7-Lunet/T7 cells (left column of panels) are shown for reference. Nuclear DNA was stained with DAPI (blue). Images were acquired with a confocal microscope. White arrows point to a regular Golgi-like distribution of PI4P.
Finally, we analyzed the effect of the PI4KIIIα inhibitor PIK93 on the subcellular distribution of PI4P in HCV-containing cells. PIK93 treatment inhibited the HCV-mediated PI4P induction and in addition, caused a clustering phenotype of NS5A very much alike the clustering observed upon knock-down of PI4KIIIα expression (Fig. 7C). Since PIK93 is not specific for PI4KIIIα and inhibits primarily the beta-isoform (Borawski et al., 2009), we analyzed the distribution of PI4P also by specific silencing of the alpha-isoform. Importantly, we found no co-localization of PI4P with membrane clusters induced by silencing of PI4KIIIα (Fig. 7C). PI4P levels were increased 6-fold in naïve cells expressing the HCV replicase proteins, but remained unaltered upon silencing of PI4KIIIα expression (data not shown). These results confirmed that kinase activity was required for elevated PI4P levels.
Taken together, the data suggest that PI4KIIIα is recruited to the membranous replication compartment by NS5A, which in turn stimulates kinase activity. Elevated PI4P amounts generated at these sites appear to directly contribute to membranous web integrity.
DISCUSSION
Focussing on kinases that often play key roles in regulating viral replication in this study, we used an RNAi-based screen targeting the human kinome and identified 13 kinases promoting HCV RNA replication. Up to now, 9 limited and two genome-wide siRNA screens based on different HCV replication models have been performed (Tai et al., 2009; Ng et al., 2007; Supekova et al., 2008; Vaillancourt et al., 2009; Borawski et al., 2009; Trotard et al., 2009; Randall et al., 2007; Berger et al., 2009; Li et al., 2009; Coller et al., 2009; Jones et al., 2010). However, the overlap of identified genes is very low, which may be due to different experimental conditions, reagents or hit calling criteria. Nevertheless, bioinformatic analysis of our results in the context of published HCV dependency factors provides a comprehensive overview about kinase-linked pathways involved in HCV entry and replication (Fig. 2). Eight of our 13 hits were each directly connected to at least one previously reported dependency factor, arguing for a high quality of our screen. Most notably, a comparison of host cell factors involved in HCV, DENV and West Nile virus replication revealed three commonly involved pathways (MAPK signalling pathway, focal adhesion and ErbB signalling pathway) demonstrating the close evolutionary relationship of these three viruses (Li et al., 2009; Krishnan et al., 2008).
Kinases reported earlier to be involved in HCV replication and identified also in our screen include casein kinases I and II (Neddermann et al., 2004; Kim et al., 1999; Tellinghuisen et al., 2008; Masaki et al., 2008) and choline kinase alpha (CHKA) (Li et al., 2009). The latter is required for the biosynthesis of phosphatidylethanolamine and phosphatidylcholine, a major component of lipid membranes arguing that this enzyme contributes to the formation of the membranous web. However, no change in the localization pattern of CHKA after HCV infection and no colocalization of this kinase with HCV replication sites were found arguing that the kinase may contribute to replication more indirectly (data not shown).
The most consistent hit identified in our screen was PI4KIIIα. This enzyme is one of four kinases (PI4KIIα, PI4KIIβ, PI4KIIIα and PI4KIIIβ) in mammalian cells that catalyze the synthesis of PI4P (Balla and Balla, 2006). All four enzymes have different subcellular localizations and regulation mechanisms of their activity state, thus producing distinct PI4P pools inside the cell (reviewed in (D'Angelo et al., 2008)). PI4KIIIα has also been identified by others as HCV dependency factor (Tai et al., 2009; Vaillancourt et al., 2009; Borawski et al., 2009; Trotard et al., 2009; Berger et al., 2009; Li et al., 2009). In agreement with these studies, we found that silencing of PI4KIIIα inhibits HCV RNA replication. Conflicting results have been obtained for the beta-isoform of this lipid kinase. We found that PI4KIIIβ promotes replication of HCV genotype 1b (Con1), but not of a genotype 2a (JFH-1) genome, arguing for genotype-specific dependency.
PI4KIIIα appears to localize primarily at the ER (Kakuk et al., 2006; Wong et al., 1997), assumed to be the origin of HCV replication sites. Indeed, we observed a clear colocalization of endogenous PI4KIIIα with NS5A, although the subcellular distribution of the kinase was not dramatically changed in HCV-containing cells (Fig. 3D). PI4KIIIβ is primarily localized to the Golgi compartment (Fig. 3E) (De Matteis et al., 2005), hampering kinase recruitment by HCV proteins and explaining the lack of colocalization with viral proteins (Fig. 3E). The different subcellular localization of both isoforms might also explain why silencing of the expression of the alpha-isoform can not be rescued by expression of the beta-isoform (Tai et al., 2009). However, since our results suggest that PI4P might have a central role in mediating the function of PI4 kinases in HCV replication, subtle differences in the subcellular localization and dynamics of PI4P pools in different cell lines may account for the controversial knock-down phenotypes observed for the PI4K isoforms.
Results obtained by immunohistochemical staining of serial sections of frozen liver tissues from patients with chronic hepatitis C were in full support of our in vitro data. This technique that allowed us to detect the viral antigens core and NS5A is superior to the use of paraffin-embedded tissues where detection of HCV antigens frequently fails (Shiha et al., 2005). We observed varying proportions of cells (up to a maximum of about 5%) staining positive for HCV in different patient samples (Table S4). Liang and colleagues, who used two-photon microscopy in combination with virus-specific, fluorescent, semiconductor quantum dot probes recently reported that 7 to 20% of hepatocytes in infected liver tissue are HCV positive (Liang et al., 2009). The lower frequency of HCV antigen-positive cells identified by our method might point to lower sensitivity, thus detecting only cells with high antigen load. Nevertheless, we found that HCV positive regions in the infected livers were always enriched in PI4P, thus supporting our in vitro data.
So far we do not know whether HCV replication depends on PI4KIIIα itself or on its reaction product, PI4P. On one hand, PI4P might be a direct constituent of the membranes forming the HCV replication complex; on the other hand, PI4P might specifically recruit host cell proteins required for HCV RNA replication. One potential candidate is the oxysterol binding protein, which binds to PI4P and is essential for HCV RNA replication (Amako et al., 2009). We note that a similar model has recently been proposed for generation of the replication compartment of enteroviruses (Hsu et al., 2010). Upon infection with coxsackie virus, secretory pathway organelles are reorganized leading to the formation of a PI4P lipid-enriched microenvironment in a PI4KIIIβ-dependent manner. Interestingly, the viral RNA-dependent RNA polymerase of coxsackie virus specifically binds to PI4P supporting the notion that PI4P serves as recruitment factor required for the assembly of the membrane-associated replication complex. In agreement with our observation, Hsu and coworkers also reported a slight increase of PI4P in HCV replicon cells and colocalization of PI4P with NS5A. However, in contrast to what we found, these authors described that knock-down of PI4KIIIβ reduced HCV RNA replication much stronger as compared to reducing PI4KIIIα expression. This discrepancy might be due, at least in part, to the use of only a genotype 1b replicon for their experiments.
Our knowledge on the biogenesis of the membranous web is still incremental. In principle, NS4B is capable of inducing membrane alterations reminiscent of the membranous web (Egger et al., 2002; Gosert et al., 2003) and might provide the protein scaffold of these vesicles. However, more recent results (Ferraris et al., 2010) and the data presented in our study point to a far more complex biogenesis that includes the formation of heterogeneous DMVs and MMVs. Although NS5A does not trigger vesicle formation by itself (Egger et al., 2002), our data point to an essential role of NS5A for membranous web integrity by activating PI4KIIIα leading to an accumulation of PI4P at sites of HCV replication. Whether the altered lipid composition induced in this way contributes to membranous web formation or whether this is due to viral or host proteins binding to these lipids remains speculative and will be the subject of more detailed studies.
By using stable knock-down of PI4KIIIα expression and reconstitution experiments, we demonstrate that kinase activity of PI4KIIIα is required for efficient HCV RNA replication. Although earlier studies performed with the kinase inhibitors Wortmannin and PIK93 lead to the same conclusion, the pleiotropic effects imposed by these drugs and their cytotoxicity preclude rigorous confirmation of this assumption. Both inhibitors are not exclusively targeting PI4KIIIα (Tai et al., 2009; Borawski et al., 2009). Importantly, we demonstrate elevated levels of PI4P in HCV-containing cells both in cell culture and in vivo and provide convincing evidence that NS5A binds to and activates PI4KIIIα activity. Based on these results, we propose a model in which infecting RNA genomes are translated at the rER, giving rise to high amounts of polyprotein (Quinkert et al., 2005). NS5A generated by polyprotein cleavage binds to the kinase in a domain I-dependent manner and recruits the enzyme to ER-derived membranes. Binding of NS5A to the kinase stimulates its activity, resulting in high levels of PI4P at these membrane sites. Those membranes thus obtain a PIP-signature, which might contribute directly to membrane properties or recruit viral or host factors required for proper architecture and functionality of the membranous web.
In conclusion, we have identified kinases and pathways involved in HCV replication, most notably the PIP pathway. Our data provide a molecular mechanism by which PI4KIIIα contributes to HCV RNA replication and how elevated PI4P amounts are induced by viral infection. Given the crucial role of PI4kinases in HCV replication, they represent possible targets for development of antiviral therapy. Moreover, the perturbation of the PIP pathway induced by HCV infection may impact cell cycle control and thus contribute to the formation of hepatocellular carcinoma.
EXPERIMENTAL PROCEDURES
Cell lines, viruses and plasmid constructs
For infection experiments the highly permissive cell lines Huh7.5 (Blight et al., 2002), LH86 (Zhu et al., 2007), Lunet/CD81 (Koutsoudakis et al., 2007) or Huh7.5 FLuc were used. The latter was derived from Huh7.5 cells by stable retroviral transduction of the gene encoding for the Firefly luciferase (FLuc) using a transduction approach described elsewhere (Koutsoudakis et al., 2007). PTM vectors allowing expression of the HCV non-structural proteins NS3 to 5B with or without deletions of NS5A subdomains as well as individual HCV NS-proteins have been described recently (Backes et al., 2010). All sequences were derived from the JFH-1 isolate. The gene encoding the ~230 kDa full-length PI4KIIIα isoform 2 was obtained from Kazusa DNA Research Institute, Chiba, Japan (product ID: FXC00322, corresponding to GenBank accession # AB384703). Further details about cell lines and constructs are given in the Supplemental Data.
siRNA screening protocols
The primary screen targeted 719 human kinases with 3 siRNAs each and using reverse transfection on chambered coverglass slides (Lab-Tek chamber-slides, Thermo-Scientific) with an image-based read-out. The validation screen (entry and replication) targeted 178 hit candidates with 3 siRNAs each and using reverse transfection in a 96-well plate format. The HCVpp-based validation screen targeted 13 hit candidates with 3 siRNAs each and using reverse transfection in a 96-well plate format. Detailed information can be found in the Supplemental Experimental Procedures.
Immunoblot assay for PI4P quantification
Huh7-Lunet cells were electroporated with 10 µg of the respective viral RNA transcript and seeded into 6-well plates. After 48 h cells were lysed and PI4P was extracted using the PI4P-Mass-Kit (Echelon) according to the manufacturer’s instructions. Extracted PI4P was spotted onto membranes and detected using the PI4P-Mass-Kit and the ECL-plus reagent (Amersham). Quantification of PI4P amounts was carried out with the Quantity One software package (Bio-Rad). PI4P staining observed by immunofluorescence was quantified using the Image J program. A Z-projection of confocal Z-stacks was generated using the “sum slices” option. After thresholding the signal intensity of PI4P staining, PI4P amount was determined by defining cell areas and taking the IntDen-value obtained with the “analyze particles” functions of Image J.
Immunohistochemistry
IHC was performed on serial sections obtained from snap-frozen liver tissue using the DAKO EnVison System HRP (DAKO, K4004). Cryosections were fixed for 20 min at RT with prewarmed 4 % PFA and washed two times in PBS. Blocking and antigen detection were performed according to the manufacturer’s instructions with the following modifications: PBS was used for all washing steps, primary antibodies were incubated 1 h at RT and secondary antibodies for 45 min at RT. NS5A mouse monoclonal antibody 9E10 (generous gift of C.M. Rice) was used at a final concentration of 0.5 µg/ml, core monoclonal antibody C7-50 (Moradpour et al., 1996) at 2 µg/ml and the PI4P specific IgM monoclonal antibody (Echelon, Z-P004) at 1 µg/ml. Sections were counterstained with hematoxylin and mounted with Aqua-Tex (Merck). Human Tissue samples were provided by the Tissue bank of the National Center for Tumor Diseases Heidelberg after approval by the ethics committee.
Phosphatidylinositol-Kinase Assay
The lipid kinase assay measured the incorporation of phosphate into a phosphatidylinositol containing substrate. Fifty µl reactions were supplied with 1 µCi γ-[32P] ATP (Perkin Elmer), 10 µM non-radioactive ATP, 10 µM PI:PS (Invitrogen; PV5122), 1× Kinase buffer T (20 mM Tris/HCl pH7.4; 5 mM MgCl2; 0.5 mM EGTA; 0.4% Triton X 100), 2 mM DTT, 5 µl DMSO and 1 µg PI4KIIIα (generous gift of K. Lin and B. Wiedmann; Borawski et al., 2009). As a control, the PI4KIIIα inhibitor Wortmannin (100 µM; Sigma-Aldrich; W1628) was added. Increasing amounts of NS5A, NS3 or NS5B were added to the reactions. After 45 min at RT the assay was stopped by addition of 200 µl 1 M HCl and lipids were extracted with 400 µl of a 1:1 chloroform:methanol suspension. After rigorous mixing and centrifugation (13,000 rpm for 2min in a table top centrifuge), 125 µl of the organic phase were transferred to a scintillation vial and mixed with 4 ml UltimaGold® (Perkin Elmer). Incorporation of radioactive phosphate was measured by liquid scintillation counting.
Supplementary Material
Highlights.
13 human kinases, including PI4KIIIα, are required for HCV replication
HCV NS5A recruits PI4KIIIα to replication sites & stimulates its kinase activity
PI4P levels are elevated in HCV-containing cells in vitro and in vivo
Activity of PI4KIIIα is critical for the integrity of viral replication sites
ACKNOWLEDGEMENTS
We are grateful to Ulrike Herian, Stephanie Kallis, Rahel Klein, Jürgen Beneke and Nina Beil for excellent technical assistance. We thank Wolfgang Fischl for providing the DENV reporter construct and Charles M. Rice for the Huh7.5 cells and the NS5A monoclonal antibody 9E10. We are grateful to Kai Lin and Brigitte Wiedmann for providing purified PI4KIIIα and PIK93. We thank Jeremy Luban and Thomas Pertel for stable knock-down vector and Mark Harris for the NS5A polyclonal sheep serum. We thank Rossella Pellegrino for her assistance with IHC. We are grateful to the Nikon Imaging Center at the University of Heidelberg for providing access to their facility. This work was supported in part by the research program RNS/RNAi of the Landesstiftung Baden-W rttemberg (P-LS-RNS/30 to R.B.), a grant from the FORSYS ViroQuant project (BMBF, FKZ 0313923 to R.B. and V.L.) and the German Research Foundation DFG (SFB/TRR77, Teilprojekt A1 to R.B. and V.L., Teilprojekt B5 to T.L. and P.S.; grant Lo 1556/1-1 to V.L and KFO 129/1-2 to T.L. and M.A.). The ViroQuant-CellNetworks RNAi Screening core facility was supported by CellNetworks - Cluster of Excellence (EXC81). The work directed by M.A. was supported by the German National Genome Research Network NGFN (BMBF, contract number 01GR0453) and the DFG-funded Cluster of Excellence for Multimodal Computing and Interaction. The work of P.M. was funded by the FORSYS ViroQuant project and The Ministry of Education of the Czech Republic, grant numbers 2B06052 and MSM0021622419. M.P. is supported by an intramural grant of the Medical Faculty of the University of Heidelberg and M.-S. H. is supported by a Marie-Curie fellowship from the EU (EI-HCV). The work related to liver biopsy analysis was supported by the Tissue bank of the National Center for Tumor Diseases Heidelberg.
R.B. and members of the Molecular Virology unit in Heidelberg would like to dedicate this study to the memory of Professor Heinz Schaller, a wonderful mentor and colleague and a generous sponsor, who made this unit and the work conducted therein possible and who shaped Heidelberg Life Sciences so much.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amako Y, Sarkeshik A, Hotta H, Yates J, III, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J. Virol. 2009;83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS. Pathog. 2008;4 doi: 10.1371/journal.ppat.1000035. e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 2010;84:5775–5789. doi: 10.1128/JVI.02343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla T, Downing GJ, Jaffe H, Kim S, Zolyomi A, Catt KJ. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J. Biol. Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I, Baryza J, Tallarico J, Joberty G, Bantscheff M, Schirle M, Bouwmeester T, Mathy JE, Lin K, Compton T, Labow M, Wiedmann B, Gaither LA. Class III phosphatidylinositol-4 kinase alpha & beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS. Pathog. 2009;5 doi: 10.1371/journal.ppat.1000702. e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P -- not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Di Campli A, Godi A. The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protoc. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der SH, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DM, Domingues P, Targett-Adams P, McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J. Gen. Virol. 2010;91:2238–2248. doi: 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
- Kakuk A, Friedlander E, Vereb G, Jr, Kasa A, Balla A, Balla T, Heilmeyer LM, Jr, Gergely P, Vereb G. Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytometry A. 2006;69:1174–1183. doi: 10.1002/cyto.a.20347. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee D, Choe J. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem. Biophys. Res. Commun. 1999;257:777–781. doi: 10.1006/bbrc.1999.0460. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Tellinghuisen T. Purification and crystallization of NS5A domain I of hepatitis C virus. Methods Mol. Biol. 2009;510:85–94. doi: 10.1007/978-1-59745-394-3_7. [DOI] [PubMed] [Google Scholar]
- Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 2008;82:7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matula P, Kumar A, Worz I, Erfle H, Bartenschlager R, Eils R, Rohr K. Single-cell-based image analysis of high-throughput cell array screens for quantification of viral infection. Cytometry A. 2009;75:309–318. doi: 10.1002/cyto.a.20662. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Wakita T, Tokushige K, Carlson RI, Krawczynski K, Wands JR. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J. Med. Virol. 1996;48:234–241. doi: 10.1002/(SICI)1096-9071(199603)48:3<234::AID-JMV4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 2004;78:13306–13314. doi: 10.1128/JVI.78.23.13306-13314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, Packer J, Schurdak M, Molla A. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- Poenisch M, Bartenschlager R. New insights into structure and replication of the hepatitis C virus and clinical implications. Semin. Liver Dis. 2010;30:333–347. doi: 10.1055/s-0030-1267535. [DOI] [PubMed] [Google Scholar]
- Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 2007;81:4591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiha GE, Zalata KR, Abdalla AF, Mohamed MK. Immunohisto-chemical identification of HCV target antigen in paraffin-embedded liver tissue: reproducibility and staining patterns. Liver Int. 2005;25:254–260. doi: 10.1111/j.1478-3231.2005.01101.x. [DOI] [PubMed] [Google Scholar]
- Supekova L, Supek F, Lee J, Chen S, Gray N, Pezacki JP, Schlapbach A, Schultz PG. Identification of human kinases involved in HCV replication by siRNA library screening. J. Biol. Chem. 2008;283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host. Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Grise H. Cellular and molecular biology of HCV infection and hepatitis. Clin. Sci. (Lond) 2009;117:49–65. doi: 10.1042/CS20080631. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS. Pathog. 2008;4 doi: 10.1371/journal.ppat.1000032. e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotard M, Lepere-Douard C, Regeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB. J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Wong K, Meyers d, Cantley LC. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- Zhu H, Dong H, Eksioglu E, Hemming A, Cao M, Crawford JM, Nelson DR, Liu C. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133:1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.