Abstract
Chromosome maintenance protein 1 (CRM1) is a nuclear export receptor involved in the active transport of tumor suppressors (eg, p53 and nucleophosmin) whose function is altered in cancer because of increased expression and overactive transport. Blocking CRM1-mediated nuclear export of such proteins is a novel therapeutic strategy to restore tumor suppressor function. Orally bioavailable selective inhibitors of nuclear export (SINE) that irreversibly bind to CRM1 and block the function of this protein have been recently developed. Here we investigated the antileukemic activity of KPT-SINE (KPT-185 and KPT-276) in vitro and in vivo in acute myeloid leukemia (AML). KPT-185 displayed potent antiproliferative properties at submicromolar concentrations (IC50 values; 100-500nM), induced apoptosis (average 5-fold increase), cell-cycle arrest, and myeloid differentiation in AML cell lines and patient blasts. A strong down-regulation of the oncogene FLT3 after KPT treatment in both FLT3-ITD and wild-type cell lines was observed. Finally, using the FLT3-ITD–positive MV4-11 xenograft murine model, we show that treatment of mice with oral KPT-276 (analog of KPT-185 for in vivo studies) significantly prolongs survival of leukemic mice (P < .01). In summary, KPT-SINE are highly potent in vitro and in vivo in AML. The preclinical results reported here support clinical trials of KPT-SINE in AML.
Introduction
The nucleocytoplasmic exchange of proteins (macromolecules > 40 kDa) is a spatially and temporally regulated process that involves a number of nucleocytoplasmic shuttling proteins.1 Chromosome maintenance protein 1 (CRM1; Exportin-1, XPO1), a member of the karyopherin β family of transport receptors, is an important nuclear protein export receptor that recognizes hydrophobic, leucine-rich nuclear export signal (NES) and transports target proteins across a Ran-GTP gradient.2–5 CRM1 is involved in the active transport of a number of cargo proteins, including transcription factors, tumor suppressor proteins (TSPs), and cell-cycle regulators, such as p53,6 p21, p27,7 nucleophosmin 1 (NPM1),8,9 as well as RNA molecules.2 Recent data indicate that TSPs, such as p53, can be excluded from the nucleus and thereby inactivated in cancer by hyperactive nuclear export.10–12 Overexpression of CRM1 protein has been described in several cancers (glioblastoma, ovarian and cervical cancer), and it has been associated with a worse outcome.13–15 In addition, deregulated oncogenic pathways in cancer, such as aberrant AKT or BCR/ABL signaling, have been shown to cause posttranscriptional changes (in particular, phosphorylation) of TSPs, such as p27 and FOXO3, promoting their nuclear export through CRM1.16,17 Thus, prevention of CRM1-mediated nuclear export of TSP presents itself as an attractive antineoplastic therapeutic modality. However, progress in this direction has been limited because of the severe clinical toxicity of the anti-CRM1 drugs developed so far, the most well-known of which is leptomycin B.18 Karyopharm Therapeutics has developed novel, oral, bioavailable, small-molecule selective inhibitors of nuclear export (KPT-SINE) that specifically and irreversibly bind to CRM1 and block the function of this protein by binding to the reactive site Cys 528 residue.19–21 These compounds have been shown to induce apoptosis and block proliferation in several cancer cell lines, including colon,19,22 pancreas,22 and breast cancer23 as well as chronic myeloid24 and lymphocytic leukemias.25
Acute myeloid leukemia (AML) is a heterogeneous clonal disorder characterized by the accumulation of immature myeloid progenitors (blasts) in the bone marrow and peripheral blood.26 Nonrandom chromosomal abnormalities (eg, deletions, translocations, duplications, and inversions) are identified in approximately 55% of all adult primary AML patients.26 In contrast, approximately 40% to 50% of all AML cases are cytogenetically normal (CN-AML) when assessed using conventional banding analysis.26 Recent work has identified novel molecular abnormalities in CN-AML that has improved the classification and risk stratification of this large subgroup of patients.27 Among them, mutations of the NPM1 gene, usually occurring at exon 12 and more rarely at exon 11, represent the most common genetic alteration in CN-AML (50%-60% of cases) and account for approximately one-third of all adult AML.26,27 This gene encodes for a ubiquitously expressed nucleolar protein that shuttles between the nucleus and cytoplasm in a CRM1-dependent manner.28,29 NPM1 protein is implicated in multiple functions, including ribosomal protein assembly and transport, control of centrosome duplication, and regulation of Arf tumor suppressor gene integrity.29–32 NPM1 mutations specifically result in the inappropriate relocalization of NPM1 from the nucleus into the cytoplasm,29,33,34 hence the term NPMc+ (cytoplasmic-positive) AML. Overexpression of NPMc+ in mice progenitors induces myeloid proliferation, supporting a critical role in leukemogenesis.35 Remarkably, the aberrant nuclear export of NPM1 mutants is dependent on CRM1, as the NPM1 mutations lead to the creation of a novel CRM1 binding site in the NPM1. The cytoplasmic accumulation of NPM1 mutants is blocked by specific but toxic CRM1 inhibitors, such as leptomycin B.28 In addition to NPM1, internal tandem duplications in the juxtamembrane domain or mutations in the second tyrosine kinase domain of the FLT3 gene have been found in 30% to 45% of CN-AML and frequently coexist with NPM1 mutations.27 Both types of mutations constitutively activate FLT3, and FLT3-ITD mutations have been associated with increased risk of relapse.27 Therefore, therapies that restore mutant NPMc+ to the nucleus and/or suppress FLT3 abnormalities should be highly effective in AML.
In this work, we characterize the biologic and pharmacologic activity of KPT-CRM1 inhibitors in AML cell lines, patient blasts, and in a xenograft AML model. KPT-SINE show potent antiproliferative and proapoptotic properties against AML cell lines and patient blasts, including those from patients with NPM1 and FLT3-ITD mutations. As expected, KPT-SINE treatment restored the localization of cytoplasmic mutant NPM1 into the nucleus. Furthermore, KPT treatment results in cell-cycle arrest and blast differentiation. Finally, we also show in vivo antileukemia activity of KPT-SINE in a murine AML xenograft model bearing the FLT3-ITD abnormality.
Methods
Cell lines
AML cell lines, MV4-11, Kasumi-1, MOLM-13, THP-1, and KG-1a were purchased from ATCC. OCI-AML3 was purchased from DSMZ. All cell lines were cultured in RPMI supplemented with 10% FBS and 100 U/mL penicillin and 100 μg/mL streptomycin.
Primary AML samples
Newly diagnosed untreated and frozen bone marrow AML patient samples were obtained from The Ohio State University Leukemia Tissue Bank after getting informed consent approved by the cancer institution review board in accordance with the Declaration of Helsinki. Primary cells were cultured in StemSpan SFEM supplemented with 20% FBS and StemSpan CC100 cytokine cocktail (StemCell Technologies).
Cell viability assay and establishment of IC50 values and growth curves
Cells were seeded into 96-well plates and treated for 24, 48, and 72 hours with KPT-SINE at various concentrations ranging from 10nM to 10μM. Cell viability was evaluated using the cell proliferation reagent WST-1 (Roche Diagnostics) according to the manufacturer's protocol. The absorbance of wells at 450 nm (reference wavelength, 650 nm) was measured with a microplate reader (SoftMax Pro; Molecular Devices).
Cell-cycle analysis
Cells were treated with indicated KPT-SINE for 24 hours, harvested, washed in PBS, and fixed at a final concentration of 70% ice-cold ethanol. Cells were stored at −20°C, washed in PBS, and then stained with propidium iodide/RNase A/0.01% Trizol buffer for 40 minutes at 37°C. Cells were washed again and resuspended in PBS, for analysis by flow cytometry with a FACSCalibur (BD Biosciences). Cell-cycle events were analyzed by incorporation of propidium iodide. Results are representative of 4 independent experiments performed in duplicate.
Apoptosis assay
Cells were treated with KPT-SINE for 24, 48, and 72 hours. Annexin V staining was done using the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instructions. Analysis by flow cytometry was done with a BD LSRII, and data analysis was performed using FlowJo Version 9.4.5 (TreeStar).
Western blot
Western blot was performed according to standard protocol. Briefly, cells were collected by centrifuge and then rinsed with ice-cold PBS and lysed in protease inhibitor containing buffer for 30 minutes at 4°C. Total cell lysates were centrifuged, and the soluble supernatant was collected. Protein concentration was quantified by BCA microprotein assay kit (Pierce Biotechnology). Protein lysates (∼ 40 μg) were resolved by SDS-PAGE, transferred to Immobilon-P PVDF membrane (Millipore), and membranes were blocked for 1 hour with TBS containing 5% nonfat dry milk and 0.5% Tween 20 (TBST). Membranes were incubated with TBST containing 5% nonfat dry milk and primary antibodies against CEBPA (Cell Signaling Technology, 2295), FLT3 (sc-480; Santa Cruz Biotechnology), CRM1 (sc-5595; Santa Cruz Biotechnology), p53 (sc-126, Santa Cruz Biotechnology), p21 (sc-817; Santa Cruz Biotechnology), c-Kit (sc-168; Santa Cruz Biotechnology), and β-actin (Cell Signaling Technology). After washing with TBS, membranes were probed with HRP-conjugated anti–rabbit or anti–mouse secondary antibody (Cell Signaling Technology) for 1 hour at room temperature. Membranes were developed using ECL (Pierce Biotechnology) or ECL plus chemiluminescence detection reagent (GE Healthcare). Results are representative of 3 independent experiments.
Colony formation assay
Cells were treated with KPT-SINE for the indicated specified time, after which they were washed with PBS. Approximately 3000 cells were mixed with Methocult medium (StemCell Technologies) and plated into a 30-mm dish. Colonies (> 20 μm) were counted under microscope after 14 days.
siRNA transfection
p53 or scramble siRNA transfections were performed using the Amaxa Nucleofector (Amaxa). Briefly, OCI-AML3 cells were suspended in Amaxa Nucleofector Solution T supplemented with 100 pmol p53 or scramble ON_TARGETplus Smartpool siRNAs (Dharmacon), and the nucleofection was performed using cell-type specific protocol (X-001). KPT was introduced to cell culture 3 hours after transfection. Assays were performed at the indicated time points.
Immunofluorescent staining
Cells were first counted and diluted to 1 × 106/mL. Approximately 200 000 cells were cytospun to slide. The cells were then fixed in 3.7% formaldehyde for 15 minutes followed by permeabilization with 0.25% Triton. The slides were blocked by 1% goat serum for 30 minutes. The cells were incubated with primary antibody NPM1 (sc-56503; Santa Cruz Biotechnology) for 1 hour at room temperature. After washing, the sample was incubated with fluorochrome conjugated secondary antibody for 1 hour at room temperature. Nucleus was stained by 4,6-diamidino-2-phenylindole (DAPI). Samples were analyzed by confocal microscope (The Ohio State University Image facility).
Flow cytometry
Cell lines MV4;11, Kasumi-1, and OCI-AML3 were treated with DMSO control or KPT-SINE at the predetermined IC50 concentration for 24, 48, and 72 hours and stained with CD11b antibody (eBioscience). Cells were analyzed on LSR II (BD Biosciences), and data analysis was performed using FlowJo Version 9.4.5 (TreeStar).
Morphologic examination
Cells were treated with KPT-SINE or DMSO for indicated time points and cytospun onto glass sildes. Cells were fixed with methanol and Wright-Giemsa stained for morphologic examination.
Real-time PCR
Cellular RNA was extracted using TRIzol (Invitrogen) and reverse transcribed to cDNA using superscript III first-strand synthesis system for RT-PCR (Invitrogen). Gene expression levels of CEBPA, G-CSFR, and lysozyme were detected using TaqMan Gene expression assays (Applied Biosystems). Normalization was performed using 18s RNA expression levels. Comparative real-time quantitative PCR was performed in triplicate, and relative expression was calculated using the comparative Ct method.
Mice
Female nonobese diabetic severe combined immunodeficient-γ (NSG) mice which lack mature T cells, B cells, or functional NK cells and are deficient in cytokine signaling, were purchased from Jackson ImmunoResearch Laboratories. All mice used in the experiments were between 4 and 6 weeks of age. All animal studies were conducted in accordance to the rules and regulations of the Institutional Animal Care and Use Committee at The Ohio State University.
MV4-11 xenograft mouse model
Spleen cells (0.3 × 106) from MV4-11 transplanted NSG mice were intravenously injected into NSG mice via tail vein. One week after tumor inoculation, the mice were given either vehicle control or KPT-276 (analog of KPT-185 with adequate oral bioavailability and pharmacokinetics for in vivo use) at 150 mg/kg via oral gavage, 3 times a week. Mice were monitored closely for clinical signs of leukemia, such as weight loss and hindlimb paralysis. Expected median survival for untreated animals in this model is 28 days. Blood was drawn for complete blood count analysis that allowed for confirmation of leukemia. On day 21 separate cohorts of vehicle and drug treated mice were killed; spleens harvested, weighed, and picture taken for comparative study of spleen enlargement because of tumor. Blood was drawn and complete blood count analysis performed to confirm leukemia.
Statistical analysis
Survival data were analyzed using Kaplan-Meier and long-rank test methods (GraphPad Prism Version 5.0). Differences between continuous variables (eg, RNA expression, spleen weights) were analyzed using t tests. All P values are 2-sided.
Results
KPT-SINE significantly inhibits proliferation and induces cell-cycle arrest and apoptosis of AML cell lines and primary AML blasts
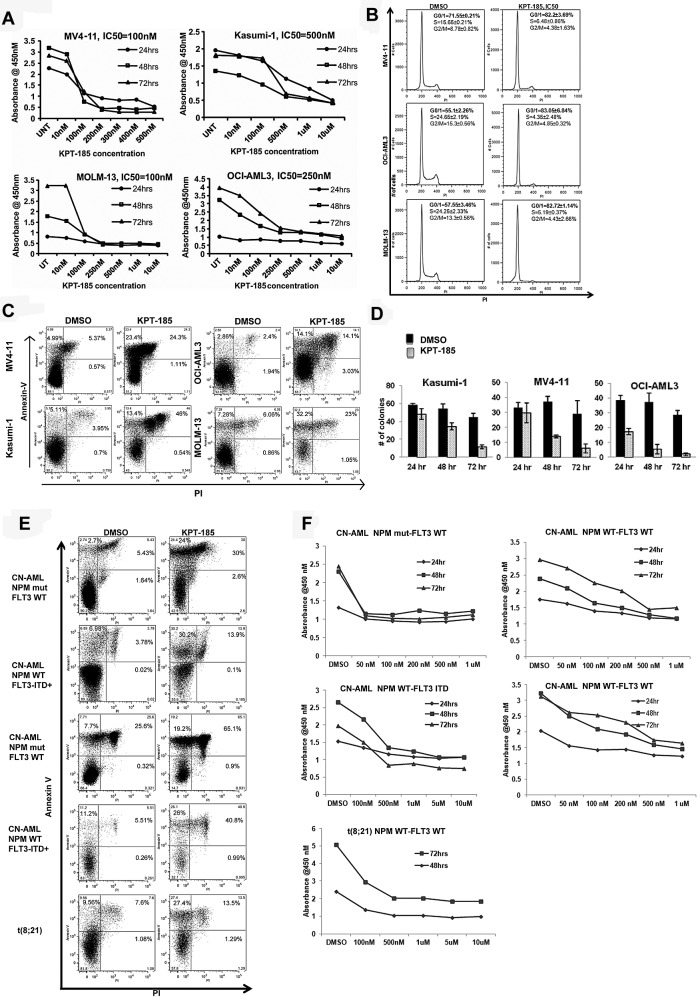
To assess the biologic activity of KPT-SINE CRM1 inhibitors, we treated a panel of AML cell lines (MV4-11, Kasumi-1, OCI/AML3, MOLM-13, KG1a, and THP-1), well characterized for cytogenetic and molecular features (Table 1). Submicromolar concentrations of KPT-185 inhibited leukemia cell proliferation, with IC50 values ranging from 100nM to 500nM (Table 1; Figure 1A).
Table 1.
IC50 values for AML cell lines
| Cell line | Relevant cytogenetic/molecular data | IC50, nM |
|---|---|---|
| MV4-11 | XY, +8, +18, +19, −21, t(4;11)(q21;q23); FLT3-ITD+ | 100 |
| MOLM-13 | XY, +8, +13, del(8), ins(11;9)(q23;p22p23); FLT3-ITD+ | 100 |
| OCI-AML3 | X/XY, +1, +5, +8, t(1;18)(p11;q11), i(5p), del(13) (q13q21), dup(17)(q21q25); NPM1 mutation (type A) and the DNMT3A R882C mutation | 250 |
| Kasumi-1 | X, −Y, −9, −13, −16, t(8;21)(q22;q22); KIT mutation (Asn822Lys) | 500 |
| KG1a | X/XY, −4, +8, +8, −12, −17, −20, del(5)(q?11q?13), dup(7)(q12q33), del(7)(q22q35) | 250 |
| THP-1 | XY/XXY, −Y, +1, +3, +6, +6, −8, −13, −19, −22, −22, , del(1)(q42.2), del(6)(p21), t(9;11)(p22;q23) | 250 |
Figure 1.
KPT-SINE significantly inhibits proliferation and induces cell-cycle arrest and apoptosis of AML cell lines and primary AML blasts. (A) WST-1 assays in MV4-11, Kasumi-1, MOLM-13, and OCI-AML-3. (B) Cell-cycle assessment using propidium iodine (PI) detection by flow cytometry at 24 hours. (C) Apoptosis as measured by annexin V/PI staining using FACS at 48 hours. (D) Colony assays in Kasumi-1, MV4-11, and OCI-AML3 cells after KPT-185 treatment at 24, 48, and 72 hours. (E) Apoptosis as measured by annexin V/PI staining using FACS at 48 hours in primary AML samples. (F) WST-1 assays in primary AML samples.
Because CRM1 is involved in the export of several proteins involved in cell-cycle control (eg, p21) to the cytoplasm, we assessed the impact of KPT-185 treatment on the cell cycle of AML cells. KPT-185 at the predetermined IC50 value induced cell-cycle arrest at G1 with respect to vehicle-treated-control (DMSO) in MV4-11 (82.2% ± 3.69% vs 71.55% ± 0.21%, P < .01), OCI/AML3 (83.05% ± 6.84% vs 55.1% ± 2.26%, P < .01), and MOLM-13 (82.72% ± 1.14% vs 57.55 ± 3.46%, P < .01) cells at 24 hours (Figure 1B). A concomitant decrease in the percentage of KPT-185–treated cells with respect to controls in S phase and G2/M phase was observed at 24 hours (Figure 1B). Interestingly, treatment of Kasumi-1 cells with KPT-185 did not cause an arrest at G1; rather, there was a significant accumulation of apoptotic cells in sub-G1 (24.32% ± 6.01% vs 5.44% ± 2%, KPT-185 vs DMSO, P < .01), with a decrease in percentage of cells in the S (7.67% ± 1.21% vs 18.6% ± 1.2%, P < .01) and G2M phase (6.35% ± 2.79% vs 15.91% ± 4.44%, P < .01).
Treatment with KPT-185 at the predetermined IC50 value induced apoptosis in AML cell lines compared with DMSO treated controls at 48 hours as follows: MV4-11, 5.7-fold increase (P < .01); Kasumi-1, 5.41-fold increase (P < .01); OCI-AML3, 4.91-fold (P < .01) and MOLM-13, 4-fold increase (P < .01; Figure 1C). Cell colony formation was reduced significantly after KPT-185 treatment at the IC50 for 72 hours in AML cell lines (Kasumi-1: 44.67 ± 4.16 vs 11.33 ± 2.08; MV4-11: 29 ± 8.88 vs 6 ± 3; OCI/AML3: 28.66 ± 3.05 vs 2 ± 1; respectively, P < .01, Figure 1D). Next, we validated our findings using primary AML blasts (Table 2 for cytogenetic and molecular abnormalities). KPT-185 inhibited cell proliferation and induced apoptosis in primary AML blasts across a variety of genotypes (Table 2; Figure 1E-F). The IC50 values were in the 500nM range, except for CN-AML patients with NPM1 mutations that were more sensitive to the KPT-SINE (IC50 100nM; Table 2).
Table 2.
IC50 for primary AML patient samples
| Patient no. | Age, y | WHO classification | WBCs* | Cytogenetics | NPM1 | FLT3 | IC50, nM |
|---|---|---|---|---|---|---|---|
| 1 | 27 | Acute myelomonocytic leukemia | 39 | 46,XX(20) | Mut (A) | WT | 100 |
| 2 | 42 | AML with maturation | 26 | 46,XY(20) | Mut (A) | WT | 100 |
| 3 | 62 | AML without maturation | 199 | 46,XX(20) | Mut (A) | WT | 100 |
| 4 | 77 | AML with maturation | 85 | 46,XY(20) | Mut (A) | WT | 50 |
| 5 | 62 | AML with MDS-related changes | 8.8 | 46,XY(20) | WT | WT | 500 |
| 6 | 52 | AML with maturation | 75 | 46,XY(20) | WT | WT | 500 |
| 7 | 45 | Acute myelomonocytic leukemia | 53 | 46,XX(20) | WT | WT | 500 |
| 8 | 56 | Acute myelomonocytic leukemia | 69 | 46,XX(20) | WT | ITD+ | 500 |
| 9 | 20 | AML with inv(16) | 45 | 46,XX,inv(16) | WT | WT | 500 |
| 10 | 53 | Acute myelomonocytic leukemia | 79 | 46,XX(20) | WT | ITD+ | 500 |
| 11 | 85 | AML without maturation | 66 | 46,XY(20) | WT | ITD+ | 500 |
| 12 | 52 | AML with t(8;21) | 2.9 | 45,X,−X,t(8;21) | WT | WT | 500 |
| 13 | 50 | AML with t(8;21) | 15.6 | 45,X,−Y,t(8;21) | WT | WT | 500 |
Mut indicates mutated; and WT, wild-type.
× 105.
KPT-SINE treatment causes decrease of CRM1 protein level and accumulation of CRM1 cargo proteins in the nucleus
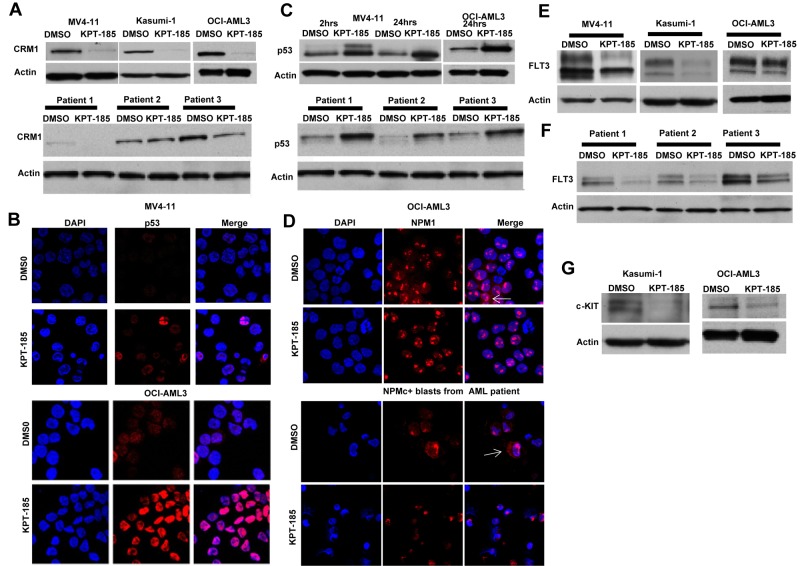
The effect of CRM1 inhibitor, KPT-185, on the level of CRM1 protein was investigated in AML cell lines and primary AML blasts. We observed a significant decrease in the level of CRM1 protein by Western blot, as shown in Figure 2A. Similar findings have been observed in multiple myeloma cell lines after using a different CRM1 inhibitor.36 In addition, we also measured the expression levels of 2 known CRM1 cargo proteins, p53 and NPM1, after treatment with KPT-SINE. A significant accumulation of p53 in the nucleus of MV4-11 and OCI-AML3 was observed after treatment with KPT-185 (Figure 2B). This was confirmed by Western blotting in both cell lines (Figure 2C top panel) and in primary AML blasts (Figure 2C bottom panel). KPT-185 treatment of the NPM1 mutated OCI/AML3 cell line (which exhibits cytoplasmic localization of the protein37) and of primary AML blasts from a patient with NPM1 mutation resulted in the accumulation of NPM1 into the nucleolus with respect to the controls (Figure 2D).
Figure 2.
KPT-SINE treatment causes decrease of CRM1 protein level, accumulation of CRM1 cargo proteins in the nucleus, and down-regulation of FLT3 and KIT oncoproteins. (A) Top panel: CRM1 protein expression as measured by Western blotting in MV4-11, Kasumi-1, and OCI-AML3 cells after KPT-185 treatment or control (DMSO) after 24 hours. Loading control is actin. Bottom panel: CRM1 protein expression as measured by Western blotting in 3 primary AML blasts after KPT-185 treatment or control. (B) Confocal microscopy of p53 in MV4-11 and OCI-AML3 cells treated with KPT-185 or control at 24 hours. Left panel: DAPI staining (cell nucleus). Middle panel: p53 staining. Right panel: merge of p53 and DAPI staining. Note the increase in the p53 expression in the nucleolus. (C) Whole-cell p53 protein expression in MV4-11 and OCI-AML3 cells (top panel) or primary AML blasts (n = 3) after KPT-185 or control treatment at 2 and 24 hours. (D) Confocal microscopy of NPM1 in OCI-AML3 cells and in a primary AML blast from a patient with CN-AML and NPM1 mutation treated with KPT-185 or control at 24 hours. Left panel: DAPI staining (cell nucleus). Middle panel: NPM1 staining. Right panel: merge of NPM1 and DAPI staining. The arrows indicate the localization of NPM1 in the cytoplasm (NPMc+) in the untreated samples and the elimination of the cytoplasmic signal on treatment with the drug, being detected exclusively in the nucleus. (E) FLT3 protein expression in MV4-11, Kasumi-1, and OCI-AML3 cells as measured by Western blotting after KPT-185 treatment or control at 24 hours. (F) FLT3 protein expression in primary AML blasts as measured by Western blotting after KPT-185 treatment or control at 24 hours: patient 1, CN-AML NPM1 WT, FLT3 ITD+; patient 2, CN-AML, NPM1 mutated, FLT3 WT; and patient 3, CN-AML, NPM1 mutated, FLT3 WT. (G) c-KIT expression in Kasumi-1 and OCI-AML3 cells after KPT-185 treatment or control at 24 hours.
FLT3 and KIT oncogene proteins are down-regulated after CRM1 inhibition at the posttranscriptional level
Next, we asked whether critical oncogenic proteins involved in myeloid leukemogenesis are affected by CRM1 inhibition. In particular, we were interested in FLT3 because this tyrosine kinase receptor is overexpressed in the majority of AML patients and mutated (FLT3-ITD) frequently in patients with CN-AML and NPM1 mutations.26,27 Remarkably, we found a strong down-regulation of total FLT3 protein expression in AML cell lines and primary AML samples (Figure 2E-F), irrespective of the mutation status. To dissect the possible mechanism of such an effect, we measured FLT3 mRNA expression after CRM1 inhibition and found no changes in FLT3 mRNA levels after KPT-185 treatment (data not shown). This result suggests that FLT3 protein expression down-regulation occurs at the posttranscriptional level. Next, we asked whether c-KIT (another tyrosine kinase receptor), which is also up-regulated in the great majority of AML cases and mutated in a fraction of patients with core binding factor leukemia, could be a target for CRM1 inhibition.38,39 Interestingly, similarly to FLT3, we found that c-KIT protein, but not mRNA expression levels, was decreased in Kasumi-1 and OCI-AML3 cells after KPT-185 treatment (Figure 2G). Thus, our data indicate that KPT-SINE decreases FLT3 and KIT protein expression posttranscriptionally in AML blasts.
CRM1 inhibition induces differentiation of AML cell lines
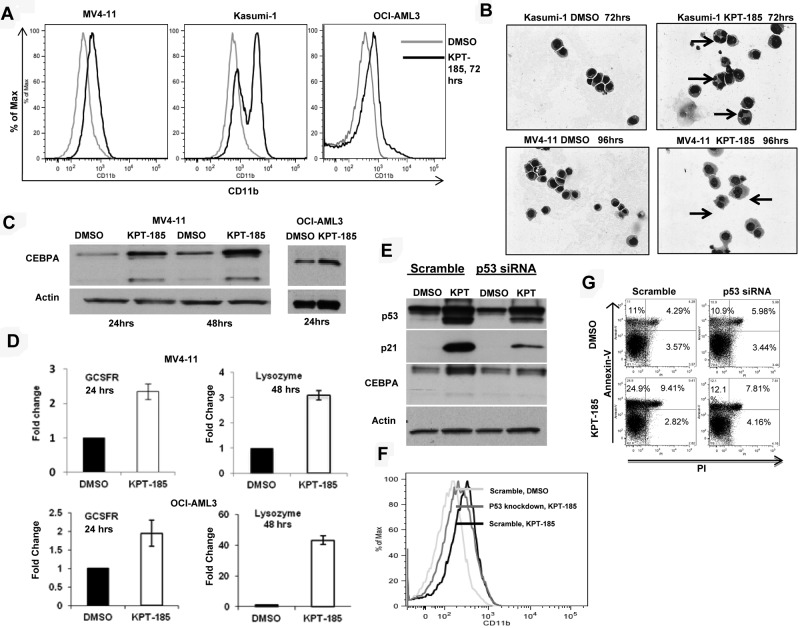
Next, we investigated whether CRM1 inhibition could result in AML blast differentiation. Indeed, we observed a significant increase in the myeloid differentiation marker CD11b expression as determined by FACS in MV4-11, Kasumi-1, and OCI-AML3 after KPT-185 treatment with respect to controls (Figure 3A). In addition, morphologic changes characteristic of differentiation, such as the appearance of granules and condensation of the nucleus, were confirmed by Giemsa staining of cytospin preparations of KPT-185-treated cells (Figure 3B).
Figure 3.
CRM1 inhibition induces differentiation of AML cell lines. (A) CD11b measurement by FACS in AML cell lines after KPT-185 treatment at 72 hours. (B) Giemsa stain of cytospins of Kasumi-1 and MV4-11 cells treated with KPT-185 or controls. Magnification, 40×. Arrows indicate nuclear condensation. (C) CEBPA protein expression in MV4-11 cells and OCI-AML3 cells after KPT-185 treatment or controls for 24 and 48 hours. Loading control is actin. (D) GCSFR and lysozyme mRNA expression after KPT-185 treatment or controls in MV4-11 cells and OCI-AML3 cells. Results are shown as fold change after normalization with 18s and 2ΔCt calculations. (E) CEBPA protein expression after p53 siRNA or scramble oligonucleotide transfection in OCI-AML3 cells subsequently treated with KPT-185 or controls for 24 hours. Protein expression of p21 was measured as control for successful p53 inhibition. (F) CD11b expression in OCI-AML3 cells after transfection with scramble or p53 antisense oligonucleotides and treatment with DMSO or KPT-185 at 72 hours. (G) Apoptosis as measured by annexin V/PI staining using FACS at 48 hours after scramble or p53 antisense oligonucleotides and treatment with DMSO or KPT-185.
To obtain insights about the mechanisms by which KPT-SINE induces blast differentiation we measured the expression of CEBPA, a member of the basic region leucine zipper family of transcription factors, which has been shown to be indispensible for granulocytic differentiation of myeloid progenitors.40,41 Remarkably, we observed a significant increase in the protein levels of CEBPA, with little change in mRNA levels, in MV4-11 and OCI-AML3 cell lines (Figure 3C). We did not see any changes in the CEBPA protein levels when we treated the Kasumi-1 cell line with the CRM1 inhibitor (data not shown). This result could be explained by the fact that the AML1-ETO fusion protein encoded by the t(8;21) is known to suppress the both CEBPA mRNA and protein levels.42 Because CEBPA is known to induce the transcription of differentiation-related genes, such as the G-CSFR and lysozyme,41,43 we measured the mRNA expression of these genes by RT-PCR in the MV4-11 and the OCI-AML3 cell lines and confirmed that both genes were induced after KPT-185 treatment with respect to control (Figure 3D).
Based on previous reports that indicate that CEBPA is a p53 regulated DNA damage-inducible gene44 and that p53 induction is involved in myeloid differentiation,45 we asked whether the CEBPA up-regulation observed after KPT-185 treatment is mediated through p53 and/or whether p53 increased expression is responsible for the phenotypic effects observed. To answer this question, we repressed endogenous p53 expression using siRNA in OCI-AML3 cells, treated the cells with KPT-185 or DMSO for 24 hours, and measured CEBPA, p53 protein, CD11b, and apoptosis over time. As shown in Figure 3E, dampening p53 expression in OCI-AML3 cells abrogates CEBPA up-regulation induced by KPT-185, decreases the expression of CD11b, and induces less apoptosis (Figure 3F-G). These results suggest that, in this model, CEBPA is a p53-regulated gene44 that may be involved along with other p53 target genes in the differentiation program activated by CRM1 inhibition.
CRM1 inhibitor increases survival in a human leukemia xenograft model
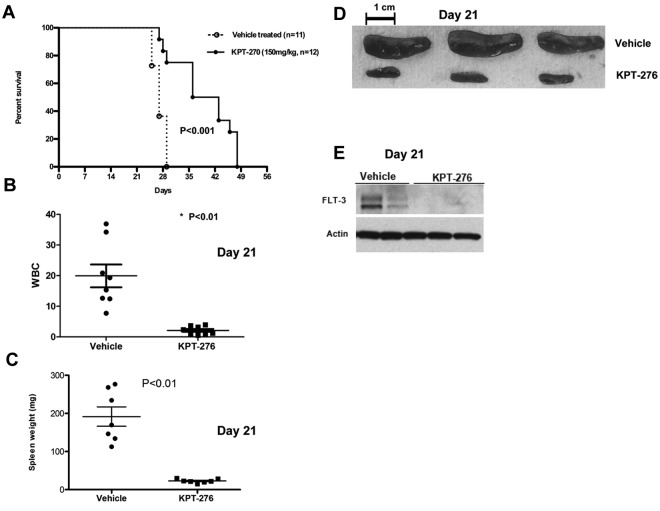
To establish the activity of KPT-SINE in AML in vivo, we used a xenograft human AML murine model (MV4-11). In this model, NOD/SCID γ mice were intravenously inoculated (tail vein) with human MV4-11 AML cells, which carry FLT3-ITD. One week after leukemic cell inoculation, the mice were given KPT-276 at 150 mg/kg via oral gavage, 3 times a week, or vehicle control. KPT-276 has the identical CRM1 binding warhead and specificity as KPT-185, similar biologic activity in vitro, but superior oral bioavailability and pharmacokinetics, which allow it to be used in vivo. Mice were monitored for survival. Some mice were killed at day 21 to assess the effects of KPT-276 on leukemia burden by measuring spleen weight and white blood cell count. KPT-SINE treatment significantly increased the survival of the mice (median survival vehicle vs drug-treated, 27 vs 39.5 days, respectively; P < .01, log-rank test, n = 12 per group, Figure 4A). The average white blood cell count was 19 950 ± 3.7 in the control group versus 2120 ± 0.48 in the KPT-276 group (P = .0003; Figure 4B) at 21 days. The average spleen weight was also significantly higher in the control group versus KPT-276 arm at 21 days (191.7 mg ± 25.3 vs 22.81 mg ± 1.9, respectively, P < .01; Figure 4C-D). FLT3 protein expression was significantly lower in the spleen of leukemic mice treated with KPT-276 with respect to the vehicle controls at 21 days (Figure 4D). Altogether, our data show that KPT-276 is active in vivo and prolongs the survival of the leukemic mice.
Figure 4.
CRM1 inhibitor increases survival in a human leukemia xenograft model. (A) Survival of MV4-11 xenograft mice after treatment with KPT-276 150 mg/kg (n = 12) or vehicle control (n = 11). Survival comparison was made with log-rank test. (B) White blood cell count (in thousands) in KPT-treated mice versus vehicle control (n = 8) at 21 days. P values obtained using t test. (C) Spleen weights (mg) in KPT-treated mice versus vehicle control (n = 7). P values obtained using t test. (D) Spleen photographs of 3 representative cases (KPT-276 n = 3; vehicle n = 3) at 21 days. (E) FLT3 protein expression in mouse spleen cells as measured by Western blotting after KPT-276 treatment or vehicle control at 21 days.
Discussion
In this manuscript, we show the in vitro and in vivo antitumor efficacy of KPT-SINE CRM1 antagonists in AML. Our results indicate that KPT-185 inhibits cell proliferation and induces cell-cycle arrest and apoptosis of AML cell lines and primary blasts. Treatment of AML blasts with KPT-185 causes a reduction in the amount of CRM1 protein and shows a significant nuclear accumulation of CRM1 cargo proteins, such as p53 and NPM1. Using a xenograft AML mouse model, we show that in vivo treatment of leukemic mice with oral KPT-276 (analog of KPT-185 for in vivo studies) significantly prolongs survival of leukemic mice and reduces leukemic burden.
Interestingly, we found that primary AML blasts harboring NPM1 mutations were very responsive to CRM1 inhibition. NPM1 is a nucleolar tumor suppressor phosphoprotein that continuously shuttles between the nucleolus and cytoplasm and regulates the p53-ARF tumor suppressor pathway.30–32,46 Mutations in the exon 12 of NPM1 have been described in 25% to 35% of AML and are one of the most frequent mutations described to date in this disease.27,29,33,47 These mutations alter the NPM1 protein at the C-terminus, leading to the formation of a novel and efficient CRM1 binding site, thus causing its aberrant cytoplasmic localization through a CRM1-dependent transport.28 Supporting an oncogene role for mislocalized mutant NPM1, it has been reported that overexpression of a human NPMc+ mutation in murine myeloid progenitors resulted in cytoplasmic NPM1 localization and myeloproliferation in the bone marrow and spleen.35 Based on these data, we reasoned that blocking NPMc+ nuclear export by inhibiting CRM1 may restore NPM1 tumor suppressor functions and prevent leukemogenesis. Here we have shown that blocking CRM1 does indeed restore nuclear localization of NPM1 mutants and induces potent antileukemic effects in cell lines and primary human AML blasts. Indeed, NPMc+ AML blasts from patients were the most sensitive to KPT inhibition (IC50 100 nm) among all primary samples tested. Altogether, our results indicate that redirecting mutated NPM1 to the nucleus through CRM1 inhibition is a potential targeted therapy for this frequent subtype of AML. However, the fact that other AML cytogenetics groups harboring wild-type NPM1 were also sensitive to KPT-SINE indicate that other TSPs and, in particular p53, are involved in the antileukemic effects of the KPT-SINE.
Disruption of terminal differentiation is a salient feature in the pathogenesis of AML,48 and differentiation-based anticancer treatments, such as all-trans retinoic acid, have been developed to overcome this block, thereby inducing apoptosis.49 Here we show that treatment of AML cancer cells with CRM1 inhibitors induces blast differentiation, as shown by the surface expression of the myeloid differentiation marker CD11b, as well as morphologic changes associated with differentiation. To investigate further possible mechanisms by which CRM1 inhibition using KPT-SINE resulted in blast differentiation, we measured the expression of proteins known to be involved in myeloid differentiation, such as CEBPA. Interestingly, we found a significant increase in the protein levels of CEBPA in MV4-11 and OCI-AML3 cells, whereas the levels of CEBPA mRNA were mildly increased only for MV4-11 cells. CEBPA has been shown to induce myeloid differentiation via transcriptional activation of several genes that are critical for myeloid granulocytic differentiation.41,43 Based on work by Yoon and Smart, who reported that CEBPA expression is up-regulated by p53 induced by ultraviolet radiation in keratinocytes,44 we reasoned that CEBPA up-regulation after CRM1 inhibition could be mediated by p53, a direct CRM1 target. Indeed, we show that blocking p53 expression using siRNA in OCI-AML3 cell lines abrogates CEBPA protein induction and differentiation by KPT-185. It is important to mention that CEBPA protein expression did not change in Kasumi-1 cells despite exhibiting blast differentiation after KPT treatment. This argues that CEBPA alone is unlikely to be the primary mechanism. Our results point to p53 as a critical mediator not only for the KPT induced apoptosis but also differentiation.
In this work, we also show that growth-promoting c-KIT and FLT-3 tyrosine kinase receptor proteins are down-regulated after KPT-185 treatment of AML cell lines and primary samples. This is relevant to AML because both tyrosine kinase receptors are overexpressed in the majority of AML and/or mutated in some cases.26,27,38,39 c-KIT gain-of-function mutations appear to be restricted to core binding factor AML, which confers poor prognosis.38,39 In contrast, activating mutations in the JM domain (FLT3-ITD) and in the tyrosin kinase domain of FLT3 are found in 30% to 35% of patients with AML and represent the most frequent genetic alterations in AML.26,50 Aberrant activation of either c-KIT or FLT3 signaling resulting from mutations and/or overexpression promotes cell proliferation and contributes to leukemogenesis.27,38,39 Thus, the ability of KPT-SINE CRM1 inhibitors to down-regulate KIT and FLT3 protein expression in AML constitutes an important antileukemic mechanism for these compounds. Because patients with NPM1 mutations frequently have FLT3-ITD mutations,27 CRM1 inhibitors have the potential to target 2 critical mutated signaling pathways simultaneously while increasing levels and nuclear localizations of TSP. Last, our discovery has an immediate translational impact because FLT3 expression could be used as a novel pharmacodynamic endpoint for testing of CRM1 inhibitors in phase 1 clinical trials for AML.
In conclusion, here we report the biologic and pharmacologic activity of KPT-SINE CRM1 inhibitors in both AML cells, primary AML samples, and in a murine AML xenograft model. Our results indicate that KPT-SINE CRM1 inhibitors dampen cell proliferation and induce cell-cycle arrest, apoptosis, and cell differentiation through the p53-CEBPA pathway. We have identified that both KIT and FLT3 protein are down-regulated after KPT-CRM1 inhibitor treatment. The preclinical in vitro and in vivo results reported here support further study of KPT-SINE CRM1 inhibitors as a novel therapeutic strategy for AML.
Acknowledgments
The authors thank Donna Bucci for providing primary AML samples from The Ohio State University Leukemia Tissue Bank, Rosa Lapalombella for helpful discussions, and Dr John C. Byrd for reviewing the final manuscript.
This work was supported by the National Institutes of Health (grant P50CA140158 and Pelotonia Fellowship award, P.R.).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.R., X.Y., C.N., and R.S. performed all the experiments; S.S., M.K., M.C., C.M.C., G.M., and R.G. designed, supervised, and analyzed research performed in their laboratories; A.W., R.K., and W.B. and collected primary AML samples; and P.R. and R.G. wrote the manuscript.
Conflict-of-interest disclosure: M.K. and S.S. are employees of Karyopharm Therapeutics, a clinical-stage biopharmaceutical company that develops selective inhibitors of nuclear export-targeted therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Ramiro Garzon, Division of Hematology and the Comprehensive Cancer Center, The Ohio State University, 1084 BRT, 460 West 12th Ave, Columbus, OH 43210; e-mail: ramiro.garzon@osumc.edu.
References
- 1.Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4(3):127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 2.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17(4):193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr Med Chem. 2008;15(26):2648–2655. doi: 10.2174/092986708786242859. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda M, Asano S, Nakamura T, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390(6657):308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 5.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278(5335):141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 6.Kanai M, Hanashiro K, Kim SH, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9(10):1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 7.Connor MK, Kotchetkov R, Cariou S, et al. CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol Biol Cell. 2003;14(1):201–213. doi: 10.1091/mbc.E02-06-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinmura K, Tarapore P, Tokuyama Y, George KR, Fukasawa K. Characterization of centrosomal association of nucleophosmin/B23 linked to Crm1 activity. FEBS Lett. 2005;579(29):6621–6634. doi: 10.1016/j.febslet.2005.10.057. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005;7(8):823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 10.Mutka SC, Yang WQ, Dong SD, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69(2):510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stommel JM, Marchenko ND, Jimenez GS, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18(6):1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vousden KH, Woude GF. The ins and outs of p53. Nat Cell Biol. 2000;2(10):E178–E180. doi: 10.1038/35036427. [DOI] [PubMed] [Google Scholar]
- 13.Noske A, Weichert W, Niesporek S, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112(8):1733–1743. doi: 10.1002/cncr.23354. [DOI] [PubMed] [Google Scholar]
- 14.Shen A, Wang Y, Zhao Y, et al. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65(1):153–159. doi: 10.1227/01.NEU.0000348550.47441.4B. discussion 159-160. [DOI] [PubMed] [Google Scholar]
- 15.van der Watt PJ, Maske CP, Hendricks DT, et al. The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124(8):1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeshan K, Cotter TG, McKenna SL. Bcr-Abl upregulates cytosolic p21WAF-1/CIP-1 by a phosphoinositide-3-kinase (PI3K)-independent pathway. Br J Haematol. 2003;123(1):34–44. doi: 10.1046/j.1365-2141.2003.04538.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, Liao Y, Xia W, et al. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3(3):245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 18.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74(4):648–649. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draetta G. F., Shacham S., Kauffman M., et al. Cytotoxicity of novel, small molecule, CRM1-selective inhibitors of nuclear export (SINE) in colorectal cancer (CRC) cells [abstract]. J Clin Oncol. 2011;29 Abstract e14091. [Google Scholar]
- 20.Shacham S, Joel G, Turner P, Nir R, et al. Preclinical development of small-molecule CRM1 inhibitors as novel therapy for the treatment of myeloma and other hematological malignancies [abstract]. Blood (ASH Annual Meeting Abstracts) 2010;116 Abstract 3012. [Google Scholar]
- 21.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83(8):1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azmi AS, Kauffman M, McCauley D, Shacham S, Mohammad RM. Novel small-molecule CRM-1 inhibitor for GI cancer therapy [abstract]. J Clin Oncol. 2012;30(Suppl 4) Abstract 245. [Google Scholar]
- 23.McCauley D, Landesman Y, Senapedis W, et al. Preclinical evaluation of selective inhibitors of nuclear export (SINE) in basal-like breast cancer (BLBC) [abstract]. J Clin Oncol. 2012;30(Suppl) Abstract 1055. [Google Scholar]
- 24.Walker C, Oaks JJ, Santhanam R, et al. Nuclear export (karyopherin) inhibitors: a novel therapeutic strategy for treating Philadelphia-positive (Ph+) acute leukemias.. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; March 31 to April 4, 2012; Chicago, IL. Abstract 3839. [Google Scholar]
- 25.Lapalombella R, Berglund C, Mahoney E, et al. CRM1/XPO1 represents a promising therapeutic target for treatment of chronic lymphocytic leukemia [abstract]. Blood (ASH Annual Meeting Abstracts) 2011;116 Abstract 232. [Google Scholar]
- 26.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 27.Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109(2):431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falini B, Bolli N, Shan J, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107(11):4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 29.Falini B, Nicoletti I, Martelli MF, Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood. 2007;109(3):874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 30.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Hann SR. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8(17):2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meani N, Alcalay M. Role of nucleophosmin in acute myeloid leukemia. Expert Rev Anticancer Ther. 2009;9(9):1283–1294. doi: 10.1586/era.09.84. [DOI] [PubMed] [Google Scholar]
- 33.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 34.Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009;21(6):573–581. doi: 10.1097/CCO.0b013e3283313dfa. [DOI] [PubMed] [Google Scholar]
- 35.Cheng K, Sportoletti P, Ito K, et al. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood. 2010;115(16):3341–3345. doi: 10.1182/blood-2009-03-208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakakibara K, Saito N, Sato T, et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118(14):3922–3931. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- 37.Quentmeier H, Martelli MP, Dirks WG, et al. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia. 2005;19(10):1760–1767. doi: 10.1038/sj.leu.2403899. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda H, Kanakura Y, Tamaki T, et al. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991;78(11):2962–2968. [PubMed] [Google Scholar]
- 39.Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24(24):3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 40.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273(44):28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 41.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90(2):489–519. [PubMed] [Google Scholar]
- 42.Pabst T, Mueller BU, Harakawa N, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7(4):444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 43.Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPalpha is required for induction of granulocytic differentiation. Blood. 2003;102(4):1267–1275. doi: 10.1182/blood-2003-02-0477. [DOI] [PubMed] [Google Scholar]
- 44.Yoon K, Smart RC. C/EBPalpha is a DNA damage-inducible p53-regulated mediator of the G1 checkpoint in keratinocytes. Mol Cell Biol. 2004;24(24):10650–10660. doi: 10.1128/MCB.24.24.10650-10660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer M, Rubsamen D, Slany R, et al. Oncogenic RAS enables DNA damage- and p53-dependent differentiation of acute myeloid leukemia cells in response to chemotherapy. PLoS One. 2009;4(11):e7768. doi: 10.1371/journal.pone.0007768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4(7):529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 47.Falini B, Bolli N, Liso A, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23(10):1731–1743. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- 48.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3(2):89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 49.Bruserud O, Gjertsen BT. New strategies for the treatment of acute myelogenous leukemia. Differentiation induction: present use and future possibilities. Stem Cells. 2000;18(3):157–165. doi: 10.1634/stemcells.18-3-157. [DOI] [PubMed] [Google Scholar]
- 50.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100(1):59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]