Abstract
Acute myeloid leukemia (AML) is the result of a multistep transforming process of hematopoietic precursor cells (HPCs) which enables them to proceed through limitless numbers of cell cycles and to become resistant to cell death. Increased proliferation renders these cells vulnerable to acquiring mutations and may favor leukemic transformation. Here, we review how deregulated cell cycle control contributes to increased proliferation in AML and favors genomic instability, a prerequisite to confer selective advantages to particular clones in order to adapt and independently proliferate in the presence of a changing microenvironment. We discuss the connection between differentiation and proliferation with regard to leukemogenesis and outline the impact of specific alterations on response to therapy. Finally, we present examples, how a better understanding of cell cycle regulation and deregulation has already led to new promising therapeutic strategies.
Keywords: Acute myeloid leukemia (AML), cell cycle, genetic instability, proliferation, differentiation
Leukemogenesis and cell cycle – differentiation block, continuous proliferation and genome instability
AML is characterized by the clonal expansion of immature myeloid cells. At initial diagnosis approximately half of the patients harbor at least one cytogenetic aberration [1,2]. Moreover, aberrations at the molecular genetic level have increasingly been identified in AML during the last decade [3]. Several genetic aberrations have been associated with the hallmark of AML: the combination of a differentiation block and hyperproliferation [4].
In the normal hematopoietic system, proliferation is tightly linked to differentiation. In an asymmetrical cell division, one daughter cell retains stem cell properties to guarantee a pool of stem cells while the other one undergoes differentiation to respond to the high and permanent demand for granulocytes, monocytes, erythrocytes and platelets [5].
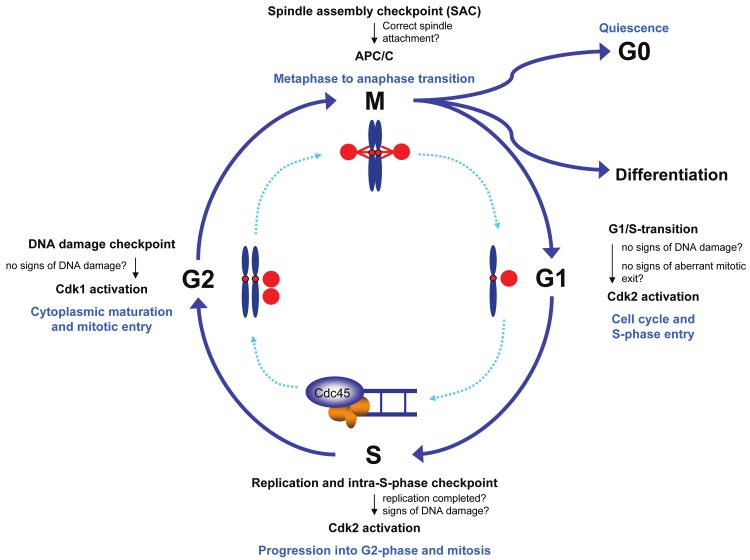
Hyperproliferation bears an enhanced risk of genetic damage. In leukemia, compromised DNA damage response pathways and weakened checkpoints add to the inherent risk and pave the way for further genetic aberrations promoting progression of the disease [4,6]. Thus, to fully understand leukemogenesis, it is crucial to determine the role of proto-oncogenic pathways in the regulation of the cell division cycle in myeloid malignancies. The cell cycle itself is divided into four major phases: the G1-, S-, G2- and M-phase [7]. From G1 a cell can exit the cell cycle into a state of quiescence (G0) from where it can undergo differentiation or reenter the cell cycle to proliferate. If the cell continues cell division the genomic DNA will be replicated during S-phase. Once cells have completed DNA replication, they proceed through the G2-phase to prepare for the subsequent chromosome separation and cytokinesis during M-phase (mitosis) (Figure 1) [7].
Figure 1.
The cell cycle and its checkpoints. The human cell cycle can be divided into four phases - G1-phase, S-phase, G2-phase and M-phase (mitosis). Cells must proceed through the cell cycle in a unidirectional manner and cell cycle progression is restricted to cells that have fulfilled specific requirements to enter the next phase of the cell cycle. Whether requirements for cell cycle progression are met is supervised by checkpoints which hold back cells at cell cycle transitions.
G1-phase - at the crossroad of proliferation and differentiation
The reproductive rate of the myeloid system depends on the number of cells that are actively cycling. In healthy individuals, myeloid precursor cells are mostly in a quiescent state [8,9] and can be recruited to enter the cell cycle in case of demand, for example when a severe infection occurs. Actively cycling progenitors proceed through multiple cell division cycles and give rise to daughter cells that can differentiate. Short intervals between cell division cycles and a large pool of cells that are capable of rapid self-duplication are strategies to guarantee a sufficient cellular defense.
Quiescent cells can be activated to re-enter the cell cycle by various external stimuli including growth factors and cytokines. In the following section, pathways will be discussed that regulate whether a cell enters the S-phase, exits the cell cycle into quiescence or undergoes differentiation.
Stimuli from the microenvironment in normal hematopoiesis and AML
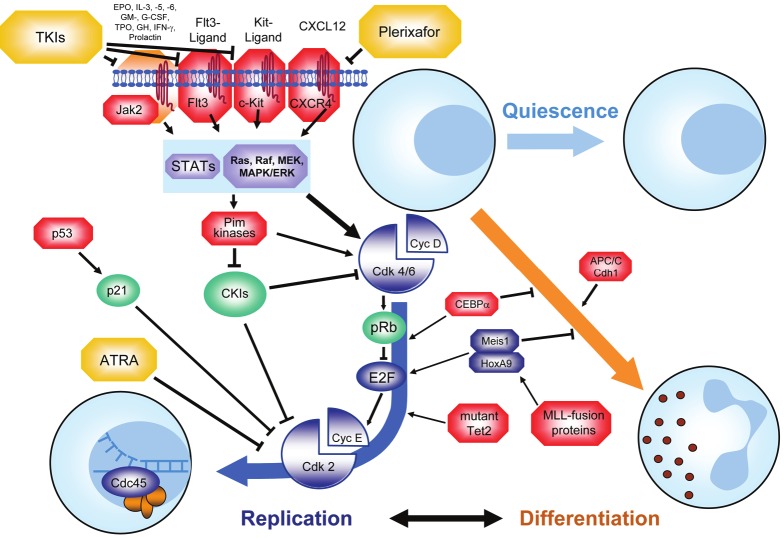
The FMS-like tyrosine kinase 3 (Flt3) [10], c-Kit (CD117 or stem cell factor receptor) [11] or Janus kinase 2 (Jak2) [12] are tyrosine kinases that translate external stimuli into pro-proliferative signaling cascades (Figure 2). Constitutive firing of mutated kinases in AML frequently causes ongoing activation of the downstream pathways and hence enhances transitioning from G1- into S-phase. Downstream of the receptor- and non-receptor kinases, the Stat-pathway (Stat = signal transducers and activators of transcription) is activated upon stimulation by various interleukins [13]. Further downstream, the serine-threonine kinase Pim1, which is activated by the Stat-pathway [14], has been shown to act as an important S-phase promoter by regulating Skp2-dependent degradation of the Cdk-inhibitor p27 [15]. In addition to signaling in the G1 phase, Pim1 was shown to enhance the transition from G2 phase into mitosis [16].
Figure 2.
To cycle or to differentiate - the G1 phase of the cell cycle. Cells that have exited mitosis into G1 can either enter a state of quiescence (G0), differentiate along a particular lineage or progress to another cell division cycle. The process of decision making between these possibilities is governed by a multitude of different proteins. Proteins that are frequently overexpressed or mutated and hence contribute to leukemic transformation are marked in red. Tumor suppressor proteins are marked in green, proteins that contribute to differentiation are shown in orange. Therapeutic agents are marked in yellow. See text for details.
As mentioned above, alterations of these proliferative regulators may confer unlimited growth during leukemogenesis [13]. For instance, certain mutations in FLT3, ie, point mutations in the tyrosine kinase domain (FLT3-TKD) and internal tandem duplications in the juxtamembrane region (FLT3-ITD), lead to constitutive activation which causes phosphorylation of substrate proteins even in the absence of external stimuli [17]. FLT3-ITD is found in approximately 20% of AML patients and FLT3-TKD in approximately 5% [18,19]. Both mutations are more frequent among patients with AML and a normal karyotype [18]. In addition to Flt3, the receptor tyrosine kinase Kit can be affected by activating mutations in AML. Such mutations, similar to Flt3, are mainly found in the tyrosine kinase domain of Kit, but are in part also observed in the extracellular domain. KIT mutations mostly occur in patients with AML and a chromosomal translocation disrupting a core binding factor, ie, t(8; 21) or inv(16) [20]. Mutations in JAK2 are rarely found in AML, but are characteristic for myeloproliferative neoplasms (MPN) [21].
Both types of FLT3 mutations activate PI3K/AKT and the MAP kinase pathway [22,23]. Activation of the MAP kinase pathway leads to upregulation of proto-oncogenic cell cycle regulators, such as the transcription factor c-Myc, and promotes premature entry into the following S-phase in AML cells [24]. FLT3-ITD also activates the Stat5 pathway and negatively regulates CEBPα and other transcription factors such as PU.1 to suppress differentiation [25,26]. Major changes in proliferative patterns can also result from deregulated chemokine signaling. In normal hematopoiesis, progenitor cells are stimulated through binding of CXCL12 (alias SDF-1) to its receptor CXCR4. Ligand binding to CXCR4 leads to activation of the MAP kinase pathway and to calcium release from the endoplasmatic reticulum which favors cell cycle progression, proliferation and survival [27]. High expression levels of CXCR4 have been observed in subsets of human AML and predict a poor prognosis [28]. Targeting CXCR4 has emerged as a promising leukemia therapy [28-30]. The use of the CXCR4-antagonist plerixafor in a mouse model of acute promyelocytic leukemia (APL) renders cells more susceptible to chemotherapy due to a mobilization of leukemia cells from their protective microenvironment [31].
Transcriptional control of cell fate in normal hematopoiesis and AML
Cell fate depends on the orchestrated action of regulatory proteins (Figure 2). The scheduled presence of such regulators can be achieved by synthesis and targeted degradation. Transcriptional activity and thus synthesis can be epigenetically controlled by cytosine hydroxymethylation of promoter DNA, such as at the p15 and p16 gene locus [32]. The ten-eleven translocation 2 (Tet2) protein is a methylcytosine dioxygenase that is important for the synthesis of 5-hydroxymethylcytosine and hence regulates the epigenetic status of the particular cell. Mutations in TET2 have been identified in various myeloid neoplasm [33-35]. In AML, they occur in approximately 10-15% of cases. They are more frequent in older AML patients and in AML with normal karyotype [36,37]. Loss of Tet2 in mice was associated with an enhanced expansion of early HPCs eventually leading to progressive myeloproliferation underscoring the transforming potential of TET2 aberrations [38].
Another important player in epigenetic regulation in AML is the mixed lineage leukemia (MLL) gene located on chromosome band 11q23 [39,40]. The wild type MLL protein is a human trithorax homologue and facilitates histone H3 lysine 4 (H3K4)-methylation. These histone marks then mediate transcriptional activation of a set of target genes [41]. Taspase1-mediated cleavage of the wild type full-length MLL-precursor protein into N-terminal (N-320) and C-terminal (C-180) fragments is the prerequisite for the characteristic MLL expression/activity peaks at the G1/S- and G2/M-boundary and for the formation of the active heterodimeric MLL complex [42,43] which mediates H3K4-methylation at the corresponding promoter sites to activate transcription of genes [44]. Importantly, this methylation process also involves the cell cycle regulator E2F, which binds processed MLL to direct the active MLL complex to the site of methylation [45]. In leukemia with 11q23 rearrangements, the N-terminal part of the MLL-protein is fused to one of more than 70 possible fusion partners [46]. All 11q23-associated MLL-aberrations lead to loss of taspase1 cleavage sites due to truncation [47]. Consequently, leukemogenic MLL fusion proteins show stable expression levels throughout the different stages of the cell cycle and this aberrant expression is believed to be an important hit in MLL leukemias with MLL rearrangements through abrogation of proper cell cycle checkpoint function [45]. MLL fusion proteins also lead to enhanced transcription of homeobox family of transcription factors. In addition to important functions in development these transcription factors regulate differentiation and cell cycle progression. For MEIS1 along with HOXA9, there is a well established role in promoting cell cycle progression and protection from apoptosis in MLL leukemia [48,49]. Meis1 induces important proto-oncogenes such as Flt3 and c-Myb and hence promotes S-phase transition [40,50]. Repression of Meis1 is associated with downregulation of cyclin D3 and a delay at the G1/S transition which can be overcome by reexpression of cyclin D3 [51].
Cell cycle progression by activation of cyclin dependent kinases (Cdks)
The MAP kinase pathway enhances cell cycle progression by activation of the cyclin-dependent kinases Cdk4, Cdk6 and Cdk2 (Figure 2). The latter is important to promote transition into S-phase (Figure 2). Activation of Cdk4 and Cdk6 through its activating subunit cyclin D leads to phosphorylation and inactivation of the retinoblastoma protein (pRb) which in turn releases the transcription factor E2F [52]. Importantly, pRb is dysfunctional in most cases of promyelocytic leukemia [53]. E2F promotes transcription of cyclin E which drives cells into S-phase [54]. A variant of E2F (E2F1) is aberrantly expressed in cases of AML and hence promotes premature entry into S-phase [55]. In addition, active Cdk4 and Cdk6 bind and thus sequester Cdk inhibitors, such as p14, p16 or p27, which otherwise hold back cells in G1-phase. Once activation of Cdk2 outbalances its inhibition by p21 and p27, cells can enter S-phase.
The classic tumor suppressor p53 is considered to be the guardian of the genome [56]. This important transcription factor can lead to cell cycle arrest, induce DNA damage response and enhance proapoptotic signaling. It has been proposed that p53 contributes to a G1 block via induction of p21 in cells that were able to exit aberrant mitoses [57-60]. A recent approach in AML cells enforced this particular G1-checkpoint by stabilizing p53 and the Cdk inhibitor p21 to counter polyploidy through induction of a stable G1-arrest [60]. The results are promising and might lead to novel therapeutic options.
Similar to p53, dephosphorylation of the pRb is required to maintain a G1-arrest. This is achieved by binding to E2F and prevention of transcription of genes that are critical for S-phase entry. This mechanism constitutes an additional barrier to ensure that error-prone cells do not enter S-phase. In malignant hematopoiesis, weakening of these barriers allows aneuploid cells to replicate and enhance genetic instability. Enhanced genetic instability is associated with a higher level of genetic variability which may confer a selective advantage, favor outgrowth of individual malignant clones and thus drive disease progression.
A way to promote differentiation is achieved through prevention of S-phase entry by downregulation of Cdk2 and cyclin E. This mechanism of action has been described for the vitamin A derivative all-trans-retinoic-acid (ATRA), which is a compound that mediates rapid differentiation of lineage-committed cells and is used as a therapeutic agent in AML with t(15; 17). The product of t(15; 17) is the PML-RARα fusion protein which leads to a differentiation block at the promyelocytic stage by functional inhibition of the retinoic acid receptor alpha (RARα). This differentiation block can be overcome by high doses of ATRA, which functions at least in part through inactivation of Cdk2 and cyclin E [61]. In neuroblastoma, ATRA induces differentiation via activation of the E3 ubiquitin ligase APC/CCdh1 [62]. Since the APC/C activator Cdh1 has been found to be repressed in some AML cell lines [63], failure of the APC/C to establish a stable G1 phase has been hypothesized to also play a role in the differentiation block in leukemogenesis [64].
Taken together, during G1 phase we observe alterations that uncouple proliferation from external stimuli, contributing to independence from the microenvironment. Most of these alterations promote a faster entry into the following S-phase, thus counteracting the establishment of a stable G1 phase, which is in somatic cells a prerequisite for both transition into G0 and differentiation or entry into an accurately prepared S phase to ensure genome stability (Figure 1). For a synopsis of the involved proteins see Supplementary Table 1.
S-phase - replicating the leukemia genome
The genome of cells is replicated during S-phase. To overcome inhibitory forces at the G1/S boundary and enter S-phase, Cdk-activity is necessary. A sophisticated surveillance network ensures exact duplication of the genetic material, with normal HPCs having a variety of mechanisms to guarantee correct replication- the so-called S-phase checkpoints - (Figure 3). The S-phase checkpoints restrict cell cycle progression into G2-phase to cells that have successfully completed DNA replication. Inaccuracies during replication cause checkpoint activation which initiates DNA repair and prevents cell cycle progression until the problem is solved. Malfunction of these checkpoints may lead to accumulation of genetic alterations which can confer a selective advantage to individual clones. Inaccuracies during S-phase are commonly considered a source of point mutations and smaller insertions or deletions [65], in contrast to errors in DNA segregation during mitosis which normally result in chromosomal aberrations.
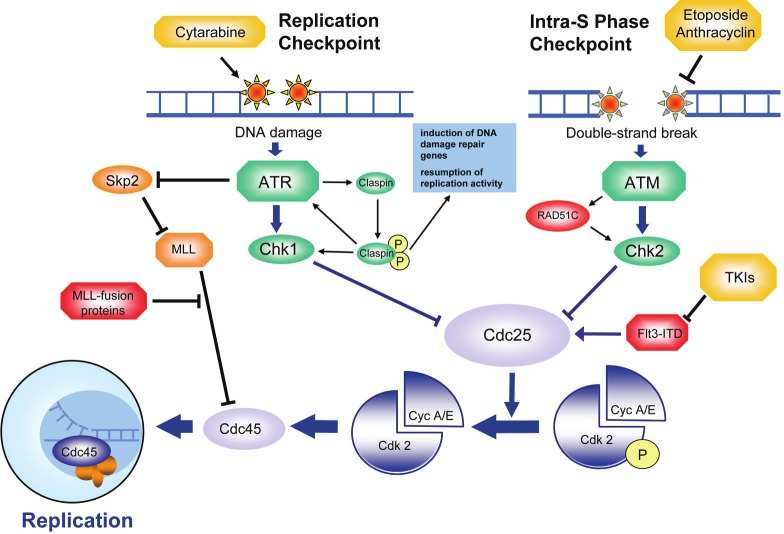
Figure 3.
DNA replication during S-phase - therapeutic target and source of mutation. Highest accuracy during DNA replication is essential to guarantee genomic integrity. In case of perturbations during the replication process or in case of DNA double-strand breakage, an ATM-driven DNA damage response (originating from DNA lesions which are shown at the top) leads to inihibiton of Cdk2 (shown in the lower part) via Cdc25 to block cell cycle progression. Interference with DNA replication by cytarabine or induction of DNA double-strand breaks by etoposide (both agents are shown in yellow) can block proliferation of leukemia. In general, inaccuracies of the DNA damage response allows accumulation of oncogenic mutations which favors the clonal outgrowth of genetically unstable clones to enhance leukemic progression. Proteins that are frequently overexpressed or mutated and hence contribute to leukemic transformation are marked in red. Tumor suppressor proteins are marked in green. Therapeutics is marked in yellow. See text for details.
S-phase checkpoints can be subdivided into the replication checkpoint, which is induced by inaccuracies during the replication process, and the intra-S-phase checkpoint, which is induced by double-strand breakage (DSB) (Figure 3). Activation of the replication checkpoint depends on the presence of replication forks, which are formed by multiprotein complexes that unwind the parental double-strand structure into single stranded DNA in order to allow replication [66]. An imbalance between DNA unwinding and replication activity, such as observed in case of errors during DNA replication, can result in naked single strand DNA. The presence of naked single strand DNA then attracts regulators such as replication protein A (RPA) which mediate activation of the replication checkpoint [67].
Response of the replication checkpoint is mainly mediated by the kinase ataxia tele-angiectasia and RAD3 related (ATR) in concert with the Chk-kinases [68]. Upon activation, ATR phosphorylates claspin which triggers downstream signaling to the effector kinases [69]. The claspin-driven checkpoint response induces pathways which coordinate DNA-replication, initiate DNA repair, stabilize replication forks, resume stalled replication forks and transcriptionally induce DNA damage repair genes [68].
Induction of DNA damage has become an important therapeutic strategy for AML because it blocks the cell division cycle of rapidly dividing blast populations in S-phase in consequence to checkpoint activation. This is achieved both by blocking the unwinding of the DNA strands and incorporation of nucleoside analoga during replication. The topoisomerase-inhibitors etoposide and anthracyclines impair DNA unwinding by inhibition of topoisomerase II and hence favor the accumulation of DNA strand breaks. Both agents are used as part of induction protocols in AML treatment since they provoke a checkpoint response in consequence to DNA damage involving the kinase ataxia teleangiectasia mutated (ATM) [65]. In contrast, the well-established nucleoside analogon cytarabine interferes with the replication process itself. Importantly, interference with DNA replication using a combination of cytarabine and anthracyclins has been the mainstay in AML therapy for more than thirty years [70,71]. The active metabolite cytosine arabinoside triphosphate is incorporated into the replicating DNA strand instead of cytosine triphosphate. This leads to replication arrest and activation of Chk1 and Chk2 kinases. Chk1 and Chk2 then inhibit Cdc25 activity through addition of inhibitory phosphate groups. The reduced activity of Cdc25 results in an accumulation of nonfunctional Cdk2 molecules which carry an inhibitory phosphorylation on tyrosine 15 [72,73]. Lack of Cdk2 dependent phosphorylation then leads to a delay in S-phase.
Leukemic cells harboring a FLT3-ITD have been shown in vitro to be deficient in inducing S-phase arrest upon DNA-damage caused by the nucleoside analogon clofarabine [74]. This work proposed that the enhanced activity of Cdc25 in the presence of mutated FLT3 might override the replication checkpoint. In accordance with this finding, longer exposure to clofarabine was efficient in killing FLT3 mutated leukemia cells. This higher therapeutic efficacy is thought to be a consequence of an improper S-phase arrest with slippage of cells out of S-phase in the presence of unsolved problems during DNA replication and subsequent cell death. In contrast, short-term exposure to clofarabine led to less efficient killing, most probably due to potent DNA repair pathways [74].
The chromatin remodeler MLL also participates in regulation of the response to DNA damage. Recent results have identified MLL as a downstream target of the ATR-kinase [75]. Phosphorylation of MLL at serine 516 by ATR in case of DNA damage disrupts binding to Skp2, an activator of an important E3 ubiquitin ligase, the Skp-Cullin-F-box-protein containing complex (SCF) leading to stabilization of wild-type MLL. Chromatin-bound MLL at damaged DNA restricts binding of Cdc45 and thus delays DNA replication [75]. Leukemogenic MLL-fusion proteins have dominant negative impact on this DNA damage response [75]. MLL-fusion proteins, such as MLL-AF4 and MLL-AF9, hinder the interaction between ATR and wild-type MLL. This favors a more rapid degradation of the MLL wild-type form and erroneously renders damaged DNA accessible for the replication machinery [75]. This leads to the duplication of damaged DNA and might give rise to potentially leukemogenic mutations.
In conclusion, recurrent alterations during S-phase found in AML lead to accelerated and enhanced replication. This drives proliferation, facilitates overriding of chemotherapy-induced checkpoint-mediated arrest and, due to interference with checkpoint signaling, helps to establish a mutator phenotype. For a synopsis of the involved proteins see Supplementary Table 2.
G2-phase - getting prepared for genomic and cytoplasmic division
During the G2-phase of the cell cycle, cells prepare for the subsequent segregation of DNA to the two developing daughter cells. Several checkpoints ensure that only cells without structural damage of the DNA are able to enter mitosis and segregate their genetic material to their daughter cells. While response to DNA damage during S-phase results in deceleration of replication, stabilization of the replisome, and prevention of homologous recombination [76], the task of the G2-checkpoint is to prevent that cells with damaged DNA enter mitosis. Major players of the "genome integrity checkpoint" in the G2-phase are ATM- and ATR-kinases and their target proteins Chk1- and Chk2-kinases [77,78]. Upon structural damage, such as DSBs, these proteins reduce Cdk1-activity via Cdc25 and various other mediators, such as p53 [78,79] (Figure 4).
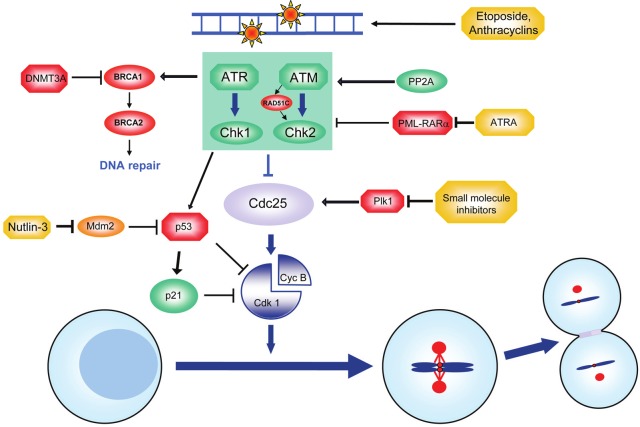
Figure 4.
G2-checkpoint - no passage for cells with damaged DNA. During the G2-phase cells prepare for genomic and cytoplasmic separation. An ATR-/ATM-driven DNA damage response (originating from DNA lesions which are shown at the top) leads to inhibition of Cdk1 (shown in the lower part) and hence arrests cells with signs of DNA damage at the G2-phase until the problem is solved. Therapeutic agents such as etoposide or anthracyclins (shown in yellow) work by induction of a cell cycle arrest in the G2-phase. Leukemia-promoting alterations, such as overexpression of Plk1, FOXM1, SET or loss of function of p53 (marked in red) allow cells to override the DNA damage response and allow the accumulation of oncogenic mutations. Different therapeutic approaches aim at reconstituting the DNA damage response by stabilization of p53. See text for details.
Recent data provide evidence of a high level of DNA damage in cells of high-risk cytogenetic AML patients accompanied by DNA damage pathway activation [80]. Malignant cells often continue to undergo limitless numbers of cell divisions despite the presence of damaged DNA. This is achieved either through silencing or uncoupling of the DNA damage response from cell cycle control [81]. Enhancing the cell cycle-restrictive and proapoptotic effects of p53 by inhibition of p53-inactivating proteins has therefore become a promising approach in the treatment of hematologic malignancies [60,82]. Inhibition of Mdm2, an ubiquitin ligase involved in p53 degradation, synergizes with classical AML therapeutics such as cytarabine and anthracyclins given an unmutated and thus functional p53 [82].
ATM- or ATR-kinases are cornerstones in DNA damage response that are often mutated in malignancies [83-86]. Various genes were found to be mutated that act in a similar way: one recent report showed that mutation of RAD51C is implicated in the pathogenesis of a Fanconi anemia-like disorder, a heterogenous disorder leading to developmental deviations, bone marrow failure and predisposition to leukemia [87]. Mutation of RAD51C abrogated the ability to arrest cells in G2 following DNA damage [87,88].
BRCA gene mutations are tightly connected to the pathogenesis of gynecological tumors. Mutations in BRCA1 and, to a lesser extent, in BRCA2, are associated with a high risk of developing ovarian- or breast cancer. BRCA1 and BRCA2 are tumor suppressors that are part of the DNA damage response and regulate non-homologous end joining and homology-directed repair following DNA double strand breaks [89,90]. BRCA mutations also put women who underwent radiochemotherapy for gynecologic malignancies at a high risk of developing secondary, therapy-associated AML [89,91], as BRCA mutations compromise the fidelity of DNA repair at sites of structural genetic alterations. BRCA mutations hence lead to a tolerance towards genetic lesions which accumulate over time. This process favors the rise of translocations with leukemogenic potential [91,92]. Of note, three out of four women with tAML and nearly one third of patients with primary AML showed suppressed BRCA1 expression levels when compared to normal bone marrow. In most cases BRCA1 was hypermethylated, a finding that was associated with overexpression of DNA methyltransferase 3A (DNMT3A) [92,93]. These findings support the notion that a compromised fidelity of the DNA damage response and repair mechanisms favor the development of AML [92]. These data also support the view that breast and ovarian cancer patients harboring BRCA-mutations should be monitored closely following completion of therapy because these mutations might add to the treatment-related risk of developing leukemia [92].
In healthy cells, activation of the DNA damage response pathway leads to a G2/M-arrest which constitutes a barrier against cellular growth in the presence of damaged DNA to guarantee genetic integrity (Figure 4) [81,94]. DNA damage can cause a mutator phenotype which favors subsequent genetic alterations and contributes to malignant transformation. In addition to the frequently observed inactivation of components of the DNA damage response pathway, there are data that claim a role for uncoupling of DNA damage-induced ATM activation and downstream activation of Chk1- and Chk2-effector kinases in patient-derived AML cells [81]. While myelodysplastic cells exhibit high levels of γ-H2AX foci, cells that had progressed to AML displayed decreased numbers of foci. This suggests that in progressive disease time-consuming DNA repair activities are skipped in favor of rapid cellular expansion [81].
While DNA damage recognition pathways remain intact, the execution of cell cycle arrest in response to these pathways may be uncoupled which allows proliferation in the presence of DNA damage and clonal outgrowth of genetically unstable cells. An example for uncoupling damage recognition from effectors is the leukemogenic fusion protein PML-RARα, which disrupts Chk2-mediated pro-apoptotic signaling in consequence to DNA damage [95].
Despite the observation of uncoupling the DNA damage sensors, i.e. ATM and ATR, from the effector cascade, measurements are being performed in AML to monitor the extent to which histone and ATM phosphorylation takes place in primary leukemia cells under chemotherapy. The intention of these measurements is to establish biomarkers that can predict response to therapy. In some cases a correlation between non-response to chemotherapy and low levels of γ-H2AX- and ATM-phosphorylation have been observed [96]. Comparable approaches might influence treatment decisions in the future since the probability of therapeutic success can be estimated at early time points.
G2/M arrest as a consequence of persistent DNA damage is achieved through a tight control of Cdk1-activity (Figure 4). Reducing Cdk1-activity delays G2/M-progression and provides time for DNA repair. Less stringent checkpoint activity was shown to be associated with milder symptoms in patients with Fanconi anemia. However, Fanconi anemia patients with an attenuated checkpoint were at a higher risk of developing MDS or leukemia [97]. Recent reports provided evidence that an accurate checkpoint control renders cells more resistant towards DNA-damage inducing agents [98]. Leukemias with a weakened checkpoint can be eradicated more efficiently by genotoxic therapeutics because they enter mitosis in the presence of damaged DNA. This circumstance can also be used for therapeutic purposes: Chk1 kinase inhibition in the presence of genotoxic therapeutics led to very promising responses in primary leukemia cells and reduced colony-forming potential in undifferentiated leukemias [98]. However, in leukemias showing more differentiated myelomonocytic morphology response rates were lower [98].
In addition to Cdc25, the multifaceted protein phosphatase 2 (PP2A) regulates the G2/M transition. At this critical point, PP2A works as a sensitizer in the response to DNA damage [99]. Lenalidomide, which is approved in the US for use in MDS with del(5q) and currently tested in AML, has been described to interfere with the interplay between phosphatases, like PP2A, Cdc25, and Cdks to modulate cell cycle progression [100]. In MDS with del(5q) lenalidomide exerts its effect in part due to the allelic haplodeficiency of Cdc25 and PP2A, both of which map to the commonly deleted region on chromosome 5 [100]. Reduced expression of these proteins, either by shRNA-mediated silencing in vitro or haplodeficiency in consequence to del (5q) in patients, causes an enhanced susceptibility to the proapoptotic effects of lenalidomide [100]. Since lenalidomide is an indirect inhibitor of the Cdc25 homologue which activates Cdk2 (Cdc25A) at the G1/S transition, a concomitant reduction/inhibition of both isoforms (Cdc25A and Cdc25C) may foster a G2 arrest und promote cell death in cell clones which harbor del (5q) [100].
Unlike in MDS with del(5q) where Cdc25 phosphatase is expressed at lower levels due to loss of one coding region, Cdc25 also underlies activating factors that enhance progression into mitosis. The Polo-like kinase 1 (Plk1) is an activator of Cdc25 phosphatase and hence plays an important role in the recovery from a DNA-damage induced G2-arrest [98,101]. Various studies suggested synergistic effects of Plk1-inhibitors and for instance spindle poisons which might prove useful especially in the treatment of elderly patients due to fewer side effects [102].
In conclusion, the myeloid precursor cell has several mechanisms to respond to DNA damage to ensure that only cells without signs of DNA damage enter mitosis (Figure 4). While normal HPCs rely on accurate checkpoint function to guarantee genetic integrity, leukemia may take advantage of genetic instability to increase cellular diversity and thus the likelihood of a clone with a major survival advantage. This is achieved by deregulations and mutations of components responsible for genome integrity which favors alterations of the genetic material and allows transition into mitosis, even in the presence of gross genetic abnormalities. That leukemias frequently override the DNA damage response, has been shown to render leukemic cells susceptible to genotoxic therapeutic approaches. Approaches that abrogate the DNA damage response and hence sensitize the cells to genotoxic agents are tested and might constitute important add-ons to therapeutic approaches in the near future. For a synopsis of the involved proteins see Supplementary Table 3.
M-phase (mitosis) - segregating chromosomes and cytokinesis
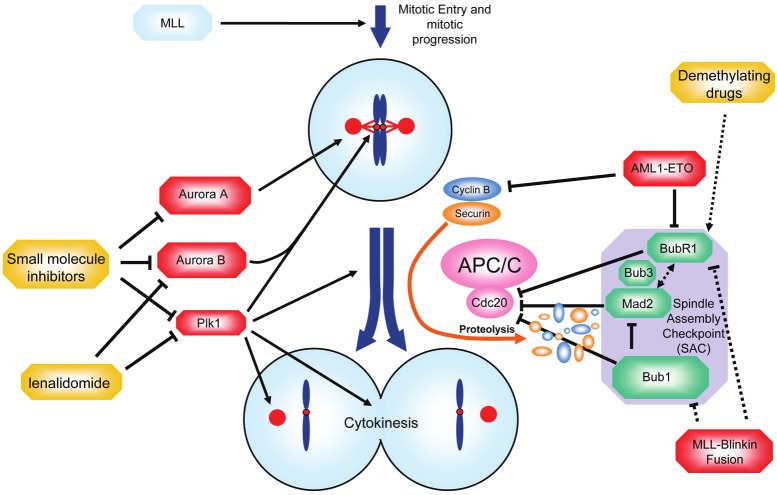
Mitosis is one of the most critical periods during the cell cycle because the cell has to separate its genome and distribute the DNA to the two developing daughter cells. To guarantee the equal distribution of the chromosomes, the spindle assembly checkpoint (SAC) senses and monitors attachment of chromosomes to the mitotic spindle and allows that chromosome separation only occurs in the absence of inaccuracies, such as chromosomal misalignment or a dysfunctional mitotic spindle (Figure 5). In essence, the SAC is an inhibitor of the anaphase-promoting complex or cyclosome (APC/C) which prevents premature mitotic exit [103]. The APC/C, activated by Cdc20, is a major ubiquitin ligase that mediates the exact timing of degradation of cyclin B and securin to trigger anaphase onset and mitotic exit. Degradation of cyclin B and securin is inhibited until all chromosomes have established a stable attachment to the mitotic spindle. Failure to satisfy the SAC, such as in case of chromosomal misalignment, leads to the formation of the mitotic checkpoint complex (MCC), a complex which consists of the proteins Mad2, Bub3 and BubR1. The MCC is able to bind to and inhibit the APC/C-activator Cdc20. This mitotic surveillance mechanism is often weakened in malignancies [104], rendering cancer cells more susceptible for gain or loss of genetic material.
Figure 5.
Mitosis - segregating the blueprint for leukemia. During mitosis, the DNA and the cytoplasm of the cell have to be segregated to the daughter cells. The spindle assembly checkpoint (SAC) is a surveillance mechanisms which monitors interactions between chromosomes and microtubules and arrests cells at metaphase until every single chromosome has properly attached to the mitotic spindle. Restriction is achieved through inhibition of the activating APC/C-subunit Cdc20. Loss of function of SAC proteins such as BubR1, Mad2 or Bub1 (shown in green) reduces the accuracy of the SAC and favors chromosomal maldistribution. Overexpression of mitotic kinases such as Plk1, Aurora A and B (shown in red) can also result in premature anaphases. Small molecule inhibitors targeting Plk1 and the Aurora kinases are currently tested in clinical studies in AML patients (shown in yellow). See text for details.
Leukemia cells use various ways to interfere with checkpoint controls to divide even in the presence of gross abnormalities. AML1-ETO is the fusion protein resulting from t(8; 21) which is the most common structural chromosome aberration in AML and has been described to promote AML in mice if expressed in a C-terminally truncated form [105,106]. Such a C-terminally truncated AML1-ETO construct has also been shown to compromise the integrity of the SAC and to associate with a higher incidence of aneuploid cells in vitro [107]. Cells expressing the truncated AML1-ETO had reduced levels of the SAC component BubR1 and of cyclin B [107]. Repression of checkpoint proteins such as BubR1 is frequently observed in cancer and has been shown to perturb the accuracy of the mitotic control favoring genetic instability and malignant transformation [104].
Bub1 is another SAC component, which shares sequence homology with BubR1. Bub1 directs the association of the MCC and inhibits APC/CCdc20 by phosphorylation. A screen for mutations in BUB1 and analysis of the expression levels of Bub1 in AML revealed recurrent repression of Bub1 while mutations appeared to be rare in AML [108]. This is in line with findings in different tumors where mutations in the sequence coding for SAC proteins are considered to be rare events while a deregulated expression might more frequently play a role in abrogation of SAC fidelity [104].
In addition to a deregulated expression of BubR1 and Bub1, interference with the SAC can occur in the presence of a leukemogenic fusion protein involving the mitotic regulator Blinkin (alias AF15q14). Here, Blinkin was described as an MLL-fusion partner in a case of AML and turned out to be of special importance for the recruitment of BubR1 and Bub1 to the kinetochore of chromosomes during the mitotic alignment process [109]. As Blinkin was identified to play a central role in regulating the attachment of kinetochores to spindle microtubules, a direct role for its MLL-fused derivative in leukemogenesis as a driver of genetic instability is conceivable [109,110].
The Aurora A kinase localizes to the centrosomes during mitosis, and its overexpression in breast, colorectal and gastric cancers has been associated with overriding the mitotic checkpoint in the presence of spindle poisons [111]. The finding that Aurora A is also frequently overexpressed in AML raised the question whether targeted inhibition might be a valuable treatment approach [112,113]. Indeed, response to cytarabine could be achieved in a priori cytarabine-resistant cell lines upon targeted Aurora A inhibition [114]. It is a matter of debate whether a highly specific kinase inhibitor should be preferred over a less specific multikinase inhibitor. Frequently, successful treatment with tyrosine kinase inhibitors may depend on the genetic context and treatment with multikinase inhibitors might be more reasonable. For example, Aurora A kinase overexpression is often accompanied by an activating FLT3 mutation [115]. Aurora kinase inhibitors which share inhibitory potential for Aurora and Flt3 kinase, such as CEP-701 or PKC-412, induced better response rates in FLT3 mutated leukemias in vitro [115]. Similar to the overexpression of Aurora A, Aurora B overexpression is frequently observed in AML [116]. Selective Aurora B kinase inhibition showed synergistic effects with vincristine and daunorubicin by enhancing the antiproliferative activity [117]. Continued exposure to AZD1152, an Aurora B inhibitor, resulted in a growing fraction of polyploid cells leading either to cell cycle arrest or apoptosis [118,119]. Due to the induction of polyploidy, there are concerns that therapeutic agents which inhibit mitotic regulators, such as Aurora kinases, also give rise to more aggressive clones with a complex karyotype due to polyploidization. Aberrant exit from mitosis, which appears to be the underlying cause of polyploidy in those cells, leads to the initiation of p53-dependent signaling in healthy cells to prevent cells from entering another cell division cycle and induce apoptosis. In malignant disease, however, p53 is frequently inactivated by mutation or deletion. The immanent need for functional p53 in order to be able to efficiently induce apoptosis following exposure to the Aurora B kinase inhibitor AZD1152 questions the use of this compound in cases of unknown p53 mutation status [120]. p53 mutations are relatively rare in AML with an estimated frequency of 2% in patients without a complex karyotype [121]. About half of the patients suffering from AML with a complex karyotype have lost one p53 allele. Almost all of those patients also carry a mutation in their remaining p53 allele [60,121]. Thus, it has been suggested to first exclude a functional biallelic loss of p53 before treatment with Aurora B kinase inhibitors [60]. Interestingly, Nutlin-3, an antagonist of the E3-ubiquitin ligase Mdm2, which targets p53, increased p53-levels and led to efficient apoptosis in p53-wild type cells upon treatment with the Aurora B kinase inhibitor [60].
Plk1 is another example of a mitotic kinase with various functions associated with the coordination of mitotic entry, chromosome segregation and cytoplasmic division [122]. Plk1 is frequently overexpressed both in leukemia cell lines and patient derived blasts [123] and leukemia blasts loose proliferative capacities along with a decrease of clonogenic potential upon treatment with the PLK1 inhibitor BI2536 while the inhibitor exerts a less dramatic effect in normal HPCs [123].
Deregulations during mitosis may also be based on epigenetic alterations, i.e. changes in promoter methylation and/or chromatin remodeling. As described before MLL is known to be expressed in a cell-cycle dependent manner reaching its first peak during G1/S-transition and its second peak at the G2/M boundary [45,124]. It has been shown that during mitosis MLL locates to promoter regions of genes whose expression is required in the subsequent interphase [125]. This distinct pattern was observed in the presence of condensed chromatin and indicates that MLL-based gene regulation governs transcriptional regulation even during one of the most vulnerable cell cycle stages [125]. It is therefore conceivable that leukemogenic MLL-translocations render mitotic control susceptible for errors during chromosomal and cytoplasmic separation.
In conclusion, since alterations of mitotic regulators are commonly observed, an aberrant mitosis might be frequent in AML and alterations of mitotic regulators might constitute an additional class of leukemogenic hits. An insufficient mitotic checkpoint allows cells to divide in the presence of unfavorable conditions, and to escape from death in mitosis. Moreover, aberrant mitotic control can cause genetic instability, a common characteristic of malignancies [6]. Genetic instability drives diversification leading to a multitude of different subclones with further enhanced malignant growth capacity in the presence of adverse conditions [6]. For a synopsis of the involved proteins see Supplementary Table 4.
Concluding remarks
AML is the result of a sequence of transforming events that hit HPCs and give rise to clonal outgrowth and uncontrolled, limitless expansion. Expansion is achieved through constitutive activation of pathways, e.g. by activating mutations or overexpression of proto-oncogenic regulators, which, in the healthy individual, drive myeloid cell expansion e.g. in case of infectious disease. The physiological response induces enhanced proliferation along with cellular differentiation in order to produce functional cells. In contrast, leukemic transformation results in an excess of immature cells that are compromised in their ability to differentiate. Limitless expansion is achieved through an endless sequence of cell division cycles and abrogation of restriction points. An accepted model of leukemogenesis suggests that two major classes of mutations cooperate to transform HPCs [4]. Class I mutations confer the ability of limitless growth and class II mutations impair hematopoietic differentiation [4]. In addition, a third class of mutations (class III mutations) which hits epigenetic modifiers and hence alters protein synthesis in favor of proteins with oncogenic characteristics has recently come into focus [126]. During disease progression, leukemia cells can become genetically unstable and experience losses and gains of genetic material which allows them to expand even more rapidly and adapt to a variable environment. Genetic instability may occur through perturbation of DNA damage response, inaccuracies during replication and chromosome segregation in mitosis. Hits causing these defects might constitute an additional class of mutations in AML.
Classic therapeutic agents for leukemia, such as anthracyclines or cytarabine, prevent leukemia cells from cycling. This is achieved through induction of DNA damage and subsequent checkpoint activation leading to cell cycle arrest. In contrast to the latter strategy, some recent tailored therapies, such as PLK1 and Aurora kinase-inhibitors, abrogate checkpoint fidelity to trigger cell death in response to an aberrant mitotic exit. In some cases of leukemia, cells are addicted to constitutive firing of mutated kinases. Here, tyrosine kinase inhibitors block proliferative signaling and can lead to favorable clinical responses. These promising results provide excellent examples how a detailed understanding of cell cycle regulation and proliferation can translate into therapeutic success.
Supporting Information
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 2.Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, Wheatley K, Burnett AK, Goldstone AH. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 3.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 4.Deguchi K, Gilliland DG. Cooperativity between mutations in tyrosine kinases and in hematopoietic transcription factors in AML. Leukemia. 2002;16:740–744. doi: 10.1038/sj.leu.2402500. [DOI] [PubMed] [Google Scholar]
- 5.Congdon KL, Reya T. Divide and conquer: how asymmetric division shapes cell fate in the hematopoietic system. Curr Opin Immunol. 2008;20:302–307. doi: 10.1016/j.coi.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 8.Renstrom J, Kroger M, Peschel C, Oostendorp RA. How the niche regulates hematopoietic stem cells. Chem Biol Interact. 2010;184:7–15. doi: 10.1016/j.cbi.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Qian H, Buza-Vidas N, Hyland CD, Jensen CT, Antonchuk J, Mansson R, Thoren LA, Ekblom M, Alexander WS, Jacobsen SE. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Kikushige Y, Yoshimoto G, Miyamoto T, Iino T, Mori Y, Iwasaki H, Niiro H, Takenaka K, Nagafuji K, Harada M, Ishikawa F, Akashi K. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J Immunol. 2008;180:7358–7367. doi: 10.4049/jimmunol.180.11.7358. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, Gurudutta GU, Satija NK, Pati S, Afrin F, Gupta P, Verma YK, Singh VK, Tripathi RP. Stem cell c-KIT and HOXB4 genes: critical roles and mechanisms in self-renewal, proliferation, and differentiation. Stem Cells Dev. 2006;15:755–778. doi: 10.1089/scd.2006.15.755. [DOI] [PubMed] [Google Scholar]
- 12.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 13.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26:6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 14.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–1991. [PubMed] [Google Scholar]
- 15.Cen B, Mahajan S, Zemskova M, Beharry Z, Lin YW, Cramer SD, Lilly MB, Kraft AS. Regulation of Skp2 levels by the Pim-1 protein kinase. J Biol Chem. 2010;285:29128–29137. doi: 10.1074/jbc.M110.137240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachmann M, Hennemann H, Xing PX, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J Biol Chem. 2004;279:48319–48328. doi: 10.1074/jbc.M404440200. [DOI] [PubMed] [Google Scholar]
- 17.Toffalini F, Demoulin JB. New insights into the mechanisms of hematopoietic cell transformation by activated receptor tyrosine kinases. Blood. 2010;116:2429–2437. doi: 10.1182/blood-2010-04-279752. [DOI] [PubMed] [Google Scholar]
- 18.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, Wermke M, Bornhauser M, Ritter M, Neubauer A, Ehninger G, Illmer T. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 19.Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters--an analysis of 3082 patients. Blood. 2008;111:2527–2537. doi: 10.1182/blood-2007-05-091215. [DOI] [PubMed] [Google Scholar]
- 20.Paschka P, Marcucci G, Ruppert AS, Mrozek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ, Kolitz JE, Larson RA, Bloomfield CD. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8; 21): a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 21.Theocharides A, Boissinot M, Girodon F, Garand R, Teo SS, Lippert E, Talmant P, Tichelli A, Hermouet S, Skoda RC. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood. 2007;110:375–379. doi: 10.1182/blood-2006-12-062125. [DOI] [PubMed] [Google Scholar]
- 22.Pratz KW, Levis MJ. Bench to bedside targeting of FLT3 in acute leukemia. Curr Drug Targets. 2010;11:781–789. doi: 10.2174/138945010791320782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agrawal S, Koschmieder S, Baumer N, Reddy NG, Berdel WE, Muller-Tidow C, Serve H. Pim2 complements Flt3 wild-type receptor in hematopoietic progenitor cell transformation. Leukemia. 2008;22:78–86. doi: 10.1038/sj.leu.2404988. [DOI] [PubMed] [Google Scholar]
- 24.Kojima K, Konopleva M, Samudio IJ, Ruvolo V, Andreeff M. Mitogen-activated protein kinase kinase inhibition enhances nuclear proapoptotic function of p53 in acute myelogenous leukemia cells. Cancer Res. 2007;67:3210–3219. doi: 10.1158/0008-5472.CAN-06-2712. [DOI] [PubMed] [Google Scholar]
- 25.Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140–2150. [PubMed] [Google Scholar]
- 26.Zheng R, Friedman AD, Levis M, Li L, Weir EG, Small D. Internal tandem duplication mutation of FLT3 blocks myeloid differentiation through suppression of C/EBPalpha expression. Blood. 2004;103:1883–1890. doi: 10.1182/blood-2003-06-1978. [DOI] [PubMed] [Google Scholar]
- 27.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 28.Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2007;109:786–791. doi: 10.1182/blood-2006-05-024844. [DOI] [PubMed] [Google Scholar]
- 29.Konoplev S, Rassidakis GZ, Estey E, Kantarjian H, Liakou CI, Huang X, Xiao L, Andreeff M, Konopleva M, Medeiros LJ. Overexpression of CXCR4 predicts adverse overall and event-free survival in patients with unmutated FLT3 acute myeloid leukemia with normal karyotype. Cancer. 2007;109:1152–1156. doi: 10.1002/cncr.22510. [DOI] [PubMed] [Google Scholar]
- 30.Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood. 2004;104:550–557. doi: 10.1182/blood-2004-02-0566. [DOI] [PubMed] [Google Scholar]
- 31.Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, Prior JL, Piwnica-Worms D, Bridger G, Ley TJ, DiPersio JF. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 33.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 34.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 36.Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, Lee MC, Ko BS, Tang JL, Yao M, Tsay W, Wu SJ, Huang SY, Hsu SC, Chen YC, Chang YC, Kuo KT, Lee FY, Liu MC, Liu CW, Tseng MH, Huang CF, Tien HF. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 37.Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP, Wu YZ, Blum W, Powell BL, Carter TH, Wetzler M, Moore JO, Kolitz JE, Baer MR, Carroll AJ, Larson RA, Caligiuri MA, Marcucci G, Bloomfield CD. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 44.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EH, Hsieh JJ. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamai H, Inokuchi K. 11q23/MLL acute leukemia: update of clinical aspects. J Clin Exp Hematop. 2010;50:91–98. doi: 10.3960/jslrt.50.91. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Cheng EH, Hsieh JJ. MLL fusions: pathways to leukemia. Cancer Biol Ther. 2009;8:1204–1211. doi: 10.4161/cbt.8.13.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, Ji H, Thirman MJ. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Argiropoulos B, Yung E, Xiang P, Lo CY, Kuchenbauer F, Palmqvist L, Reindl C, Heuser M, Sekulovic S, Rosten P, Muranyi A, Goh SL, Featherstone M, Humphries RK. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115:4071–4082. doi: 10.1182/blood-2009-06-225573. [DOI] [PubMed] [Google Scholar]
- 52.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 53.Paggi MG, de Fabritiis P, Bonetto F, Amadio L, Santarelli G, Spadea A, Gentile FP, Floridi A, Felsani A. The retinoblastoma gene product in acute myeloid leukemia: a possible involvement in promyelocytic leukemia. Cancer Res. 1995;55:4552–4556. [PubMed] [Google Scholar]
- 54.Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4:855–864. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- 55.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 57.Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- 58.Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a "tetraploidy checkpoint". J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kojima K, Konopleva M, Tsao T, Nakakuma H, Andreeff M. Concomitant inhibition of Mdm2-p53 interaction and Aurora kinases activates the p53-dependent postmitotic checkpoints and synergistically induces p53-mediated mitochondrial apoptosis along with reduced endoreduplication in acute myelogenous leukemia. Blood. 2008;112:2886–2895. doi: 10.1182/blood-2008-01-128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang Y, Zhou X, Lin M, Jing H, Zhong L, Ying M, Luo P, Yang B, He Q. The ubiquitin-proteasome pathway plays essential roles in ATRA-induced leukemia cells G0/G1 phase arrest and transition into granulocytic differentiation. Cancer Biol Ther. 2010;10:1157–1167. doi: 10.4161/cbt.10.11.13556. [DOI] [PubMed] [Google Scholar]
- 62.Cuende J, Moreno S, Bolanos JP, Almeida A. Retinoic acid downregulates Rae1 leading to APC(Cdh1) activation and neuroblastoma SH-SY5Y differentiation. Oncogene. 2008;27:3339–3344. doi: 10.1038/sj.onc.1210987. [DOI] [PubMed] [Google Scholar]
- 63.Engelbert D, Schnerch D, Baumgarten A, Wäsch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 2008;27:907–917. doi: 10.1038/sj.onc.1210703. [DOI] [PubMed] [Google Scholar]
- 64.Wäsch R, Robbins JA, Cross FR. The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene. 2010;29:1–10. doi: 10.1038/onc.2009.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heisig P. Type II topoisomerases--inhibitors, repair mechanisms and mutations. Mutagenesis. 2009;24:465–469. doi: 10.1093/mutage/gep035. [DOI] [PubMed] [Google Scholar]
- 66.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 67.Recolin B, Van der Laan S, Maiorano D. Role of replication protein A as sensor in activation of the S-phase checkpoint in Xenopus egg extracts. Nucleic Acids Res. 2012;40:3431–3442. doi: 10.1093/nar/gkr1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol Cell. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka K. Multiple functions of the S-phase checkpoint mediator. Biosci Biotechnol Biochem. 2010;74:2367–2373. doi: 10.1271/bbb.100583. [DOI] [PubMed] [Google Scholar]
- 70.Vogler WR, Velez-Garcia E, Weiner RS, Flaum MA, Bartolucci AA, Omura GA, Gerber MC, Banks PL. A phase III trial comparing idarubicin and daunorubicin in combination with cytarabine in acute myelogenous leukemia: a Southeastern Cancer Study Group Study. J. Clin. Oncol. 1992;10:1103–1111. doi: 10.1200/JCO.1992.10.7.1103. [DOI] [PubMed] [Google Scholar]
- 71.Wiernik PH, Banks PL, Case DC Jr, Arlin ZA, Periman PO, Todd MB, Ritch PS, Enck RE, Weitberg AB. Cytarabine plus idarubicin or daunorubicin as induction and consolidation therapy for previously untreated adult patients with acute myeloid leukemia. Blood. 1992;79:313–319. [PubMed] [Google Scholar]
- 72.Sampath D, Cortes J, Estrov Z, Du M, Shi Z, Andreeff M, Gandhi V, Plunkett W. Pharmacodynamics of cytarabine alone and in combination with 7-hydroxystaurosporine (UCN-01) in AML blasts in vitro and during a clinical trial. Blood. 2006;107:2517–2524. doi: 10.1182/blood-2005-08-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valdez BC, Li Y, Murray D, Champlin RE, Andersson BS. The synergistic cytotoxicity of clofarabine, fludarabine and busulfan in AML cells involves ATM pathway activation and chromatin remodeling. Biochem Pharmacol. 2011;81:222–232. doi: 10.1016/j.bcp.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seedhouse C, Grundy M, Shang S, Ronan J, Pimblett H, Russell N, Pallis M. Impaired S-phase arrest in acute myeloid leukemia cells with a FLT3 internal tandem duplication treated with clofarabine. Clin Cancer Res. 2009;15:7291–7298. doi: 10.1158/1078-0432.CCR-09-1222. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, Cheng EH, Hsieh JJ. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 77.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 78.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chakraborty J, Banerjee S, Ray P, Hossain DM, Bhattacharyya S, Adhikary A, Chattopadhyay S, Das T, Sa G. Gain of cellular adaptation due to prolonged p53 impairment leads to functional switchover from p53 to p73 during DNA damage in acute myeloid leukemia cells. J Biol Chem. 2010;285:33104–33112. doi: 10.1074/jbc.M110.122705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cavelier C, Didier C, Prade N, Mansat-De Mas V, Manenti S, Recher C, Demur C, Ducommun B. Constitutive activation of the DNA damage signaling pathway in acute myeloid leukemia with complex karyotype: potential importance for checkpoint targeting therapy. Cancer Res. 2009;69:8652–8661. doi: 10.1158/0008-5472.CAN-09-0939. [DOI] [PubMed] [Google Scholar]
- 81.Boehrer S, Ades L, Tajeddine N, Hofmann WK, Kriener S, Bug G, Ottmann OG, Ruthardt M, Galluzzi L, Fouassier C, Tailler M, Olaussen KA, Gardin C, Eclache V, de Botton S, Thepot S, Fenaux P, Kroemer G. Suppression of the DNA damage response in acute myeloid leukemia versus myelodysplastic syndrome. Oncogene. 2009;28:2205–2218. doi: 10.1038/onc.2009.69. [DOI] [PubMed] [Google Scholar]
- 82.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, Ruvolo V, Tsao T, Zeng Z, Vassilev LT, Andreeff M. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schaffner C, Stilgenbauer S, Rappold GA, Dohner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- 84.Corbo V, Beghelli S, Bersani S, Antonello D, Talamini G, Brunelli M, Capelli P, Falconi M, Scarpa A. Pancreatic endocrine tumours: mutational and immunohistochemical survey of protein kinases reveals alterations in targetable kinases in cancer cell lines and rare primaries. Ann Oncol. 2012;23:127–134. doi: 10.1093/annonc/mdr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 86.Zighelboim I, Schmidt AP, Gao F, Thaker PH, Powell MA, Rader JS, Gibb RK, Mutch DG, Goodfellow PJ. ATR mutation in endometrioid endometrial cancer is associated with poor clinical outcomes. J. Clin. Oncol. 2009;27:3091–3096. doi: 10.1200/JCO.2008.19.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 88.Somyajit K, Subramanya S, Nagaraju G. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis. 2010;31:2031–2038. doi: 10.1093/carcin/bgq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shah NP. Bench to bedside: BRCA: from therapeutic target to therapeutic shield. Nat Med. 2008;14:495–496. doi: 10.1038/nm0508-495. [DOI] [PubMed] [Google Scholar]
- 90.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7:152. doi: 10.1186/1471-2407-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cole M, Strair R. Acute myelogenous leukemia and myelodysplasia secondary to breast cancer treatment: case studies and literature review. Am J Med Sci. 2010;339:36–40. doi: 10.1097/MAJ.0b013e3181bedb74. [DOI] [PubMed] [Google Scholar]
- 93.Scardocci A, Guidi F, D'Alo F, Gumiero D, Fabiani E, Diruscio A, Martini M, Larocca LM, Zollino M, Hohaus S, Leone G, Voso MT. Reduced BRCA1 expression due to promoter hypermethylation in therapy-related acute myeloid leukaemia. Br J Cancer. 2006;95:1108–1113. doi: 10.1038/sj.bjc.6603392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 95.Yang S, Jeong JH, Brown AL, Lee CH, Pandolfi PP, Chung JH, Kim MK. Promyelocytic leukemia activates Chk2 by mediating Chk2 autophosphorylation. J Biol Chem. 2006;281:26645–26654. doi: 10.1074/jbc.M604391200. [DOI] [PubMed] [Google Scholar]
- 96.Halicka HD, Ozkaynak MF, Levendoglu-Tugal O, Sandoval C, Seiter K, Kajstura M, Traganos F, Jayabose S, Darzynkiewicz Z. DNA damage response as a biomarker in treatment of leukemias. Cell Cycle. 2009;8:1720–1724. doi: 10.4161/cc.8.11.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ceccaldi R, Briot D, Larghero J, Vasquez N, Dubois d'Enghien C, Chamousset D, Noguera ME, Waisfisz Q, Hermine O, Pondarre C, Leblanc T, Gluckman E, Joenje H, Stoppa-Lyonnet D, Socie G, Soulier J. Spontaneous abrogation of the GDNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J Clin Invest. 2011;121:184–194. doi: 10.1172/JCI43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Didier C, Cavelier C, Quaranta M, Galcera MO, Demur C, Laurent G, Manenti S, Ducommun B. G2/M checkpoint stringency is a key parameter in the sensitivity of AML cells to genotoxic stress. Oncogene. 2008;27:3811–3820. doi: 10.1038/sj.onc.1211041. [DOI] [PubMed] [Google Scholar]
- 99.Yan Y, Cao PT, Greer PM, Nagengast ES, Kolb RH, Mumby MC, Cowan KH. Protein phosphatase 2A has an essential role in the activation of gamma-irradiation-induced G2/M checkpoint response. Oncogene. 2010;29:4317–4329. doi: 10.1038/onc.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei S, Chen X, Rocha K, Epling-Burnette PK, Djeu JY, Liu Q, Byrd J, Sokol L, Lawrence N, Pireddu R, Dewald G, Williams A, Maciejewski J, List A. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci USA. 2009;106:12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Vugt MA, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 102.Ikezoe T, Yang J, Nishioka C, Takezaki Y, Tasaka T, Togitani K, Koeffler HP, Yokoyama A. A novel treatment strategy targeting polo-like kinase 1 in hematological malignancies. Leukemia. 2009;23:1564–1576. doi: 10.1038/leu.2009.94. [DOI] [PubMed] [Google Scholar]
- 103.Wäsch R, Engelbert D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene. 2005;24:1–10. doi: 10.1038/sj.onc.1208017. [DOI] [PubMed] [Google Scholar]
- 104.Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM, Lee HS, Yoo SH, Chung DH, Sung YC, McKeon F, Lee CW. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell. 2003;4:483–497. doi: 10.1016/s1535-6108(03)00302-7. [DOI] [PubMed] [Google Scholar]
- 105.Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci USA. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, Zhang DE. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 107.Boyapati A, Yan M, Peterson LF, Biggs JR, Le Beau MM, Zhang DE. A leukemia fusion protein attenuates the spindle checkpoint and promotes aneuploidy. Blood. 2007;109:3963–3971. doi: 10.1182/blood-2006-09-045583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin SF, Lin PM, Yang MC, Liu TC, Chang JG, Sue YC, Chen TP. Expression of hBUB1 in acute myeloid leukemia. Leuk Lymphoma. 2002;43:385–391. doi: 10.1080/10428190290006206. [DOI] [PubMed] [Google Scholar]
- 109.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 110.Kiyomitsu T, Murakami H, Yanagida M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol. 2011;31:998–1011. doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 112.Ye D, Garcia-Manero G, Kantarjian HM, Xiao L, Vadhan-Raj S, Fernandez MH, Nguyen MH, Medeiros LJ, Bueso-Ramos CE. Analysis of Aurora kinase A expression in CD34(+) blast cells isolated from patients with myelodysplastic syndromes and acute myeloid leukemia. J Hematop. 2009;2:2–8. doi: 10.1007/s12308-008-0019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ikezoe T, Yang J, Nishioka C, Tasaka T, Taniguchi A, Kuwayama Y, Komatsu N, Bandobashi K, Togitani K, Koeffler HP, Taguchi H. A novel treatment strategy targeting Aurora kinases in acute myelogenous leukemia. Mol Cancer Ther. 2007;6:1851–1857. doi: 10.1158/1535-7163.MCT-07-0067. [DOI] [PubMed] [Google Scholar]
- 114.Cheong JW, Jung HI, Eom JI, Kim SJ, Jeung HK, Min YH. Aurora-A kinase inhibition enhances the cytosine arabinoside-induced cell death in leukemia cells through apoptosis and mitotic catastrophe. Cancer Lett. 2010;297:171–181. doi: 10.1016/j.canlet.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 115.Moore AS, Blagg J, Linardopoulos S, Pearson AD. Aurora kinase inhibitors: novel small molecules with promising activity in acute myeloid and Philadelphia-positive leukemias. Leukemia. 2010;24:671–678. doi: 10.1038/leu.2010.15. [DOI] [PubMed] [Google Scholar]
- 116.Lucena-Araujo AR, de Oliveira FM, Leite-Cueva SD, dos Santos GA, Falcao RP, Rego EM. High expression of AURKA and AURKB is associated with unfavorable cytogenetic abnormalities and high white blood cell count in patients with acute myeloid leukemia. Leuk Res. 2011;35:260–264. doi: 10.1016/j.leukres.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 117.Yang J, Ikezoe T, Nishioka C, Tasaka T, Taniguchi A, Kuwayama Y, Komatsu N, Bandobashi K, Togitani K, Koeffler HP, Taguchi H, Yokoyama A. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood. 2007;110:2034–2040. doi: 10.1182/blood-2007-02-073700. [DOI] [PubMed] [Google Scholar]
- 118.Oke A, Pearce D, Wilkinson RW, Crafter C, Odedra R, Cavenagh J, Fitzgibbon J, Lister AT, Joel S, Bonnet D. AZD1152 rapidly and negatively affects the growth and survival of human acute myeloid leukemia cells in vitro and in vivo. Cancer Res. 2009;69:4150–4158. doi: 10.1158/0008-5472.CAN-08-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Walsby E, Walsh V, Pepper C, Burnett A, Mills K. Effects of the aurora kinase inhibitors AZD1152-HQPA and ZM447439 on growth arrest and polyploidy in acute myeloid leukemia cell lines and primary blasts. Haematologica. 2008;93:662–669. doi: 10.3324/haematol.12148. [DOI] [PubMed] [Google Scholar]
- 120.Ikezoe T, Yang J, Nishioka C, Yokoyama A. p53 is critical for the Aurora B kinase inhibitor-mediated apoptosis in acute myelogenous leukemia cells. Int J Hematol. 2010;91:69–77. doi: 10.1007/s12185-009-0462-7. [DOI] [PubMed] [Google Scholar]
- 121.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 122.Wäsch R, Hasskarl J, Schnerch D, Lübbert M. BI_2536--targeting the mitotic kinase Polo-like kinase 1 (Plk1) Recent Results Cancer Res. 2010;184:215–218. doi: 10.1007/978-3-642-01222-8_15. [DOI] [PubMed] [Google Scholar]
- 123.Renner AG, Dos Santos C, Recher C, Bailly C, Creancier L, Kruczynski A, Payrastre B, Manenti S. Polo-like kinase 1 is overexpressed in acute myeloid leukemia and its inhibition preferentially targets the proliferation of leukemic cells. Blood. 2009;114:659–662. doi: 10.1182/blood-2008-12-195867. [DOI] [PubMed] [Google Scholar]
- 124.Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–435. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
- 125.Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dombret H. Gene mutation and AML pathogenesis. Blood. 2011;118:5366–5367. doi: 10.1182/blood-2011-09-379081. [DOI] [PubMed] [Google Scholar]
- 127.Lee JW, Kim YG, Soung YH, Han KJ, Kim SY, Rhim HS, Min WS, Nam SW, Park WS, Lee JY, Yoo NJ, Lee SH. The JAK2 V617F mutation in de novo acute myelogenous leukemias. Oncogene. 2006;25:1434–1436. doi: 10.1038/sj.onc.1209163. [DOI] [PubMed] [Google Scholar]
- 128.Frohling S, Lipka DB, Kayser S, Scholl C, Schlenk RF, Dohner H, Gilliland DG, Levine RL, Dohner K. Rare occurrence of the JAK2 V617F mutation in AML subtypes M5, M6, and M7. Blood. 2006;107:1242–1243. doi: 10.1182/blood-2005-09-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Birg F, Courcoul M, Rosnet O, Bardin F, Pebusque MJ, Marchetto S, Tabilio A, Mannoni P, Birnbaum D. Expression of the FMS/KIT-like gene FLT3 in human acute leukemias of the myeloid and lymphoid lineages. Blood. 1992;80:2584–2593. [PubMed] [Google Scholar]
- 130.Horiike S, Yokota S, Nakao M, Iwai T, Sasai Y, Kaneko H, Taniwaki M, Kashima K, Fujii H, Abe T, Misawa S. Tandem duplications of the FLT3 receptor gene are associated with leukemic transformation of myelodysplasia. Leukemia. 1997;11:1442–1446. doi: 10.1038/sj.leu.2400770. [DOI] [PubMed] [Google Scholar]
- 131.Beghini A, Ripamonti CB, Cairoli R, Cazzaniga G, Colapietro P, Elice F, Nadali G, Grillo G, Haas OA, Biondi A, Morra E, Larizza L. KIT activating mutations: incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica. 2004;89:920–925. [PubMed] [Google Scholar]
- 132.Jacobi A, Thieme S, Lehmann R, Ugarte F, Malech HL, Koch S, Thiede C, Muller K, Bornhauser M, Ryser M, Brenner S. Impact of CXCR4 inhibition on FLT3-ITD-positive human AML blasts. Exp Hematol. 2010;38:180–190. doi: 10.1016/j.exphem.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ, Langabeer SE, Kottaridis PD, Moorman AV, Burnett AK, Linch DC. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 134.Christiansen DH, Andersen MK, Desta F, Pedersen-Bjergaard J. Mutations of genes in the receptor tyrosine kinase (RTK)/RAS-BRAF signal transduction pathway in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2005;19:2232–2240. doi: 10.1038/sj.leu.2404009. [DOI] [PubMed] [Google Scholar]
- 135.Platanias LC. Map kinase signaling pathways and hematologic malignancies. Blood. 2003;101:4667–4679. doi: 10.1182/blood-2002-12-3647. [DOI] [PubMed] [Google Scholar]
- 136.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118:693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muller-Tidow C, Metzelder SK, Buerger H, Packeisen J, Ganser A, Heil G, Kugler K, Adiguzel G, Schwable J, Steffen B, Ludwig WD, Heinecke A, Buchner T, Berdel WE, Serve H. Expression of the p14ARF tumor suppressor predicts survival in acute myeloid leukemia. Leukemia. 2004;18:720–726. doi: 10.1038/sj.leu.2403296. [DOI] [PubMed] [Google Scholar]
- 138.Paul TA, Bies J, Small D, Wolff L. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood. 2010;115:3098–3108. doi: 10.1182/blood-2009-07-233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Taniguchi T, Chikatsu N, Takahashi S, Fujita A, Uchimaru K, Asano S, Fujita T, Motokura T. Expression of p16INK4A and p14ARF in hematological malignancies. Leukemia. 1999;13:1760–1769. doi: 10.1038/sj.leu.2401557. [DOI] [PubMed] [Google Scholar]
- 140.Peterson LF, Yan M, Zhang DE. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007;109:4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yokozawa T, Towatari M, Iida H, Takeyama K, Tanimoto M, Kiyoi H, Motoji T, Asou N, Saito K, Takeuchi M, Kobayashi Y, Miyawaki S, Kodera Y, Ohno R, Saito H, Naoe T. Prognostic significance of the cell cycle inhibitor p27Kip1 in acute myeloid leukemia. Leukemia. 2000;14:28–33. doi: 10.1038/sj.leu.2401640. [DOI] [PubMed] [Google Scholar]
- 142.Jamal R, Gale RE, Shaun N, Thomas B, Wheatley K, Linch DC. The retinoblastoma gene (rb1) in acute myeloid leukaemia: analysis of gene rearrangements, protein expression and comparison of disease outcome. Br J Haematol. 1996;94:342–351. doi: 10.1046/j.1365-2141.1996.d01-1804.x. [DOI] [PubMed] [Google Scholar]
- 143.Wang L, Wang J, Blaser BW, Duchemin AM, Kusewitt DF, Liu T, Caligiuri MA, Briesewitz R. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110:2075–2083. doi: 10.1182/blood-2007-02-071266. [DOI] [PubMed] [Google Scholar]
- 144.Radosevic N, Delmer A, Tang R, Marie JP, Ajchenbaum-Cymbalista F. Cell cycle regulatory protein expression in fresh acute myeloid leukemia cells and after drug exposure. Leukemia. 2001;15:559–566. doi: 10.1038/sj.leu.2402092. [DOI] [PubMed] [Google Scholar]
- 145.Iida H, Towatari M, Tanimoto M, Morishita Y, Kodera Y, Saito H. Overexpression of cyclin E in acute myelogenous leukemia. Blood. 1997;90:3707–3713. [PubMed] [Google Scholar]
- 146.Amanullah A, Hoffman B, Liebermann DA. Deregulated E2F-1 blocks terminal differentiation and loss of leukemogenicity of M1 myeloblastic leukemia cells without abrogating induction of p15(INK4B) and p16(INK4A) Blood. 2000;96:475–482. [PubMed] [Google Scholar]
- 147.Somervaille TC, Cleary ML. PU. 1 and Junb: suppressing the formation of acute myeloid leukemia stem cells. Cancer Cell. 2006;10:456–457. doi: 10.1016/j.ccr.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 148.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 2011;117:5701–5709. doi: 10.1182/blood-2010-04-280123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Muller-Tidow C, Mann M, Serve H. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell. 2009;36:326–339. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 150.Gombart AF, Hofmann WK, Kawano S, Takeuchi S, Krug U, Kwok SH, Larsen RJ, Asou H, Miller CW, Hoelzer D, Koeffler HP. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 151.Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–2918. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 152.Bisaillon R, Wilhelm BT, Krosl J, Sauvageau G. C-terminal domain of MEIS1 converts PKNOX1 (PREP1) into a HOXA9-collaborating oncoprotein. Blood. 2011;118:4682–4689. doi: 10.1182/blood-2011-05-354076. [DOI] [PubMed] [Google Scholar]
- 153.Jin G, Yamazaki Y, Takuwa M, Takahara T, Kaneko K, Kuwata T, Miyata S, Nakamura T. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 154.Taylor AM. Ataxia telangiectasia genes and predisposition to leukaemia, lymphoma and breast cancer. Br J Cancer. 1992;66:5–9. doi: 10.1038/bjc.1992.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Boultwood J. Ataxia telangiectasia gene mutations in leukaemia and lymphoma. J Clin Pathol. 2001;54:512–516. doi: 10.1136/jcp.54.7.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Joe Y, Jeong JH, Yang S, Kang H, Motoyama N, Pandolfi PP, Chung JH, Kim MK. ATR, PML, and CHK2 play a role in arsenic trioxide-induced apoptosis. J Biol Chem. 2006;281:28764–28771. doi: 10.1074/jbc.M604392200. [DOI] [PubMed] [Google Scholar]
- 157.Rossi R, Lidonnici MR, Soza S, Biamonti G, Montecucco A. The dispersal of replication proteins after Etoposide treatment requires the cooperation of Nbs1 with the ataxia telangiectasia Rad3-related/Chk1 pathway. Cancer Res. 2006;66:1675–1683. doi: 10.1158/0008-5472.CAN-05-2741. [DOI] [PubMed] [Google Scholar]
- 158.Montecucco A, Biamonti G. Cellular response to etoposide treatment. Cancer Lett. 2007;252:9–18. doi: 10.1016/j.canlet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 159.Mesa RA, Loegering D, Powell HL, Flatten K, Arlander SJ, Dai NT, Heldebrant MP, Vroman BT, Smith BD, Karp JE, Eyck CJ, Erlichman C, Kaufmann SH, Karnitz LM. Heat shock protein 90 inhibition sensitizes acute myelogenous leukemia cells to cytarabine. Blood. 2005;106:318–327. doi: 10.1182/blood-2004-09-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Aktas D, Arno MJ, Rassool F, Mufti GJ. Analysis of CHK2 in patients with myelodysplastic syndromes. Leuk Res. 2002;26:985–987. doi: 10.1016/s0145-2126(02)00075-9. [DOI] [PubMed] [Google Scholar]
- 161.Gautier EF, Picard M, Laurent C, Marty C, Villeval JL, Demur C, Delhommeau F, Hexner E, Giraudier S, Bonnevialle N, Ducommun B, Recher C, Laurent G, Manenti S, Mansat-De Mas V. The cell cycle regulator CDC25A is a target for JAK2V617F oncogene. Blood. 2012;119:1190–1199. doi: 10.1182/blood-2011-01-327742. [DOI] [PubMed] [Google Scholar]
- 162.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 163.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Muller-Tidow C, Wang W, Idos GE, Diederichs S, Yang R, Readhead C, Berdel WE, Serve H, Saville M, Watson R, Koeffler HP. Cyclin A1 directly interacts with B-myb and cyclin A1/cdk2 phosphorylate B-myb at functionally important serine and threonine residues: tissue-specific regulation of B-myb function. Blood. 2001;97:2091–2097. doi: 10.1182/blood.v97.7.2091. [DOI] [PubMed] [Google Scholar]
- 165.Ekberg J, Landberg G, Holm C, Richter J, Wolgemuth DJ, Persson JL. Regulation of the cyclin A1 protein is associated with its differential subcellular localization in hematopoietic and leukemic cells. Oncogene. 2004;23:9082–9089. doi: 10.1038/sj.onc.1208090. [DOI] [PubMed] [Google Scholar]
- 166.Yang R, Nakamaki T, Lübbert M, Said J, Sakashita A, Freyaldenhoven BS, Spira S, Huynh V, Muller C, Koeffler HP. Cyclin A1 expression in leukemia and normal hematopoietic cells. Blood. 1999;93:2067–2074. [PubMed] [Google Scholar]
- 167.Cristobal I, Garcia-Orti L, Cirauqui C, Alonso MM, Calasanz MJ, Odero MD. PP2A impaired activity is a common event in acute myeloid leukemia and its activation by forskolin has a potent anti-leukemic effect. Leukemia. 2011;25:606–614. doi: 10.1038/leu.2010.294. [DOI] [PubMed] [Google Scholar]
- 168.Seliger B, Papadileris S, Vogel D, Hess G, Brendel C, Storkel S, Ortel J, Kolbe K, Huber C, Huhn D, Neubauer A. Analysis of the p53 and MDM-2 gene in acute myeloid leukemia. Eur J Haematol. 1996;57:230–240. doi: 10.1111/j.1600-0609.1996.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 169.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Smith PJ, Soues S, Gottlieb T, Falk SJ, Watson JV, Osborne RJ, Bleehen NM. Etoposide-induced cell cycle delay and arrest-dependent modulation of DNA topoisomerase II in small-cell lung cancer cells. Br J Cancer. 1994;70:914–921. doi: 10.1038/bjc.1994.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S, Ohnishi K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31:2012–2021. doi: 10.1093/carcin/bgq185. [DOI] [PubMed] [Google Scholar]
- 172.Didier C, Cavelier C, Quaranta M, Demur C, Ducommun B. Evaluation of Polo-like Kinase 1 inhibition on the G2/M checkpoint in Acute Myelocytic Leukaemia. Eur J Pharmacol. 2008;591:102–105. doi: 10.1016/j.ejphar.2008.06.062. [DOI] [PubMed] [Google Scholar]
- 173.Cerveira N, Correia C, Bizarro S, Pinto C, Lisboa S, Mariz JM, Marques M, Teixeira MR. SEPT2 is a new fusion partner of MLL in acute myeloid leukemia with t(2; 11)(q37; q23) Oncogene. 2006;25:6147–6152. doi: 10.1038/sj.onc.1209626. [DOI] [PubMed] [Google Scholar]
- 174.Damm F, Oberacker T, Thol F, Surdziel E, Wagner K, Chaturvedi A, Morgan M, Bomm K, Gohring G, Lubbert M, Kanz L, Fiedler W, Schlegelberger B, Heil G, Schlenk RF, Dohner K, Dohner H, Krauter J, Ganser A, Heuser M. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J. Clin. Oncol. 2011;29:682–689. doi: 10.1200/JCO.2010.31.1118. [DOI] [PubMed] [Google Scholar]
- 175.Liu J, Wang XN, Cheng F, Liou YC, Deng LW. Phosphorylation of mixed lineage leukemia 5 by CDC2 affects its cellular distribution and is required for mitotic entry. J Biol Chem. 2010;285:20904–20914. doi: 10.1074/jbc.M109.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Krapf G, Kaindl U, Kilbey A, Fuka G, Inthal A, Joas R, Mann G, Neil JC, Haas OA, Panzer-Grumayer ER. ETV6/RUNX1 abrogates mitotic checkpoint function and targets its key player MAD2L1. Oncogene. 2010;29:3307–3312. doi: 10.1038/onc.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mundt KE, Golsteyn RM, Lane HA, Nigg EA. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.