Abstract
RanBPM is a ubiquitous protein that has been reported to regulate several cellular processes through interactions with various proteins. However, it is not known whether RanBPM may regulate gene expression patterns. As it has been shown that RanBPM interacts with a number of transcription factors, we hypothesized that it may have wide ranging effects on gene expression that may explain its function. To test this hypothesis, we generated stable RanBPM shRNA cell lines to analyze the effect of RanBPM on global gene expression. Microarray analyses were conducted comparing the gene expression profile of Hela and HCT116 RanBPM shRNA cells versus control shRNA cells. We identified 167 annotated genes significantly up- or down-regulated in the two cell lines. Analysis of the gene set revealed that down-regulation of RanBPM led to gene expression changes that affect regulation of cell, tissue, and organ development and morphology, as well as biological processes implicated in tumorigenesis. Analysis of Transcription Factor Binding Sites (TFBS) present in the gene set identified several significantly over-represented transcription factors of the Forkhead, HMG, and Homeodomain families of transcription factors, which have previously been demonstrated as having important roles in development and tumorigenesis. In addition, the combined results of these analyses suggested that several signaling pathways were affected by RanBPM down-regulation, including ERK1/2, Wnt, Notch, and PI3K/Akt pathways. Lastly, analysis of selected target genes by quantitative RT-qPCR confirmed the changes revealed by microarray. Several of the genes up-regulated in RanBPM shRNA cells encode proteins with known oncogenic functions, such as the RON tyrosine kinase, the adhesion molecule L1CAM, and transcription factor ELF3/ESE-1, suggesting that RanBPM functions as a tumor suppressor to prevent deregulated expression of these genes. Altogether, these results suggest that RanBPM does indeed function to regulate many genomic events that regulate embryonic, tissue, and cellular development as well as those involved in cancer development and progression.
Keywords: RanBPM, ERK, Wnt, Notch, microarray, cancer, development
Introduction
Cancer development is driven by alterations in cellular pathways leading to the evasion from mechanisms that normally restrict growth, migration and invasion [1]. Tumorigenesis is associated with changes in gene expression and the progression of cell transformation from normal to tumor cells [2,3]. Altered gene expression can elicit cancer development, and may also occur as the result of downstream signaling from pathways deregulated during cancer progression [4]. These changes in expression have been linked to specific phases of cancer development often reflecting either the type or the stage of cancer progression. To this end, gene expression profiling has greatly contributed to our understanding of cancer progression, and the identification of key genes and pathways which when deregulated promote cancer development [3].
RanBPM (Ran-binding protein M, also called RanBP9) is a ubiquitous nucleo-cytoplasmic protein. RanBPM was initially identified as a binding partner for the Ran GTPase that localized to the microtubule-organizing center (MTOC) [5], although both of these observations were later dismissed [6]. Several roles for RanBPM have subsequently been proposed in cellular processes including the regulation of cell morphology [7-9], cell adhesion [10-12], cell cycle progression [13] and regulation of neurological functions [13-15]. Most of these functions result from interaction of RanBPM with various proteins, which have been reported to occur both in the cytoplasm and the nucleus. In the cytoplasm, RanBPM has been suggested to function as a scaffold for receptor signaling pathways through interactions with the neuronal cell adhesion molecule L1 [11], the Met receptor (Met proto-oncogene, also called Hepatocyte growth factor receptor) [16], and tropomyosin related kinase (Trk) TrkA [17] and TrkB [14] receptors. The interaction of RanBPM with these receptors is believed to regulate the activation of downstream signaling pathways including the extra-cellular signal regulated kinase 1/2 (ERK1/2) [11,16], Akt [14], and nuclear factor-kappa B (NF-κB) pathways [18]. In addition, RanBPM was suggested to modulate the stability of proteins, such as the pro-apoptotic transcription factor p73 [19]. In the nucleus, RanBPM has been shown to interact with the transcriptional regulator TAF4 (TBP-associated factor 4) [15] and the viral early-immediate transcriptional regulator Rta [20]. RanBPM has also been reported to modulate the transcriptional activity of the androgen receptor (AR) [21], the glucocorticoid receptor (GR) [21], and the thyroid hormone receptor (TR) [22]. These data suggest that RanBPM could have wide ranging and important influences on gene expression, either directly through interaction with transcriptional regulators, or indirectly through the modulation of intracellular signaling pathways.
We previously established that RanBPM functions to promote apoptosis in response to DNA damage [23]. We showed that down-regulation of RanBPM in Hela and HCT116 cells prevented the activation of the mitochondrial apoptotic pathway and promoted cell survival in response to ionizing radiation (IR) treatment. Therefore in the present study, we sought to gain further insight into the pathways and cellular functions that are regulated by RanBPM. Using microarray analyses that compared RanBPM downregulated cells to those in which RanBPM is expressed at physiological levels, global changes in gene expression elicited by RanBPM down-regulation were investigated. Our analyses reveal that RanBPM causes wide spread perturbances in gene expression that indicate it may be an important mediator in the control of many tumorigenic processes.
Materials and methods
Cell culture
Hela and HCT116 control shRNA and RanBPM shRNA stable cell lines were previously generated [23]. Cells were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Burlington, ON) supplemented with 10% fetal bovine serum (FBS) (Wisent Bioproducts, St. Bruno, QC), 2mM L-Glutamine, and 1mM Sodium Pyruvate (Life Technologies) at 37°C in 5% CO2. Control shRNA and RanBPM shRNA stable cell lines were maintained in media supplemented with 0.35 mg/ml G418 (Geneticin, Bioshop Canada, Burlington, ON).
Plasmid expression constructs and transfection assays
The pCMV-RanBPM shRNA mutant expression construct (RanBPM si-mt) has been previously described [23]. Transfection assays were carried out using ExGen 500™ (Thermo Scientific, Fermentas, Burlington, ON) as per the manufacturer's instructions.
Western blotting
Whole cell extracts were prepared as previously described [23]. Briefly, cells were collected in ice-cold PBS and lysed in buffer containing 150mM NaCl, 1mM EDTA, 50mM HEPES (pH 7.4), 10% Glycerol, 0.5% NP40, and supplemented with 1mM PMSF, 1mM DTT, 1μg/ml Leupeptin, 10μg/ml Aprotinin, 1μg/ml Pepstatin, 2mM Sodium fluoride, and 2mM Sodium orthovanadate. For western blot analysis, 20μg of protein extracts were resolved on 10% SDS-PAGE, transferred onto PVDF membranes (Bio-Rad, Burlington, ON), and blots were hybridized with antibodies against RanBPM (5M, BioAcademia, Japan) and β-actin (I-19, Santa Cruz Biotechnology, Santa Cruz, CA).
RNA extraction, quantitative reverse-transcriptase PCR and statistical analysis
Total RNA was isolated from Hela and HCT116 control shRNA, RanBPM shRNA, and RanBPM shRNA re-expressing RanBPM si-mt cells using the Qiagen RNeasy RNA isolation kit (Qiagen, Mississauga, ON). For quantitative reverse-transcriptase PCR (RT-qPCR) analyses, cDNA was prepared from 2.5μg of total RNA using the Superscript II Reverse Transcriptase kit (Life Technologies), and gene expression was determined using 10-100ng of cDNA incubated with primers described in Supplementary Table S1, using SYBR green (Bio-Rad) and the BioRad MyiQ single-color real-time PCR detection system. Relative gene expression was quantified via the ΔΔC(t) method with candidate gene values normalized to that of controls. Statistical significance was analyzed using a student's t-test, with a P < 0.05 indicating significant results.
RNA quality assessment, probe preparation and genechip hybridization
All sample labeling and GeneChip processing was performed at the London Regional Genomics Centre (Robarts Research Institute, London, ON; http://www.lrgc.ca).
RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA) and the RNA 6000 Nano kit (Caliper Life Sciences, Mountain View, CA).
Single stranded complementary DNA (sscDNA) was prepared from 200ng of total RNA as per the Ambion WT Expression Kit for Affymetrix GeneChip Whole Transcript WT Expression Arrays (http://www.ambion.com/techlib/prot/fm_4411973.pdf, Applied Biosystems, Carlsbad, CA) and the Affymetrix GeneChip WT Terminal Labeling kit and Hybridization User Manual (http://media.affymetrix.com/support/downloads/manuals/wt_term_label_ambion_user_manual.pdf, Affymetrix, Santa Clara, CA). Total RNA was first converted to cDNA, followed by in vitro transcription to make cRNA. 5.5 μg of single stranded cDNA was synthesized, end labeled, and hybridized, for 16 hours at 45°C to Human Gene 1.0 ST arrays.
All liquid handling steps were performed by a GeneChip Fluidics Station 450 and GeneChips were scanned with the GeneChip Scanner 3000 7G (Affymetrix) using Command Console v1.1.
Bioinformatics and data analysis
Probe level (.CEL file) data was generated using Affymetrix Command Console v1.1. Probes were summarized to gene level data, background subtraction was performed, and expression values were normalized to log base-2 in Partek Genomics Suite v6.6 (Partek, St. Louis, MO) using the Robust Multiarray Averaging (RMA) algorithm [24]. Partek was used to determine gene level ANalysis Of VAriance (ANOVA) p-values, fold-changes, and Gene Ontology (GO) enrichment, using a Chi-square test. Partek Pathway was also used to find enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, using a Fisher’s exact test.
For bioinformatics analyses, a list of genes exhibiting a minimum of 1.2-fold increase or decrease in expression in RanBPM shRNA cell lines compared to control shRNA cell lines was first generated (target gene list). Analysis of genes differentially expressed in RanBPM shRNA cells was performed using Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com) and the Protein Analysis Through Evolutionary Relationships (PANTHER) database [25]. For IPA, the target gene list was uploaded alongside the respective HUGO (HUman Genome Organization) gene symbols and fold-change values, and analyzed using Ingenuity Pathway Core Analysis, which generated a list of focus genes. IPA Functional Analysis of this gene list was performed to identify top biological processes affected by decreased RanBPM expression, based upon GO terms and curator-defined ontology terms (Ingenuity® Systems, www.ingenuity.com). IPA was also used to generate cellular networks affected by RanBPM down-regulation. Significance of identified cellular networks was determined by assigning a score to each network. A score is assigned based upon a p-value calculation that calculates the likelihood that genes appear within a given network by random chance, and is the negative exponent of the right-tailed Fisher's exact test result (Ingenuity® Systems, www.ingenuity.com). A score of ≥3 is considered significant, as it indicates a 1/1000 chance that genes appear within a network by chance.
The PANTHER database uses protein sequences to group proteins into functional families and subfamilies, and uses ontology terms to classify proteins according to molecular functions, biological processes, and protein classes. For PANTHER analyses, the target gene list and HUGO gene symbols were uploaded to the PANTHER website and the top biological processes, molecular functions, and protein classes affected in response to RanBPM down-regulation were identified.
The oPOSSUM analysis system was utilized to identify over-represented Transcription Factor Binding Sites (TFBS) in the list of genes affected by RanBPM down-regulation. Briefly, oPOSSUM compares the occurrence of TFBS within a set of co-expressed genes (target gene list) to a predetermined background set of genes, in order to identify over-represented sites in the target list [26]. The significance of any identified binding sites is calculated using a Z-score and Fisher score, with a Z-score of ≥10 indicating a significantly over-represented TFBS, in agreement with database publisher's recommendations [27].
Results
Identification of gene targets regulated by RanBPM expression
We have previously generated clonal stable cell lines in Hela and HCT116 cells expressing either a control or RanBPM shRNA (Supplementary Figure 1S) [23]. To identify gene targets that are regulated by RanBPM expression, RNA samples were prepared in triplicate from normally proliferating Hela control shRNA and RanBPM shRNA (clone 2-7) cells, and gene expression profiling was performed using Affymetrix human gene expression arrays. The mean fold-change in gene expression in RanBPM shRNA (2-7) cells compared to control shRNA cells was calculated, and using a 1.2-fold change cut-off, we identified 2621 genes for which expression was altered by RanBPM down-regulation. To minimize the potential that the observed changes in gene expression arose from the derivation of clonal cell lines, RNA samples were prepared in triplicate from a second Hela RanBPM shRNA cell line [denoted RanBPM shRNA (clone 2-6)]. Gene expression was quantified using the parameters outlined above, and we identified 3952 genes differentially expressed in RanBPM shRNA (2-6) cells compared to control shRNA cells. Comparison of the list of differentially expressed genes in the two Hela RanBPM shRNA cell lines identified 1719 genes common to both cell lines (Supplementary Figure 1S). Further, to limit possible cell-type specific effects of RanBPM downregulation on gene expression patterns, RNA samples were also prepared in triplicate from HCT116 cells expressing either a control or RanBPM shRNA [denoted HCT116 RanBPM shRNA (clone 2-8)]. The mean fold-change in expression was calculated, and using a 1.2-fold change cut-off we identified 2226 genes with altered expression in response to RanBPM down-regulation. Combining the list of differentially expressed genes obtained for each cell line we identified a total 187 genes common to all three cell lines, for which expression was changed by RanBPM down-regulation (Supplementary Figure 1S and Supplementary Table S2). Of these, 167 were annotated genes, with 74 genes down-regulated and 93 genes up-regulated upon RanBPM down-regulation. Our analysis also confirmed a strong decrease in RanBPM expression in all three RanBPM shRNA cell lines, as we observed a 2.46-fold decrease in Hela RanBPM shRNA (2-6), 2.92-fold decrease in Hela RanBPM shRNA (2-7), and 2.76-fold decrease in HCT116 RanBPM shRNA (2-8) cells respectively (Supplementary Table S2).
IPA and PANTHER analyses
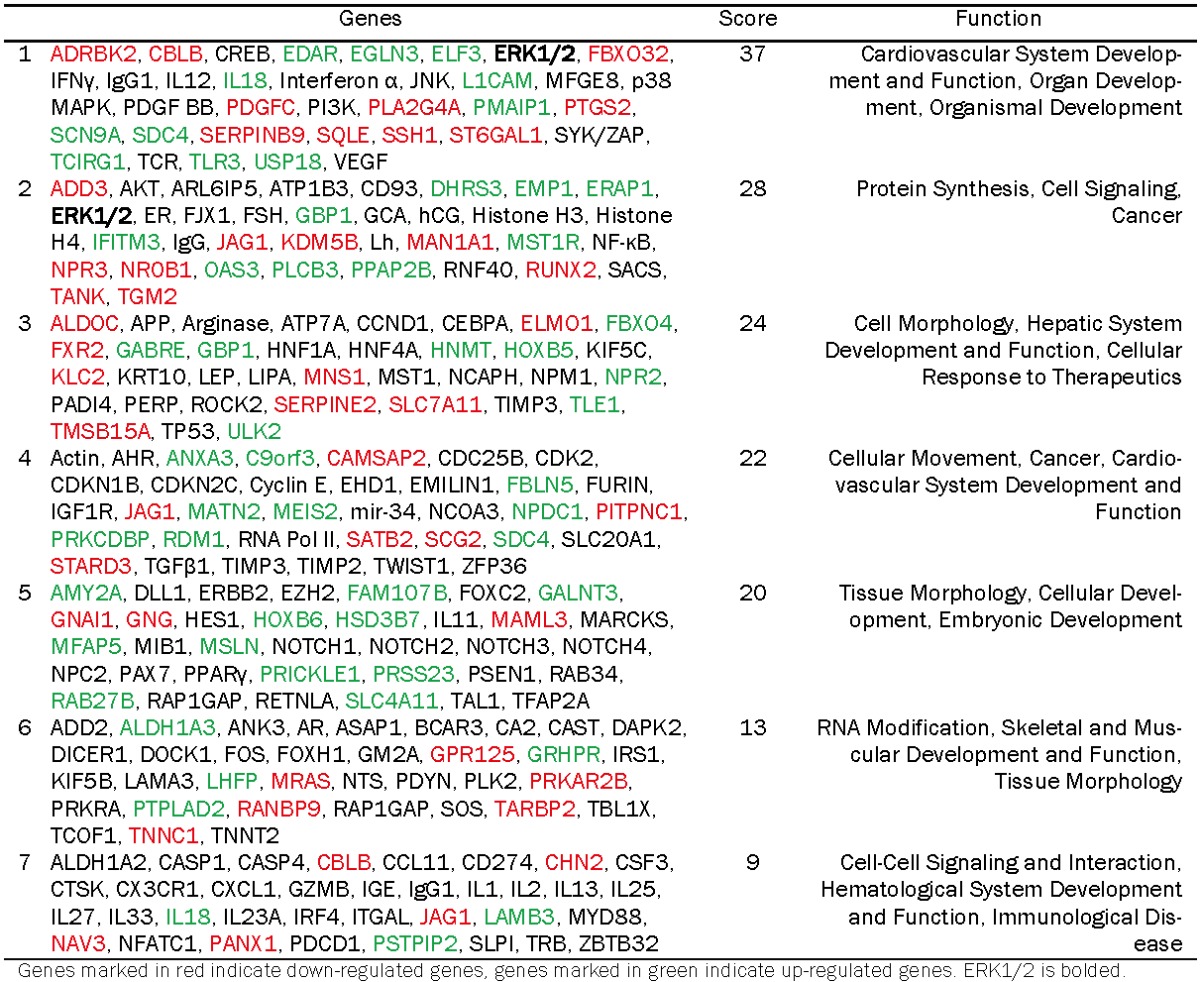
To gain insight into the functional and biological consequences associated with decreased RanBPM expression, we performed Ingenuity Pathway Analysis on the 167 genes identified above. IPA is a web-based tool that utilizes information from the literature to analyze data obtained from gene expression arrays. This analysis allows for modeling biological interactions and building networks of cellular processes to determine the effects of a given experimental treatment. IPA Functional Analysis of genes that were differentially expressed in RanBPM shRNA cells revealed that down-regulation of RanBPM expression most significantly affects cellular processes associated with cancer, tissue development, and cellular function and maintenance (Table 1). Notably, over one-third of the 167 genes analyzed were associated with cancer, suggesting a potentially important role for RanBPM in regulating cellular processes associated with tumorigenesis. To characterize pathways that are affected by down-regulation of RanBPM, we next performed Network Analyses in IPA. This analysis uses a given gene list to build networks of cellular processes, and assigns a score based upon the number of genes from that list that are found within a particular cellular network. Using the gene list from above and a cut-off score of 3 (see methods) we identified seven cellular networks that were significantly affected by the decrease in RanBPM expression. These networks encompass cellular processes such as organ development, tissue morphology, cancer, cell motility, cell signaling, RNA modification, protein synthesis, and molecular transport (Table 2). ERK1/2 was a component of the top two cellular networks affected by the down-regulation of RanBPM expression. Additionally, phosphoinositide-3 kinase (PI3K)/Akt, Notch, and NF-κB signaling were components of the most highly affected cellular networks in RanBPM shRNA cells. Together these analyses reveal that down-regulation of RanBPM leads to gene expression changes that affect regulation of cell, tissue, and organ development and morphology, as well as biological processes implicated in tumorigenesis.
Table 1.
Identification of top biological functions altered in RanBPM shRNA cells using IPA.
| Diseases and Disorders | ||
|---|---|---|
| Name | p-value | # of molecules |
| Cancer | 2.43E-04 - 3.79E-02 | 57 |
| Reproductive System and Disease | 2.43E-04 - 2.86E-02 | 19 |
| Gastrointestinal Disease | 5.43E-04 - 3.79E-02 | 16 |
| Organismal Injury and Abnormalities | 5.43E-04 - 3.79E-02 | 10 |
| Inflammatory Response | 1.34E-03 - 3.79E-02 | 19 |
|
| ||
| Molecular and Cellular Functions | ||
|
| ||
| Name | p-value | # of molecules |
| Cellular Function and Maintenance | 2.60E-04 - 3.79E-02 | 22 |
| Cell Morphology | 2.73E-04 - 3.14E-02 | 12 |
| Cellular Assembly and Organization | 2.73E-04 - 3.79E-02 | 10 |
| Lipid Metabolism | 9.00E-04 - 3.79E-02 | 10 |
| Small Molecule Biochemistry | 9.00E-04 - 3.79E-02 | 20 |
|
| ||
| Physiological System Development and Function | ||
|
| ||
| Name | p-value | # of molecules |
| Tissue Development | 7.03E-04 - 3.79E-02 | 42 |
| Hematological System Development and Function | 2.60E-04 - 3.79E-02 | 22 |
| Organ Morphology | 9.00E-04 - 3.79E-02 | 10 |
| Reproductive System Development and Function | 9.00E-04 - 3.79E-02 | 9 |
| Connective Tissue Development and Function | 1.02E-03 - 3.79E-02 | 23 |
Top biological function in each category is bolded.
Table 2.
IPA analysis of cellular networks affected by RanBPM down-regulation.
|
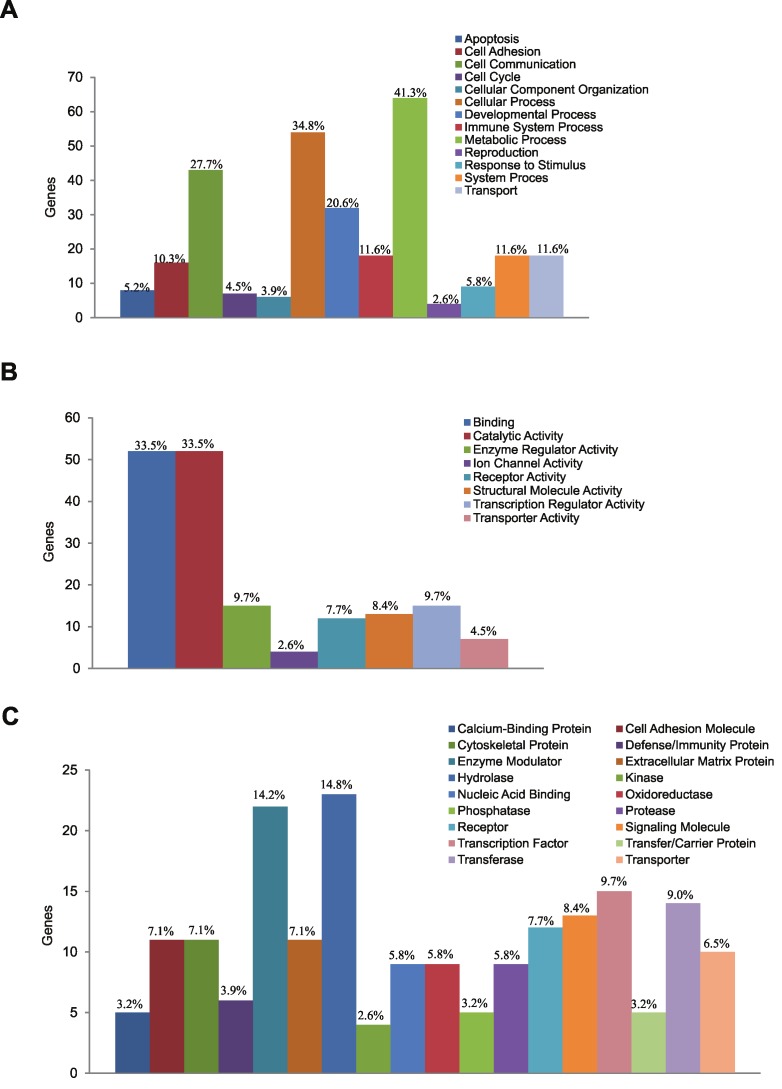
To corroborate the findings from IPA, and further characterize the molecular processes that are altered in cells with decreased RanBPM expression, we analyzed our list of differentially expressed genes using the PANTHER database [25]. Similarly to IPA, PANTHER allows for functional analysis of data gathered from gene expression profiling by using curator-defined groupings of protein sequences to build protein families. These protein families can then be used to identify biological processes, molecular functions, pathways, and protein classes to which groups of genes may be assigned. We first identified the most highly affected biological processes and molecular functions associated with reduced expression of RanBPM (Figures 1A and 1B). We determined that in RanBPM down-regulated cells, the most highly affected biological processes include cell communication, tissue development, and cellular metabolism; and the most highly affected molecular functions include receptor and protein binding, enzyme catalysis, and transcriptional regulation. PANTHER also allows for the classification of protein classes to which genes from a given gene list may belong. Using the protein class analyses, the expression of transcription factors, receptors, cell adhesion proteins, and cytoskeletal proteins were all found to be affected by the down-regulation of RanBPM expression (Figure 1C). Our analyses using PANTHER verify our findings with IPA and indicate that decreased RanBPM expression leads to changes in gene expression patterns that affect cellular processes involved in both development and cancer.
Figure 1.
PANTHER analysis of cellular processes altered by RanBPM down-regulation. A and B. Gene Ontology (GO) analyses of top biological processes (A) and molecular functions (B) affected by RanBPM down-regulation. The number of genes in target gene list that are annotated to a given function are plotted, with percentages indicating the number of genes that appear in selected gene list divided by the total number of genes assigned to that function. C. Protein classes for which expression is most significantly affected in RanBPM shRNA cells. Data is represented as in A and B.
Validation of selected gene targets
To validate the expression data obtained from our gene arrays, we selected 11 candidate genes which had previously been linked to tumorigenesis and performed RT-qPCR using RNA extracts from control shRNA and RanBPM shRNA Hela and HCT116 cells (Figures 2A and 2B). For 10 of the candidate genes selected, we confirmed the change in expression observed in the gene arrays in both the Hela and HCT116 cell lines (Figure 2B). However MFAP5 (microfibrillar associated protein 5, also known as MAGP2) mRNA expression levels were too low to be reliably measured in HCT116 cells, and therefore the change in expression of MFAP5 was only successfully confirmed in Hela cells. As might be expected we found that for the majority of candidate genes selected, the fold-change in gene expression as determined by RT-qPCR was greater than that observed using the gene arrays [28,29]. To evaluate the direct contribution of RanBPM down-regulation to the changes in gene expression observed, we ectopically re-expressed RanBPM in Hela and HCT116 RanBPM shRNA cell lines. This was achieved by transiently expressing a RanBPM cDNA containing a point mutation in the siRNA recognition sequence (RanBPM si-mt) in these cells, allowing for restoration of RanBPM expression to near endogenous levels (Figure 2A). RNA extracts were prepared 48h post-transfection from both Hela and HCT116 cells, and gene expression for the 11 candidate genes outlined above was quantified using RT-qPCR. Interestingly, our analyses revealed two categories of genes. The first category comprised genes for which re-expression of RanBPM significantly restored mRNA expression close to levels observed in control cells. These genes, grouped together to the left of the axis break in Figure 2B, C included ELF3 (E74-like factor 3, also known as ESE-1), RON [recepteur d’origine nantais/macrophage stimulating receptor 1 (MST1R)], ALDH1A3 (aldehyde dehydrogenase 1 isoform A3), Rab27B (Rab 27B member Ras oncogene family), and L1CAM (L1 cell adhesion molecule). MSLN1 (mesothelin 1) showed a statistically significant change upon RanBPM reexpression in Hela cells, but not in HCT116 cells. The second category of genes, grouped together to the right of the axis break in Figure 2B, C comprised genes for which expression was not significantly altered upon restoration of RanBPM expression. Included in this category were TG2 (transglutaminase 2, also called TGM2), PHD3 (prolyl hyrdoxylase 3, also called EGLN3), LAMB3 (laminin β3), and CHN1 (chimerin 1). Similarly, MFAP5 expression was not affected by restoration of RanBPM expression in Hela cells.
Figure 2.
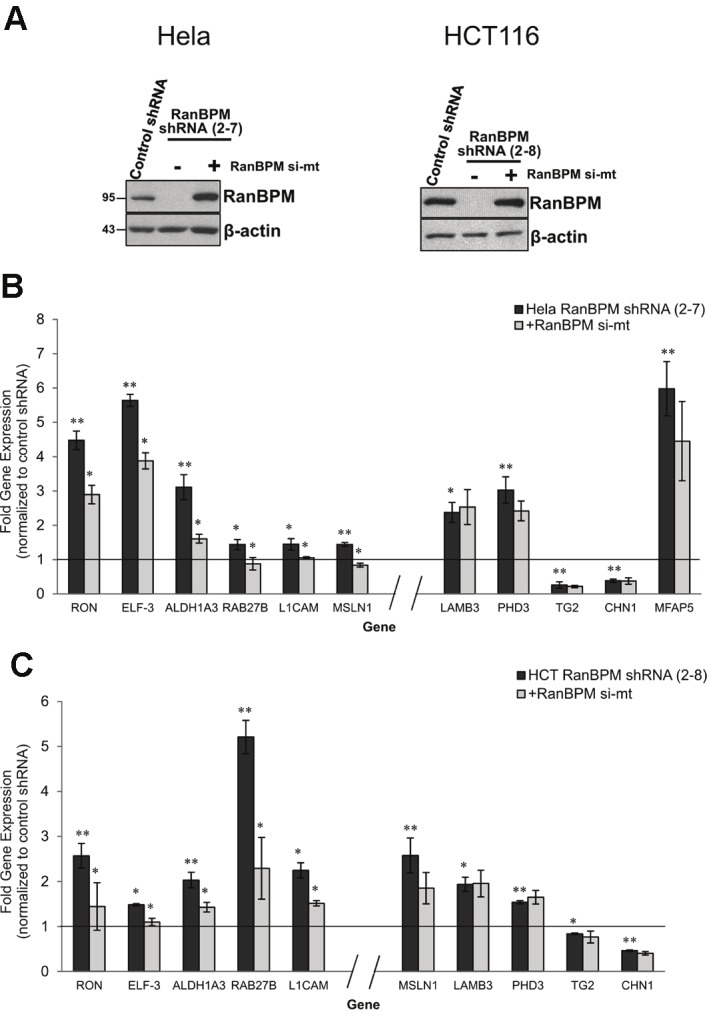
Validation of selected gene targets identified in microarray analyses. A. RanBPM shRNA Hela (left panel) and HCT116 (right panel) cells were transfected with pCMV-RanBPM si-mt construct. Forty-eight hours post-transfection, whole cell extracts were prepared and analyzed by western blotting, alongside control and untransfected RanBPM shRNA extracts. Restoration of RanBPM expression was verified by hybridizing with a RanBPM antibody. B. cDNA from control shRNA, RanBPM shRNA, and RanBPM shRNA +RanBPM si-mt Hela (top panel) and HCT116 (bottom panel) was analyzed by RT-qPCR analyses using primers specific to the indicated genes. Fold gene expression was normalized to control shRNA cells. Genes appearing to the left of x-axis break are genes whose expression was responsive to restoration of RanBPM expression, while genes appearing to the right of axis break were not responsive to restored RanBPM expression. Data represents the mean of a minimum of three independent experiments with error bars indicating SE, * P < 0.05, ** P < 0.005.
Identification of over-represented transcription factor binding sites
The PANTHER analyses outlined above identified transcriptional regulation and transcription factors as one of the most highly affected molecular functions and protein classes altered by the down-regulation of RanBPM. To expand upon this finding we sought to determine whether decreased RanBPM expression affects the expression of subsets of genes regulated by specific classes or families of transcription factors. To test this, we utilized oPOSSUM, a web-based tool that allows for the identification of over-represented TFBS in the promoters of sets of co-expressed genes [27]. Analysis of the 167 genes differentially expressed in RanBPM shRNA cells using oPOSSUM identified 20 transcription factors which contain significantly over-represented TFBS within the promoters of these genes (Table 3). We found that the most over-represented TFBS in our gene list was HOXA5 (homeobox A5), a member of the Homeobox family of transcription factors. Of the 167 genes analyzed, 134 contained binding sites for HOXA5 within their promoters. We also identified six members of the Forkhead box (FOX) family of transcription factors, four additional members of the Homeobox family, and four members of the High Mobility Group (HMG) family of transcription factors, all of which contained over-represented TFBS in the promoters of our differentially expressed genes.
Table 3.
oPOSSUM analysis of over-represented transcription factor binding sites.
| Transcription Factor | Family | Target Genes | Z-Score |
|---|---|---|---|
| Class: Winged Helix-Turn-Helix | |||
| FOXI1 | Forkhead | 103 | 30.93 |
| FOXO3 | Forkhead | 112 | 29.9 |
| FOXD1 | Forkhead | 108 | 23.8 |
| Foxa2 | Forkhead | 102 | 22.82 |
| FOXA1 | Forkhead | 105 | 21.82 |
| Foxd3 | Forkhead | 97 | 21.76 |
| SPI1 | ETS | 130 | 17.32 |
|
| |||
| Class: Helix-Turn-Helix | |||
| Nkx2-5 | Homeo | 129 | 31.21 |
| HOXA5 | Homeo | 134 | 29.04 |
| Pdx1 | Homeo | 122 | 25.42 |
| Prrx2 | Homeo | 117 | 24.24 |
| Nobox | Homeo | 113 | 23.05 |
| ARID3A | Arid | 130 | 28.3 |
|
| |||
| Class: Other Alpha-Helix | |||
| SRY | HMG | 116 | 35 |
| SOX5 | HMG | 111 | 22.42 |
| Sox17 | HMG | 110 | 20.15 |
| SOX9 | HMG | 110 | 18.86 |
|
| |||
| Class: Zinc-Coordinating | |||
| Gata1 | GATA | 117 | 25.85 |
| Gfi | ββα-zinc finger | 118 | 18.11 |
|
| |||
| Class: Leucine Zipper | |||
| CEBPA | Zipper-type | 109 | 17.2 |
Discussion
RanBPM has been implicated in the control of a multitude of cellular processes including regulation of development [7,8], cell motility [16], transcription [15,20-22], and apoptosis [19,23,30-32]. However, a common modality for the function of this protein remains unclear. The aim of the present study was to utilize gene expression profiling to gain further insight into the cellular functions of RanBPM, by characterizing the impact of RanBPM down-regulation on global cellular signaling events. To identify the functional consequences associated with decreased RanBPM expression we generated a list of genes whose expression was altered in RanBPM shRNA cells, and identified the most highly affected biological processes and cellular networks in these cells. Down-regulation of RanBPM expression was found to significantly affect the expression of factors associated with embryonic, cellular, and tissue development, as well as those involved in cancer development and progression.
Implications in development
Our analyses revealed several components of the Notch and Wnt signaling pathways whose expression was altered upon RanBPM downregulation. These include JAG1 (Jagged 1), which is a ligand for Notch receptors [33]; RUNX2 (runt-related transcription factor 2), which integrates signals from Notch, Wnt, and TGFβ (transforming growth factor β) to regulate bone development and differentiation [34], and RON/MST1R, a receptor tyrosine kinase known to promote phosphorylation and nuclear accumulation of β-catenin in breast and colon tumors [35]. Moreover, Notch signaling was found to be a major component of one of the top cellular networks affected by RanBPM downregulation. It is well established that signaling pathways such as the Notch/Wnt/Hedgehog pathway, which normally regulate embryonic development, can become deregulated in cancer. For example, Notch signaling normally mediates cell-cell communication in embryogenesis, as well as cell proliferation, differentiation, and apoptosis [33,36]. Deregulated Notch signaling has been linked to tumor development in the lung, ovaries, breast, and colon, and to enhanced epithelial-to-mesenchymal transition (EMT) of cancer cells [33,37,38]. Induction of Wnt signaling occurs upon the binding of Wnt proteins to cell surface receptors, leading to the stabilization and nuclear accumulation of β-catenin [39]. Within the nucleus, β-catenin mediates the expression of Wnt target genes that regulate embryonic signaling events such as proliferation, morphogenesis, and differentiation [34,39]. Similarly to Notch, Wnt signaling is deregulated in many cancers and in certain cases, such as in colorectal tumors, deregulated Wnt signaling can initiate tumor development [33,39]. Several studies have also identified an important role for RanBPM in development. RanBPM was found to be required in the Drosophila nervous system for larval behavior associated with feeding, growth, and locomotion [7]. Recent studies in RanBPM knockout mice revealed a critical role for this protein in normal gonad development and gametogenesis in both males and females [40]. Additionally, RanBPM has been linked to developmental processes occurring through Notch-dependent signaling. For example it was shown to regulate the size, shape, and organization of the germline stem cell (GSC) niche in female Drosophila [8]. The development, and capacity of this niche for stem cells, is known to be regulated through Notch expression and signaling [8,15]. RanBPM was found to regulate neuronal differentiation in Drosophila by interacting with TAF4, a transcriptional co-activator that binds transcription factors downstream of Notch signaling to regulate neural stem cell fate and differentiation [15]. Our gene expression data indicate that RanBPM regulates the expression of several factors involved in Notch signaling, further suggesting a possible role for RanBPM in the regulation of Notch-mediated signaling during development.
In addition to factors involved in Notch/Wnt signaling, we also identified several other differentially expressed genes for which a function in both development and cancer has been demonstrated. These include GBP1 (guanylate binding protein 1), a cytokine-activated small GTPase normally involved in cellular proliferation and angiogenesis [41], which is over-expressed in ovarian and oral tumors [42,43]. Another example is NR0B1 (nuclear receptor subfamily 0 Group B member 1) that acts a transcriptional co-repressor in embryonic stem cell development, pluoripotency, and differentiation [44], and is over-expressed in several tumors [44,45]. Finally, we also identified L1CAM, which is involved in neurite outgrowth and axon guidance in normal cells [46], and is over-expressed in numerous cancers, including melanoma, lung, and thyroid cancer [46,47]. Thus, these findings suggest a complex role for RanBPM in both the regulation of normal cellular processes associated with development, as well as in the progression of diseased states such as cancer.
Implications in signaling
RanBPM has previously been demonstrated to regulate several receptor-mediated signaling pathways, including the ERK1/2 and NF-κB pathways. As such, it was hypothesized to have potential functions in tumorigenesis, although the outcome of RanBPM function in this process remains controversial due to differing findings regarding its role in activation of signaling cascades such as ERK1/2. While some reports indicate that RanBPM expression promotes activation of ERK1/2 signaling and would therefore enhance cellular transformation [14,16,17], other groups including ours (Atabakhsh and Schild-Poulter, in revision) have characterized RanBPM as a repressor of ERK1/2 activation and suggest a tumor-suppressor role for this protein [11]. Our gene expression data indicated that several signaling pathways are affected by decreased RanBPM expression, including the ERK1/2 and the PI3K/Akt pathways, both of which were components of the top two cellular networks affected by down-regulation of RanBPM. These pathways are known to play critical functions in cancer development. ERK1/2 signaling regulates many cellular processes including cell cycle progression, cell proliferation, differentiation, migration, and adhesion [48], and aberrant ERK1/2 signaling has been observed in many diseased states including cardiovascular disease and cancer [48,49]. PI3K signaling is activated by cell-surface receptors and converges upon Akt, which phosphorylates various cellular targets involved in cell growth, survival, metabolism, and autophagy [50]. Similarly to ERK1/2, both PI3K and Akt are often found to be mutated and/or deregulated in cancer [51]. Our data indicate that while gene expression of ERK1/2, PI3K, and Akt is not affected by RanBPM down-regulation, the expression of several factors that regulate these signaling pathways is altered by decreased RanBPM expression, suggesting a tumor-suppressor function for RanBPM. For example, L1CAM and IL-18 have been found to promote ERK1/2 activation and enhance ERK-target gene expression, and are often over-expressed in tumor samples [46,52,53]. Our gene expression data reveal that down-regulation of RanBPM leads to increased L1CAM and IL-18 expression, indicating a potential link between the expression of these genes and deregulated ERK1/2 signaling in RanBPM shRNA cells. Similarly, gene expression of the tyrosine kinase RON was up-regulated in cells with decreased RanBPM expression. Over-expression of RON has been observed in multiple tumors, and is associated with enhanced ERK1/2 and Akt activation and signaling [54-56]. Collectively, these findings suggest a role for RanBPM in the regulation of signaling pathways that are associated with both normal cellular function and diseased states, and further implicate a potential role for RanBPM as a tumor suppressor.
Implications in transcriptional regulation
In addition to its roles in development and receptor signaling, RanBPM has been proposed to be directly involved in regulation of gene transcription. RanBPM was reported to function as a transcriptional co-activator for AR, GR, and TR, and to mediate their ligand-dependent nuclear translocation [21,22]. RanBPM was also shown to enhance the sumoylation and transactivation of the early-immediate Epstein-Barr Virus (EBV) protein Rta [20], and to interact with the TAF4 subunit of TFIID (transcription factor IID, also known as TBP) [15,57]. As our data revealed a wide range of gene targets affected by RanBPM expression, we sought to identify potential transcription factors through which RanBPM may mediate its effects on gene expression. Analysis of the over-represented TFBS in our list of differentially expressed genes revealed that the FOX, Homeobox, and HMG families of transcription factors contain the greatest number of binding sites within the promoters of genes affected by RanBPM down-regulation.
FOX proteins comprise a large family of transcriptional regulators that are divided into subclasses according to their function in modifying chromatin structure. The FOXA subclass (FOXA 1, 2, and 3) plays an important role in development, organogenesis, metabolism, and stem cell differentiation [57]. FOXA proteins have been reported to be over-expressed or amplified in human tumors, especially in breast, prostate, thyroid, lung, and esophageal cancers [57,58]. The FOXO subclass (FOXO1, 3a, 4, and 6) is involved in insulin and growth factor mediated signaling through PI3K/Akt, and is a downstream target of activated Akt [57,59]. FOXOs regulate differentiation, metabolism, cell cycle arrest, cell death, and tumor suppression [57]. Overall, we identified six FOX family transcription factors with over-represented binding sites in our list of differentially expressed genes, including FOXA1, FOXA2, and FOXO3. Interestingly, deregulation of FOXA proteins has been linked to hormone-sensitive malignancies, and is suggested to mediate tumorigenesis through regulation of steroid hormone receptors [58]. As RanBPM has been reported to function as a coactivator of AR, it is tempting to hypothesize that it may function to regulate target gene expression through an AR/FOXA1-dependent process. Additionally, FOXA1 has been reported to mediate chromatin opening and enhance the DNA binding of the GR at the mouse mammary tumor virus (MMTV) promoter [60]. RanBPM was also reported to enhance the transcriptional activity of GR [21], further suggesting a potential link between RanBPM and FOXA1 in the regulation of steroid receptor-mediated gene expression.
Our oPOSSUM analyses identified five members of the Homeobox family of transcription factors as being over-represented in the promoters of our differentially expressed genes. Homeobox transcription factors play a pivotal role in the regulation of embryonic development [61], regulate homeostasis, cell differentiation, and organ function in adult tissues [62-64] and their expression is often deregulated in cancer [65-68]. One such example is HOXA5, which during development regulates organogenesis in lung, mammary, and tracheal tissues, and in adult tissues regulates mammary gland development and function [64,69]. HOXA5 is also believed to function as a tumor suppressor by transactivating p53 to promote p53-dependent and p53-independent apoptotic signaling [65]. Consequently, HOXA5 expression is decreased in tumors of the breast, colon, and lung, and this expression is believed to be regulated at least in part through epigenetic modifications of the HOXA5 gene in these tumors [65,68]. HOXA5 binding sites are the most highly overrepresented in our list of genes, as they are found in the promoters of 134 of 167 genes analyzed. The HMG protein family consists of a unique group of transcription factors that bind to the minor groove of DNA and regulate gene expression through modifications of the DNA structure and through interaction with other factors [70]. Analyses in oPOSSUM identified four members of the HMG family with overrepresented TFBS in our gene list including SRY (sex-determining region on Y-chromosome), and the SOX (SRY-related HMG Box) proteins SOX5, SOX9, and SOX17. HMG proteins are critical in cell lineage specification and cell maturation during development, and the SOX proteins in particular have been proposed to function in determining stem cell identity, fate, and maintenance in multiple tissues [70,71]. SOX proteins were reported to enhance the DNA-binding affinity of steroid hormone receptors such as AR, and this has implications in both development and cancer. Deregulated expression of SOX9 has been observed in prostate cancer, and is linked to prostate cancer progression [72,73]. The binding sites of SOX5, SOX9, and SOX17 are significantly over-represented in our list of genes affected by decreased RanBPM expression. Collectively, our analyses of the overrepresented TFBS in our list of differentially expressed genes identified several transcription factors which regulate key processes in development, and whose function is often deregulated in cancer. These findings further implicate a role for RanBPM in the regulation of pathways that govern the critical balance between development and tumorigenesis.
All of the candidate genes selected for validation by quantitative RT-qPCR confirmed the initial results obtained in the microarray analyses. All nine genes up-regulated in RanBPM shRNA cells have previously been reported to be overexpressed in various cancers and/or tumors. For instance, over-expression of RON has been linked to human cancers such as breast, prostate, colorectal, and ovarian carcinomas [35,55,74,75]. RON hyperactivity has been shown to lead to increased cell proliferation, motility, and transformation, and to the inhibition of apoptosis and anoikis [76]. Similarly, ELF3 overexpression has been detected in breast, prostate, colon, and cervical tumors, and is associated with cell transformation [77,78]. ELF3 is believed to promote tumorigenesis through transcriptional regulation of several known oncogenes, including TGFβ [78]. Abnormal expression of L1CAM has also been observed in various cancer types and linked to cell proliferation, migration, invasion, and metastasis of cancer cells [47]; and Rab27B [79], ALDH1A3 [80], MSLN1 [38], LAMB3 [81], PHD3 [82], and MFAP5 [83] levels have all been reported to be increased in various cancer types. Further, both TG2 and CHN1, which expression is strongly down-regulated in RanBPM shRNA cells, have been linked to tumorigenesis, and CHN1 has been proposed to function as a tumor suppressor [84,85]. Overall, these findings suggest that RanBPM functions to prevent aberrant gene expression that may lead to oncogenesis. This reinforces the notion that has previously been inferred in several studies, that RanBPM may function as a tumor suppressor [10,19,23].
Potential implications in epigenetic regulation
RT-qPCR analysis of target gene expression following re-expression of RanBPM in Hela and HCT116 RanBPM shRNA cells revealed two categories of genes. The first group comprises genes which responded to RanBPM re-expression, and consists of RON, ELF3, Rab27B, L1CAM, and ALDH1A3. For these genes, re-expression of RanBPM reversed the effect observed upon RanBPM down-regulation, at least partially. Analysis of the promoters of these genes using oPOSSUM did not reveal any common TFBS. While we cannot rule out a direct effect of RanBPM at these gene promoters, an alternate possibility is that RanBPM modulates signaling pathways that regulate the expression of these genes. The second group comprises genes which did not show a transcriptional response to restoration of RanBPM expression. This group consists of LAMB3, PHD3, TG2, CHN1, and MFAP5. Analysis of samples prepared 72h post-transfection showed identical results (data not shown). This suggests the possibility that RanBPM down-regulation establishes long-term changes in gene expression programs, such as epigenetic modifications, that cannot be reversed by transient re-expression of RanBPM. Interestingly several candidate genes and transcription factors identified by oPOSSUM analysis that are affected in response to down-regulation of RanBPM are known to be regulated through epigenetic modifications. For example, LAMB3 expression, which is up-regulated in gastric cancer cells, was shown to be regulated by demethylation of its promoter [86]. Down-regulation of TG2 expression is linked to several types of cancer [84], and has been shown to result from aberrant hypermethylation of the TG2 promoter in brain and breast tumors [87,88]. Additionally, expression of the HOXA5, SOX9, and SOX17 transcription factors is regulated through epigenetic mechanisms. As discussed above, HOXA5 promoters are hypermethylated in breast and lung cancers which results in silencing of HOXA5 expression, and may correlate with decreased p53 activation and decreased apoptosis in breast tumors [68]. SOX9 has been reported to be hypermethylated in mantle cell lymphoma (MCL), and this hypermethylation is associated with decreased SOX9 expression in these tumors [89]. Hypermethylation of SOX9 in MCL tumors also correlated with higher proliferation, increased chromosomal abnormalities, and reduced overall patient survival [89]. The promoter region of SOX17 is hypermethylated in mammary, gastric, and hepatocellular carcinomas, thereby silencing SOX17 and leading to aberrant activation of Wnt signaling [90,91]. These findings suggest that RanBPM may have broad effects on gene transcription, and may function both directly on gene promoters, and indirectly through modification of epigenetic programs, to regulate gene expression.
Overall, the results of this study indicate that alterations in the expression of RanBPM has profound and wide ranging effects on genes and pathways that play important roles in the regulation of developmental programs, and are linked to tumorigenesis when deregulated. RanBPM may therefore have a central role in controlling the activity of several signaling pathways that function to coordinate cell proliferation and differentiation during mammalian development and that are tightly regulated in adult tissues to maintain homeostatic regulations and prevent tumorigenesis.
Acknowledgements
We thank Michael Poulter for critical reading of the manuscript. This work was supported by operating grants (MOP-114958 and IC1-102946) from the Canadian Institutes for Health Research (CIHR) to CSP. EA was supported by a QEIIGSST and by funds generously donated by Mrs Marilynne Fuller.
Supporting Information
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.McIntyre RE, van der Weyden L, Adams DJ. Cancer gene discovery in the mouse. Current Opinion in Genetics & Development. 2012;22:14–20. doi: 10.1016/j.gde.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Mattison J, van der Weyden L, Hubbard T, Adams DJ. Cancer gene discovery in mouse and man. Biochim Biophys Acta. 2009;1796:140–161. doi: 10.1016/j.bbcan.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura M, Masuda H, Horii J, Kuma K, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishitani H, Hirose E, Uchimura Y, Nakamura M, Umeda M, Nishii K, Mori N, Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- 7.Scantlebury N, Zhao XL, Moncalvo VGR, Camiletti A, Zahanova S, Dineen A, Xin JH, Campos AR. The Drosophila Gene RanBPM Functions in the Mushroom Body to Regulate Larval Behavior. PLoS One. 2010;5:e10652. doi: 10.1371/journal.pone.0010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dansereau DA, Lasko P. RanBPM regulates cell shape, arrangement, and capacity of the female germline stem cell niche in Drosophila melanogaster. J Cell Biol. 2008;182:963–977. doi: 10.1083/jcb.200711046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valiyaveettil M, Bentley AA, Gursahaney P, Hussien R, Chakravarti R, Kureishy N, Prag S, Adams JC. Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. J Cell Biol. 2008;182:727–739. doi: 10.1083/jcb.200801133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem. 2004;279:13027–13034. doi: 10.1074/jbc.M313515200. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L, Lemmon S, Lemmon V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J Neurochem. 2005;94:1102–1110. doi: 10.1111/j.1471-4159.2005.03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo JA, Roh SE, Lakshmana MK, Kang DE. Pivotal role of RanBP9 in integrin-dependent focal adhesion signaling and assembly. Faseb Journal. 2012;26:1672–1681. doi: 10.1096/fj.11-194423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y, Paramasivam M, Girgenti MJ, Walikonis RS, Bianchi E, LoTurco JJ. RanBPM Regulates the Progression of Neuronal Precursors through M-Phase at the Surface of the Neocortical Ventricular Zone. Developmental Neurobiology. 2010;70:1–15. doi: 10.1002/dneu.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin YX, Sun ZP, Huang SH, Zhao L, Geng Z, Chen ZY. RanBPM contributes to TrkB signaling and regulates brain-derived neurotrophic factor-induced neuronal morphogenesis and survival. Journal of Neurochemistry. 2010;114:110–121. doi: 10.1111/j.1471-4159.2010.06745.x. [DOI] [PubMed] [Google Scholar]
- 15.Brunkhorst A, Karlen M, Shi J, Mikolajczyk M, Nelson MA, Metsis M, Hermanson O. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol Cell Neurosci. 2005;29:250–258. doi: 10.1016/j.mcn.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Li Z, Messing EM, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J Biol Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y, Fu C, Chen H, Wang X, Deng W, Huang BR. The Ran binding protein RanBPM interacts with TrkA receptor. Neurosci Lett. 2006;407:26–31. doi: 10.1016/j.neulet.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Fu CB, Cui YB, Xie YF, Yuan YH, Wang X, Chen H, Huang BR. The Ran-binding protein RanBPM can depress the NF-kappa B pathway by interacting with TRAF6. Molecular and Cellular Biochemistry. 2012;359:83–94. doi: 10.1007/s11010-011-1002-3. [DOI] [PubMed] [Google Scholar]
- 19.Kramer S, Ozaki T, Miyazaki K, Kato C, Hanamoto T, Nakagawara A. Protein stability and function of p73 are modulated by a physical interaction with RanBPM in mammalian cultured cells. Oncogene. 2005;24:938–944. doi: 10.1038/sj.onc.1208257. [DOI] [PubMed] [Google Scholar]
- 20.Chang LK, Liu ST, Kuo CW, Wang WH, Chuang JY, Bianchi E, Hong YR. Enhancement of transactivation activity of Rta of Epstein-Barr virus by RanBPM. J Mol Biol. 2008;379:231–242. doi: 10.1016/j.jmb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Rao MA, Cheng H, Quayle AN, Nishitani H, Nelson CC, Rennie PS. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J Biol Chem. 2002;277:48020–48027. doi: 10.1074/jbc.M209741200. [DOI] [PubMed] [Google Scholar]
- 22.Poirier MB, Laflamme L, Langlois MF. Identification and characterization of RanBPM, a novel coactivator of thyroid hormone receptors. J Mol Endocrinol. 2006;36:313–325. doi: 10.1677/jme.1.01891. [DOI] [PubMed] [Google Scholar]
- 23.Atabakhsh E, Bryce DM, Lefebvre KJ, Schild-Poulter C. RanBPM has proapoptotic activities that regulate cell death pathways in response to DNA damage. Mol Cancer Res. 2009;7:1962–1972. doi: 10.1158/1541-7786.MCR-09-0098. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif overrepresentation. Nucleic Acids Res. 2007;35:W245–252. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in coexpressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 30.Mikolajczyk M, Shi J, Vaillancourt RR, Sachs NA, Nelson M. The cyclin-dependent kinase 11 (p46) isoform interacts with RanBPM. Biochem Biophys Res Commun. 2003;310:14–18. doi: 10.1016/j.bbrc.2003.08.116. [DOI] [PubMed] [Google Scholar]
- 31.Bai D, Chen H, Huang BR. RanBPM is a novel binding protein for p75NTR. Biochem Biophys Res Commun. 2003;309:552–557. doi: 10.1016/j.bbrc.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Marion Schneider E, Li X, Duttenhofer I, Debatin K, Hug H. HIPK2 associates with RanBPM. Biochem Biophys Res Commun. 2002;297:148–153. doi: 10.1016/s0006-291x(02)02020-x. [DOI] [PubMed] [Google Scholar]
- 33.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, notch, and Hedgehog pathways. Nature Reviews Clinical Oncology. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 34.Lin GL, Hankenson KD. Integration of BMP, Wnt, and Notch Signaling Pathways in Osteoblast Differentiation. Journal of Cellular Biochemistry. 2011;112:3491–3501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagh PK, Gray JK, Zinser GM, Vasiliauskas J, James L, Monga SP, Waltz SE. beta-Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene. 2011;30:3694–3704. doi: 10.1038/onc.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sethi N, Kang Y. Notch signalling in cancer progression and bone metastasis. British Journal of Cancer. 2011;105:1805–1810. doi: 10.1038/bjc.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shih Ie M, Davidson B. Pathogenesis of ovarian cancer: clues from selected overexpressed genes. Future Oncol. 2009;5:1641–1657. doi: 10.2217/fon.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Puverel S, Barrick C, Dolci S, Coppola V, Tessarollo L. RanBPM is essential for mouse spermatogenesis and oogenesis. Development. 2011;138:2511–2521. doi: 10.1242/dev.062505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wehner M, Kunzelmann S, Herrmann C. The guanine cap of human guanylate-binding protein 1 is responsible for dimerization and self-activation of GTP hydrolysis. Febs Journal. 2012;279:203–210. doi: 10.1111/j.1742-4658.2011.08415.x. [DOI] [PubMed] [Google Scholar]
- 42.De Donato M, Mariani M, Petrella L, Martinelli E, Zannoni GF, Vellone V, Ferrandina G, Shahabi S, Scambia G, Ferlini C. Class III betatubulin and the cytoskeletal gateway for drug resistance in ovarian cancer. Journal of Cellular Physiology. 2012;227:1034–1041. doi: 10.1002/jcp.22813. [DOI] [PubMed] [Google Scholar]
- 43.Yu CJ, Chang KP, Chang YJ, Hsu CW, Liang Y, Yu JS, Chi LM, Chang YS, Wu CC. Identification of Guanylate-Binding Protein 1 as a Potential Oral Cancer Marker Involved in Cell Invasion Using Omics-Based Analysis. Journal of Proteome Research. 2011;10:3778–3788. doi: 10.1021/pr2004133. [DOI] [PubMed] [Google Scholar]
- 44.Khalfallah O, Rouleau M, Barbry P, Bardoni B, Lalli E. Dax-1 knockdown in mouse embryonic stem cells induces loss of pluripotency and multilineage differentiation. Stem Cells. 2009;27:1529–1537. doi: 10.1002/stem.78. [DOI] [PubMed] [Google Scholar]
- 45.Kinsey M, Smith R, Iyer AK, McCabe ER, Lessnick SL. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing's sarcoma. Cancer Res. 2009;69:9047–9055. doi: 10.1158/0008-5472.CAN-09-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gast D, Riedle S, Issa Y, Pfeifer M, Beckhove P, Sanderson MP, Arlt M, Moldenhauer G, Fogel M, Kruger A, Altevogt P. The cytoplasmic part of L1-CAM controls growth and gene expression in human tumors that is reversed by therapeutic antibodies. Oncogene. 2008;27:1281–1289. doi: 10.1038/sj.onc.1210747. [DOI] [PubMed] [Google Scholar]
- 47.Raveh S, Gavert N, Ben-Ze'ev A. L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 2009;282:137–145. doi: 10.1016/j.canlet.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 48.Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 50.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nature Reviews Molecular Cell Biology. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 51.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 52.Chandrasekar B, Patel DN, Mummidi S, Kim JW, Clark RA, Valente AJ. Interleukin-18 suppresses adiponectin expression in 3T3-L1 adipocytes via a novel signal transduction pathway involving ERK1/2-dependent NFATc4 phosphorylation. J Biol Chem. 2008;283:4200–4209. doi: 10.1074/jbc.M708142200. [DOI] [PubMed] [Google Scholar]
- 53.Silletti S, Yebra M, Perez B, Cirulli V, McMahon M, Montgomery AM. Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J Biol Chem. 2004;279:28880–28888. doi: 10.1074/jbc.M404075200. [DOI] [PubMed] [Google Scholar]
- 54.Camp ER, Yang A, Gray MJ, Fan F, Hamilton SR, Evans DB, Hooper AT, Pereira DS, Hicklin DJ, Ellis LM. Tyrosine kinase receptor RON in human pancreatic cancer: expression, function, and validation as a target. Cancer. 2007;109:1030–1039. doi: 10.1002/cncr.22490. [DOI] [PubMed] [Google Scholar]
- 55.Wagh PK, Peace BE, Waltz SE. Met-related receptor tyrosine kinase Ron in tumor growth and metastasis. Adv Cancer Res. 2008;100:1–33. doi: 10.1016/S0065-230X(08)00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danilkovitch-Miagkova A. Oncogenic signaling pathways activated by RON receptor tyrosine kinase. Curr Cancer Drug Targets. 2003;3:31–40. doi: 10.2174/1568009033333745. [DOI] [PubMed] [Google Scholar]
- 57.Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012;1819:707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. Embo Journal. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochimica Et Biophysica Acta-Molecular Cell Research. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptorregulated promoter. Mol Cell Biol. 2009;29:5413–5425. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barber BA, Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Annals of Anatomy-Anatomischer Anzeiger. 2010;192:261–274. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Cillo C, Barba P, Freschi G, Bucciarelli G, Magli MC, Boncinelli E. HOX gene expression in normal and neoplastic human kidney. Int J Cancer. 1992;51:892–897. doi: 10.1002/ijc.2910510610. [DOI] [PubMed] [Google Scholar]
- 63.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 64.Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]
- 65.Kelly ZL, Michael A, Butler-Manuel S, Pandha HS, Morgan RGL. HOX genes in ovarian cancer. Journal of Ovarian Research. 2011;4:16. doi: 10.1186/1757-2215-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang Y, Xia L, Du Z, Sheng L, Chen H, Chen G, Li Q. HOXA5 inhibits keratinocytes growth and epidermal formation in organotypic cultures in vitro and in vivo. J Dermatol Sci. 2012;66:197–206. doi: 10.1016/j.jdermsci.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 67.Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS. Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer. 2010;10:89. doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 69.Gendronneau G, Boucherat O, Aubin J, Lemieux M, Jeannotte L. The Loss of Hoxa5 Function Causes Estrous Acyclicity and Ovarian Epithelial Inclusion Cysts. Endocrinology. 2012;153:1484–1497. doi: 10.1210/en.2011-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pritchett J, Athwal V, Roberts N, Hanley NA, Hanley KP. Understanding the role of SOX9 in acquired diseases: lessons from development. Trends in Molecular Medicine. 2011;17:166–174. doi: 10.1016/j.molmed.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67:528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 74.Thobe MN, Gray JK, Gurusamy D, Paluch AM, Wagh PK, Pathrose P, Lentsch AB, Waltz SE. The Ron receptor promotes prostate tumor growth in the TRAMP mouse model. Oncogene. 2011;30:4990–4998. doi: 10.1038/onc.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu XM, Wang D, Shen Q, Chen YQ, Wang MH. RNA-mediated gene silencing of the RON receptor tyrosine kinase alters oncogenic phenotypes of human colorectal carcinoma cells. Oncogene. 2004;23:8464–8474. doi: 10.1038/sj.onc.1207907. [DOI] [PubMed] [Google Scholar]
- 76.Benvenuti S, Comoglio PM. The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol. 2007;213:316–325. doi: 10.1002/jcp.21183. [DOI] [PubMed] [Google Scholar]
- 77.Prescott JD, Poczobutt JM, Tentler JJ, Walker DM, Gutierrez-Hartmann A. Mapping of ESE-1 subdomains required to initiate mammary epithelial cell transformation via a cytoplasmic mechanism. Molecular Cancer. 2011;10:103. doi: 10.1186/1476-4598-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliver JR, Kushwah R, Hu J. Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Laboratory Investigation. 2012;92:320–330. doi: 10.1038/labinvest.2011.186. [DOI] [PubMed] [Google Scholar]
- 79.Hendrix A, Westbroek W, Bracke M, De Wever O. An Ex(o)citing Machinery for Invasive Tumor Growth. Cancer Research. 2010;70:9533–9537. doi: 10.1158/0008-5472.CAN-10-3248. [DOI] [PubMed] [Google Scholar]
- 80.Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Aldehyde dehydrogenase Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 81.Guess CM, Quaranta V. Defining the role of laminin-332 in carcinoma. Matrix Biol. 2009;28:445–455. doi: 10.1016/j.matbio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su Y, Loos M, Giese N, Hines OJ, Diebold I, Gorlach A, Metzen E, Pastorekova S, Friess H, Buchler P. PHD3 regulates differentiation, tumour growth and angiogenesis in pancreatic cancer. British Journal of Cancer. 2010;103:1571–1579. doi: 10.1038/sj.bjc.6605936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, Ozbun L, Samimi G, Brady J, Randonovich M, Pise-Masison CA, Barrett JC, Wong WH, Welch WR, Berkowitz RS, Birrer MJ. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mehta K, Kumar A, Kim HI. Transglutaminase 2: A multi-tasking protein in the complex circuitry of inflammation and cancer. Biochemical Pharmacology. 2010;80:1921–1929. doi: 10.1016/j.bcp.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 85.Yang C, Kazanietz MG. Chimaerins: GAPs that bridge diacylglycerol signalling and the small G-protein Rac. Biochem J. 2007;403:1–12. doi: 10.1042/BJ20061750. [DOI] [PubMed] [Google Scholar]
- 86.Kwon OH, Park JL, Kim M, Kim JH, Lee HC, Kim HJ, Noh SM, Song KS, Yoo HS, Paik SG, Kim SY, Kim YS. Aberrant up-regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochemical and Biophysical Research Communications. 2011;406:539–545. doi: 10.1016/j.bbrc.2011.02.082. [DOI] [PubMed] [Google Scholar]
- 87.Dyer LM, Schooler KP, Ai LB, Klop C, Qiu JX, Robertson KD, Brown KD. The transglutaminase 2 gene is aberrantly hypermethylated in glioma. Journal of Neuro-Oncology. 2011;101:429–440. doi: 10.1007/s11060-010-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ai L, Kim WJ, Demircan B, Dyer LM, Bray KJ, Skehan RR, Massoll NA, Brown KD. The transglutaminase 2 gene (TGM2), a potential molecular marker for chemotherapeutic drug sensitivity, is epigenetically silenced in breast cancer. Carcinogenesis. 2008;29:510–518. doi: 10.1093/carcin/bgm280. [DOI] [PubMed] [Google Scholar]
- 89.Enjuanes A, Fernandez V, Hernandez L, Navarro A, Bea S, Pinyol M, Lopez-Guillermo A, Rosenwald A, Ott G, Campo E, Jares P. Identification of Methylated Genes Associated with Aggressive Clinicopathological Features in Mantle Cell Lymphoma. PLoS One. 2011;6:e19736. doi: 10.1371/journal.pone.0019736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu DY, Wang ZM, Chen L, Wang BL, Shen ZZ, Huang W, Shao ZM. Sox17, the canonical Wnt antagonist, is epigenetically inactivated by promoter methylation in human breast cancer. Breast Cancer Research and Treatment. 2010;119:601–612. doi: 10.1007/s10549-009-0339-8. [DOI] [PubMed] [Google Scholar]
- 91.Jia Y, Yang YS, Liu SA, Herman JG, Lu FM, Guo MZ. SOX17 antagonizes WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010;5:743–749. doi: 10.4161/epi.5.8.13104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.