Abstract
Mutation in the BRCA1 gene is associated with increased risk for hereditary breast and ovarian cancers. In sporadic ovarian tumors, BRCA1 dysfunction is thought to be common. BRCA1 is a nuclear-cytoplasm shuttling protein. Our group has previously reported that BRCA1 proteins, unlike K109R and cancer-predisposing mutant C61G BRCA1 proteins, bind the sole SUMO E2-conjugating enzyme Ubc9. In this study, we examined the result of altered Ubc9 binding and knockdown on the sub-cellular localization and growth inhibitory function of BRCA1 proteins in ovarian cancer cells. Using live imaging of YFP, RFP-tagged BRCA1 and BRCA1a proteins, our results show enhanced cytoplasmic localization of K109R and C61G mutant BRCA1 proteins in ES-2, NIHOVCAR3 and UWB 1.289 ovarian cancer cells. Down-regulation of Ubc9 in ovarian cancer cells using Ubc9 siRNA resulted in cytoplasmic localization of BRCA1 and BRCA1a proteins. These mutant BRCA1a proteins were impaired in their capacity to inhibit growth of ES-2 ovarian cancer cells. Several ovarian cancer cells, including a BRCA1-null ovarian cancer cell line, showed higher levels of expression of Ubc9. This is the first study demonstrating the physiological link between loss of Ubc9 binding and loss of growth suppression of disease-associated mutant BRCA1a proteins in ovarian cancer cells. BRCA1, by turning off or on Ubc9 binding, regulates growth of ovarian cancers.
Keywords: BRCA1, BRCA1a, Ubc9, Ovarian cancer, RING domain mutants, nuclear import, Growth suppression
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecologic cancer, and up to 10% are caused by germ line mutations in BRCA1 [1,2]. In sporadic EOC, BRCA1 mutations are rare, but reduced expression or aberrant subcellular localization of BRCA1 is common [3-5]. Sporadic EOC display BRCA1 dysfunction and have close similarities to BRCA1 germline mutation-related disease in terms of survival and sensitivity to platinum drugs, which strongly suggests a common underlying mechanism of therapies, making traditional chemotherapy the only modality of treatment. Currently, PARP inhibitors are being used in clinical trials, but their effect is limited to some BRCA1 mutated EOC, but not sporadic ovarian cancers [6]. Therefore, there is a critical need for better targeted therapies for ovarian cancers. We have identified two major splice variants of BRCA1, namely BRCA1a/p110 and BRCA1b/p100 [7,8], which are expressed at reduced levels in ovarian tumors compared to normal cells [9-12]. We found BRCA1a protein to induce apoptosis and inhibit in vivo tumor growth of hormone-independent ES-2 ovarian cancer cells, but the mechanism of tumor suppression is not yet known [13,14]. BRCA1 and its isoforms are nuclear proteins that have several functional domains, an N-terminal RING finger domain that interacts with several proteins and two BRCA1 C-terminal domains. We have found BRCA1, BRCA1a and BRCA1b proteins to be localized in the mitochondria, and their nuclear-cytoplasmic shuttling to be a regulated process [7,12,15]. BRCA1 nuclear transport is regulated by the action of nuclear localization signal (NLS) and nuclear export signals (NES) located in the RING domain that mediates nuclear export via association with BARD1 [16]. The BRCA1 delta 11 isoform, which lacks NLS, also enters the nucleus via the RING-domain mediated BARD1 import pathway [17]. The RING domain of BRCA1, in complex with BARD1, mediates an E3 Ubiquitin ligase activity on ER-α in-vitro [18,19]. Recent findings using an Ubiquitin ligase-deficient BRCA1 I26A mutant suggested that the Ubiquitin ligase activity is dispensable for both genomic stability as well as homology-directed repair of double-strand DNA breaks, but is required for repression of ER-α activity [20,21].
Post-translational modifications of proteins play an important role in regulating gene expression [22]. SUMO (Small Ubiquitin-like modifier) modification of proteins affect several functions like stability, localization, protein-protein interactions and transcriptional regulation [23-25]. The SUMO modification pathway was shown to be involved in BRCA1 response to DNA damage and transcriptional repression [26,27]. We have shown that the amino-terminal domain of BRCA1, BRCA1a and BRCA1b proteins bind to SUMO-E2-conjugating enzyme Ubc9 and regulate ER-α activity by promoting its degradation in vivo [28]. This work suggested that there is a relationship between the SUMO and Ubiquitin pathways, similar to the Ubiquitin ligase RNF4, by highlighting the biochemical function of BRCA1 as a putative SUMO-1 and Ubc9-dependent E3 Ubiquitin ligase for ER-α SUMO conjugates [29,30]. Ubc9 binding site mutations, as well as cancer-predisposing mutation in the BRCA1 RING domain (C61G), disrupted the ability to modulate Ubc9-mediated estrogen-induced ER-α transcriptional activity in breast cancer cells [28] but did not disrupt SUMO-1 binding [26] nor auto ubiquitination activity of BRCA1 [28]. Both BRCA1/BRCA1a K109R and disease associated C61G mutants, which are localized mainly in the cytoplasm, fail to inhibit the growth of breast cancer cells [31]. Ubc9 has been shown to play an important role in both cancer progression and resistance to chemotherapy [32-36]. In fact, Ubc9 was found to act as both a positive and negative regulator of proliferation and transformation of HMGA1 proteins [37]. Here, we have further investigated these findings and have studied the effect of altered Ubc9 binding and knock-down on the subcellular localization and growth inhibitory function of BRCA1 proteins in ovarian cancer cells.
Materials and methods
Expression constructs
Full length BRCA1a, BRCA1a Mut #1, BRCA1a Mut #4, BRCA1 NLS-m and Ubc9 were cloned 3’ of the RFP or YFP [12] or pCDNA3 [28] vectors as described previously [28]. Point mutations were generated as described previously [28].
Cell culture
ES-2, NIHOVCAR3, MCF10A, UWB1.289 and SKOV3 cells were obtained from American Type Culture Collection (Rockville, MD, USA) and cultivated as described previously [14,38].
Western blot analysis
The extracts from ES-2, NIHOVCAR3, MCF10A, SKOV3 and UWB1.289 cells were separated on 4-20% SDS-PAGE gradient gel and processed for western blotting using Ubc9 antibodies as previously described [31].
Antibodies and reagents
The antibodies used in this study were MS110 ascites (Ab1, EMD Chemicals), Ubc9 N-15, beta-Actin C4, antibodies (Santa Cruz Biotechnology).
Immunoflourescence microscopy and live imaging
To analyze the subcellular localization ES-2, NIHOVCAR-3 and UWB1.289 cells were seeded into 6-well plates a day before transfection with RFP or YFP tagged BRCA1, BRCA1a and their respective mutant plasmids. The DNA was stained with Hoechst dye and, after 24h, the cells were visualized under a fluorescent microscope (Olympus, 20X lens). Similar experiments were performed in the presence of control or Ubc9 siRNA as described previously [31].
Growth suppression studies
The growth suppression assay using ES-2 cells was done as previously described [14].
siRNA transfection
NIHOVCAR-3 and ES-2 cells were seeded into culture dishes. Ubc9 SMART pool siRNA (50nM) or 50nM control RISC-free siRNA (Dharmacon, Lafayette, Co, USA) was diluted in serum free medium. HiPerFect transfection reagent (Qiagen) was added to the diluted siRNA and mixed by vortexing. It was allowed to sit at room temperature for 5-10 minutes then added drop wise to the cells and incubated in normal growth medium for 24 hours. The cells were then visualized under fluorescence microscope (Olympus, 4X lens).
Results and discussions
Mutant BRCA1 proteins are localized in the cytoplasm of ovarian cancer cells
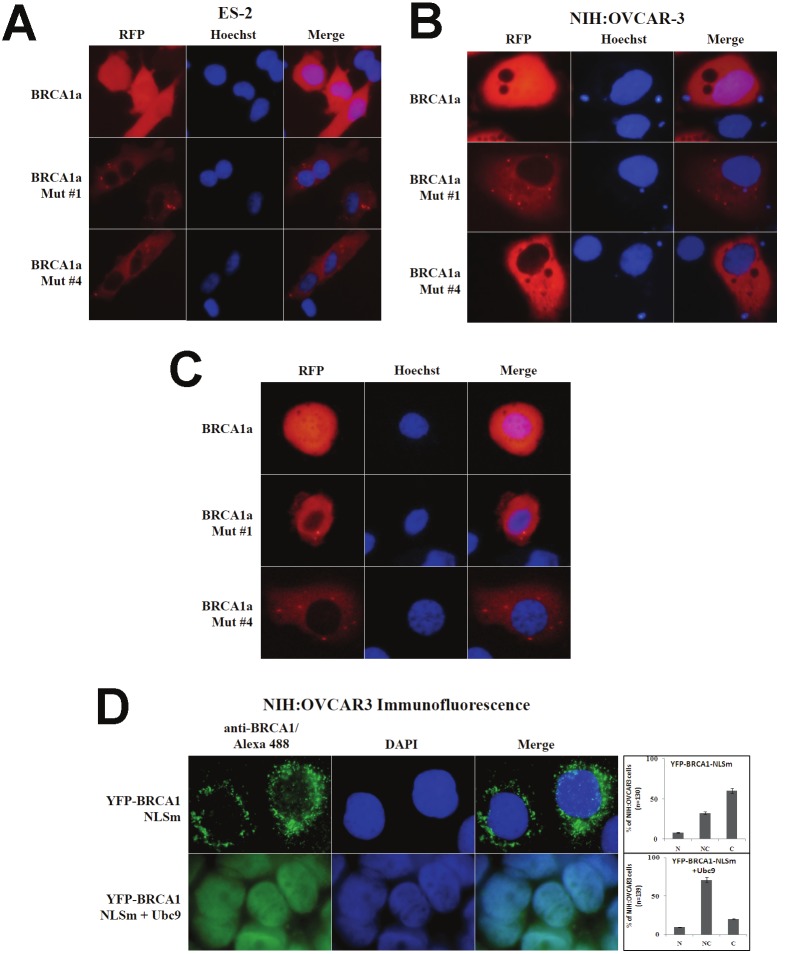
BRCA1 is a nuclear-cytoplasm shuttling protein, and many cancer-associated mutations have altered subcellular localization of BRCA1 protein [39,40]. Regulation of BRCA1 nuclear import requires the combined action of NLS [17] and interaction with BARD1, a RING domain binding protein, which masks the NES [17]. BRCA1a and BRCA1b proteins, both of which lack the two NLS, are still imported to the nucleus, suggesting an alternate mechanism for nuclear import. Due to our previous results showing the lack of binding of BRCA1/ BRCA1a Mut #1 (K 109 to R) and disease associated Mut #4 C61G to Ubc9, we wanted to study the effect of these mutations on the subcellular localization of BRCA1 and BRCA1a proteins in ovarian cancer cells. We have examined the sub-cellular localization using RFP-epitope tagged BRCA1a, BRCA1a Mut #1 and BRCA1a Mut #4 after transient transfection into ES-2 (Figure 1A) and NIHOVCAR3 ovarian cancer cells (Figure 1B) and a BRCA1 mutant cell line UWB1.289 (Figure 1C). UVB1.289 is a BRCA1-null ovarian cancer cell line obtained from a papillary serous tumor [38]. This cell line carries a germline BRCA1 mutation within exon 11 and has a deletion of wild-type allele. We observed exclusive cytoplasmic localization of both BRCA1a Mut #1 and Mut #4 unlike BRCA1a which was both in the nucleus as well as cytoplasm of ES-2, NIHOVCAR3 and UWB1.289 cells (Figure 1). These results suggest that Ubc9 may be required for nuclear import of BRCA1a proteins that lack NLS. Previous work from Dr. Henderson’s group suggested that both NLS and RING domain sequences are necessary for nuclear localization [17] of BRCA1 proteins. To demonstrate that the BRCA1 NH2-terminal domain is required for Ubc9-mediated nuclear import, we tested Ubc9 for its effect on nuclear localization of BRCA1-NLS mutants. In transfected NIHOVCAR3 cells, approximately 60% of BRCA1-NLS mutant was found in the cytoplasm (Figure 1D). Co-transfection with Ubc9 enhanced nuclear localization of BRCA1-NLS mutant and 20% was found in the cytoplasm (Figure 1D). These results indicate that Ubc9 similar to BARD1 functions as a new chaperone promoting the nuclear entry of BRCA1 and its splice variants.
Figure 1.
Ubc9 is required for nuclear localization of BRCA1 and BRCA1a proteins in ovarian cancer cells. The subcellular localization of BRCA1, BRCA1a and its mutants in various ovarian cancer cells by immunoflourescence and live imaging. A, ES-2 cells. B and D. NIHOVCAR3 cells and C. UWB1.289 cells. ES-2 and NIHOVCAR3 cells were seeded into 6-well plates and transfected with pRFP-BRCA1a or pRFP-BRCA1a Mut#1 or pRFP-BRCA1a Mut#4 or YFP-BRCA1 (Yellow fluorescent protein) NLSm with and without Ubc9 using Lipofectamine 2000 (Invitrogen) or X-tremeGENE 9 DNA transfection reagent (Roche). The nuclei were visualized with DNA staining dye Hoechst 24 hours after transfection. The live images of cells were taken by fluorescent microscope (20X) (Olympus). For immunoflourescence twenty four hours later, the cells were fixed in methanol and probed with BRCA1 antibody (Santa Cruz, Ab1 1/100) followed by Alexa Fluor 488 labeled secondary antibody (Invitrogen, 1/100) staining as described [31]. The nuclei were visualized by 4, 6-diamidino-2-phenylindole (DAPI) staining. The images were taken using fluorescent microscope (100X, oil)) (Olympus). Graphs show the proportion of transfected NIH: OVCAR3 cells showing predominantly nuclear (N), nuclear/cytoplasmic (NC), cytoplasmic (C) BRCA1 NLS (Nuclear Localization Signal) mutant. Bars represent the mean ± SE of at least two experiments.
Ubc9 knockdown results in cytoplasmic localization of endogenous and exogenous BRCA1 proteins in ovarian cancer cells
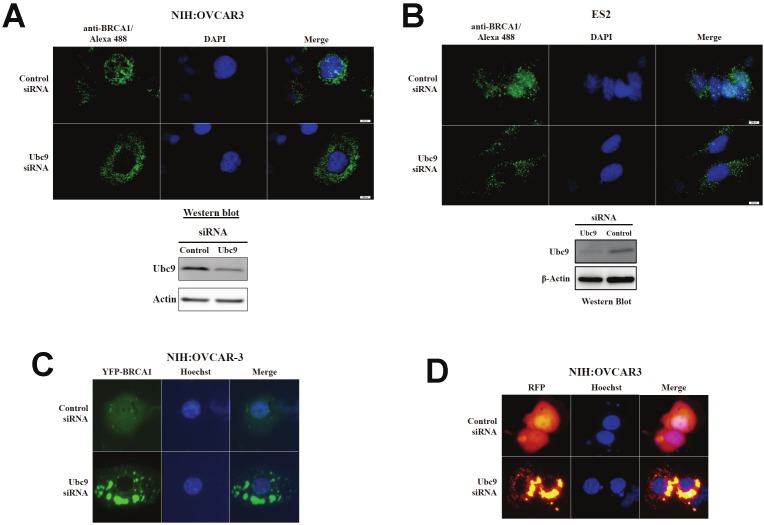
If Ubc9 is needed for nuclear localization of BRCA1 and BRCA1a proteins, then knockdown of Ubc9 in ovarian cancer cells should result in cytoplasmic localization of endogenous BRCA1 proteins. We observed cytoplasmic localization of full length RFP BRCA1a, YFP-BRCA1 and endogenous BRCA1 proteins in ES-2 and NIHOVCAR3 cells that has been treated with siRNA to Ubc9 by live imaging and immunoflourescence analysis using BRCA1 antibody (Figure 2A, B, C, D). These results confirm the requirement of Ubc9 in the nuclear localization of BRCA1 and BRCA1a proteins. These results demonstrated a role for Ubc9 in the nuclear import of BRCA1 proteins in ovarian cancer cells.
Figure 2.
Effect of Ubc9 knockdown on subcellular localization of endogenous and YFP and RFP tagged BRCA1 and BRCA1a proteins in ovarian cancer cells. Immunoflorescence (A) and (B) or live imaging using BRCA1 Ab1 antibody on ES-2 and NIHOVCAR3 cells transfected with control or Ubc9 siRNA and YFP-BRCA1 (C) RFP-BRCA1a (D). The nuclei were visualized with DAPI or blue fluorescent Hoechst dye staining using a fluorescent microscope (20X or 100X). Western blot analysis of Ubc9 expression in siRNA treated ES-2 and NIHOVCAR3 cells. The control and Ubc9 siRNA treated ES-2 and NIHOVCAR3 cells were lysed and probed with Ubc9 antibody. β-Actin was used as an internal control. The stoichiometry of Ubc9 protein levels is shown. The signal of Ubc9 protein band was quantified using software MultiGauge and the values were used to plot efficiency of Ubc9 knockdown in ES-2 and NIHOVCAR3 cells.
Ubc9 binding BRCA1a mutants are impaired in growth inhibition of ovarian cancer cells
BRCA1/1a proteins inhibit the growth of human breast and ovarian cancer cells [14,41-45]. By subjecting BRCA1a, their corresponding Mut#1, and cancer-predisposing Mut#4 C61G to colony suppression assays using ES-2, a hormone-independent ovarian cancer cell, we were able to study the consequence of altered Ubc9 binding on the growth suppressor function of BRCA1a proteins in ovarian cancer cells [14]. BRCA1a Mut#1 and BRCA1a Mut#4 lost their capacity to suppress growth of ES-2 ovarian cancer cells (Figure 3). These results are consistent with the notion that a direct association of BRCA1a proteins with Ubc9 is critical for inhibiting the growth of ovarian cancer cells.
Figure 3.
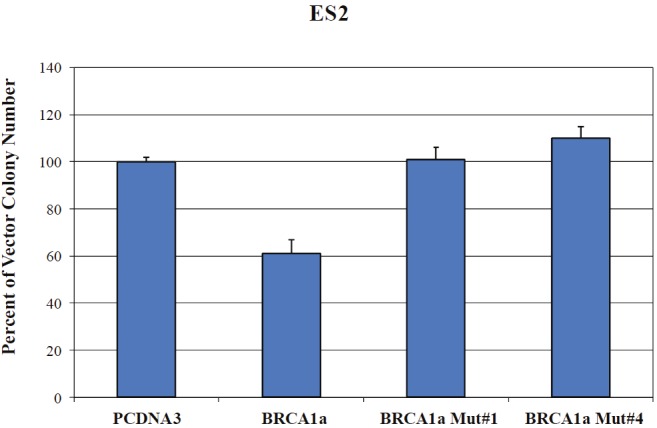
Altered Ubc9 binding results in loss of growth suppression by BRCA1a in ovarian cancer cells. ES-2 cells were transfected with pCDNA3 or pCDNA3-BRCA1a or pCDNA3-BRCA1a Mut#1 or pCDNA3-BRCA1a Mut#4 and selected with G418 and the colonies were stained with crystal violet and counted as described [14]. The number of colonies obtained by pCDNA3 was considered as 100%. Each experiment was repeated 3 times and the bars shown represent s.d.
Deregulated levels of Ubc9 in ovarian cancer cell lines
Ubc9 is the sole E2 enzyme essential for protein sumoylation and is a multifunctional protein, which is over-expressed in several cancers like colon, prostate, breast, lung, ovarian and melanomas [32-36]. Ubc9 functions as a co-activator of several nuclear receptors [46,47]. Ubc9 has also been shown to bind to HMGA1 proteins and integrate both positive and negative signals for proliferation and transformation [37]. Recently, Ubc9 was shown to promote cell invasion and metastasis of breast cancer cells [48] implicating a role in tumorigenesis. Ubc9 levels have been shown to correlate with drug resistance in some ovarian cancer cells [35]. Since BRCA1 mutant proteins do not bind Ubc9 and are transforming in breast cancer cells [38], we wanted to test if the inability to bind Ubc9 could result in aberrant Ubc9 expression as observed previously in tumors. We studied Ubc9 expression in MCF10A, a normal mammary epithelial and several other ovarian cancer cells. We observed over-expression of Ubc9 in ES-2, UWB1.289 and SKOV3 ovarian cancer cells compared to normal mammary epithelial cells (Figure 4A). This is similar to our results obtained using breast cancer cell lines [31] and previous report that documented higher levels in several cancers compared with their normal tissue counterparts [36]. These results are consistent with the model that a direct interaction of BRCA1 with Ubc9 is critical for growth/tumor suppression by BRCA1 proteins and lack of binding results in deregulated Ubc9 levels causing cancer (Figure 4B).
Figure 4.
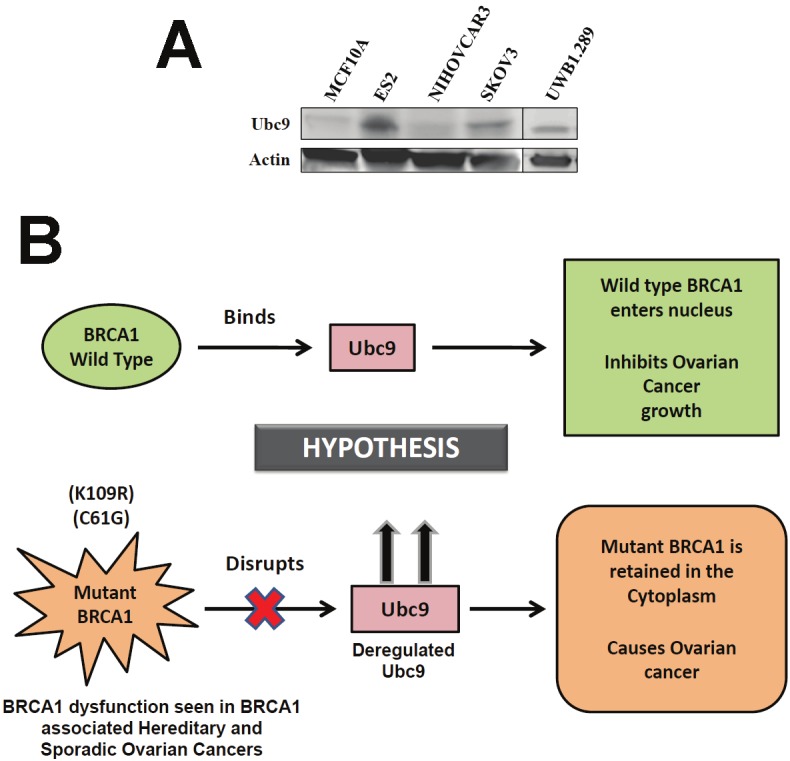
Ubc9 is expressed at high levels in several ovarian cancer cells lines and UWB1.289 ovarian cancer cells and MCF10A normal mammary epithelial cells. Expression of Ubc9 in ES-2, NIHOVCAR3, and SKOV3 cells by western blot analysis. Protein concentrations were normalized using β-actin (A). Working hypothetical model showing how BRCA1/1a, by binding to Ubc9 inhibits the growth of ovarian cancer cells. Mutant BRCA1/1a is unable to interact with Ubc9 causing deregulated levels of UBC9 resulting in ovarian cancer (B).
Discussion
Investigating the functional significance of loss of nuclear BRCA1 localization in women with ovarian cancer is critical to understanding how BRCA1 dysfunction results in ovarian cancer. BRCA1 is a tumor suppressor that undergoes active nuclear import and export, which can provide a regulatory function [49]. Knowing that somatic mutations in BRCA1 are rarely found in sporadic ovarian cancers, studying the nuclear-cytoplasm shuttling of BRCA1 may offer an important mechanism for regulating its function. The mistargeting of tumor suppressors like BRCA1 can have severe consequences on the normal function of these proteins. Nuclear transport of proteins can be altered either by mutations or through post translational modifications (including phosphorylation, ubiquitylation, glycosylation and sumoylation), which could prevent the binding of proteins like Ubc9 resulting in non-nuclear distribution of BRCA1 proteins.
Here we are demonstrating for the first time that both BRCA1/BRCA1a K109R and disease associated C61G mutants, which fail to bind Ubc9, lack E3 Ubiquitin ligase activity [28] and growth inhibitory function are localized in the cytoplasm of ovarian cancer cells. Knockdown of endogenous Ubc9 using siRNA resulted in enhanced cytoplasmic retention of BRCA1 and BRCA1a proteins. Co-expression of Ubc9 induced nuclear accumulation of NLS-deficient forms of BRCA1 proteins. These results suggest the existence of a new import pathway involving Ubc9, which transports BRCA1 proteins to the nucleus via a piggyback mechanism [38] similar to BARD1 [17] BRCA1 is known to enter the nucleus by binding to importin alpha, which subsequently binds importin beta as well as through interaction with BARD1 [49]. Ubc9 does not bind importin alpha nor beta proteins [50]. Recently, importin 13 was identified as a novel importin beta-related transport receptor that mediates nuclear import of Ubc9 via RanGTPase system [50]. We can propose the existence of a new nuclear import pathway for BRCA1 proteins involving the Ubc9: importin 13 and BRCA1a protein may use only this mechanism for nuclear transport [38]. We have found elevated levels of expression of Ubc9 in BRCA1 mutant ovarian cancer cell line UWB1.289 as well as several ovarian cancer cells compared to normal mammary epithelial cells (Figure 4A). These findings agree with others regarding over expression of Ubc9 in ovarian carcinoma and breast cancer cells where it promotes invasion and metastasis [48].
These results suggest a molecular interplay between BRCA1 and Ubc9 which maintains the balance of two opposing effects: tumor suppression or tumorigenesis. BRCA1 deficiency can tilt this balance resulting in ovarian cancers (Figure 4B). BRCA1 thus functions as a master switch which by turning off or on Ubc9 binding regulates cell growth. Post translational modification of BRCA1 proteins could prevent the binding of BRCA1 to Ubc9 resulting in ovarian cancers. We can speculate that the nuclear-cytoplasm shuttling of BRCA1 protein may provide an important mechanism for regulating its tumor suppression function in sporadic ovarian cancers where somatic mutations in BRCA1 are rarely found. Future efforts will be directed towards developing targeted drugs that can mimic the function of BRCA1 proteins to combat BRCA1-associated ovarian cancers.
Acknowledgements
We are grateful to Dr. Henderson (West Mead Institute for Cancer Research, Australia), Professor Hay and Dr. Jaffrey (University of Dundee, UK) for generously providing BRCA1 NLS-m, YFP-BRCA1, Ubc9 constructs. We thank all the members of Drs. Rao and Reddy labs for their help. We thank Mr. Abramson, research media services, Ms Wimes and Mr. Hill for editorial assistance and RCMI core facilities at Morehouse School of Medicine, for their assistance. This work was supported in part by Georgia Cancer Coalition Distinguished Cancer Scholar award, NIH-NCRR-RCMI grant G-12-RR003034, U54 RR02613, 5P20RR11104 and NIHMD research endowment grant 2S21MD000101 and U54 CA118638 to V.N.R. Georgia Cancer Coalition Distinguished Cancer Scholar award to E.S.P.R. V.N.R’s lab was also supported in part by funds from the ING foundation.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, Mcclure M, Frye C, Hattier T, PHELPS R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, LAI M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Berchuck A, Heron KA, Carney ME, Lancaster JM, Fraser EG, Vinson VL, Deffenbaugh AM, Miron A, Marks JR, Futreal PA, Frank TS. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998;4:2433–2437. [PubMed] [Google Scholar]
- 3.Merajver SD, Pham TM, Caduff RF, Chen M, Poy EL, Cooney KA, Weber BL, Collins FS, Johnston C, Frank TS. Somatic mutations in the BRCA1 gene in sporadic ovarian tumours. Nat Genet. 1995;9:439–443. doi: 10.1038/ng0495-439. [DOI] [PubMed] [Google Scholar]
- 4.Clark-Knowles KV, O'Brien AM, Weberpals JI. BRCA1 as a Therapeutic Target in Sporadic Epithelial Ovarian Cancer. J Oncol. 2010;2010:891059. doi: 10.1155/2010/891059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguewa PA, Fuertes MA, Cepeda V, Alonso C, Quevedo C, Soto M, Perez JM. Poly(ADP-ribose) polymerase-1 inhibitor 3-aminobenzamide enhances apoptosis induction by platinum complexes in cisplatin-resistant tumor cells. Med Chem. 2006;2:47–53. doi: 10.2174/157340606775197697. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Shao N, Ding QM, Cui J, Reddy ES, Rao VN. BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases. Oncogene. 1997;15:143–157. doi: 10.1038/sj.onc.1201252. [DOI] [PubMed] [Google Scholar]
- 8.Chai Y, Chipitsyna G, Cui J, Liao B, Liu S, Aysola K, Yezdani M, Reddy ES, Rao VN. c-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b-mediated growth suppression in breast cancer cells. Oncogene. 2001;20:1357–1367. doi: 10.1038/sj.onc.1204256. [DOI] [PubMed] [Google Scholar]
- 9.Wilson CA, Payton MN, Elliott GS, Buaas FW, Cajulis EE, Grosshans D, Ramos L, Reese DM, Slamon DJ, Calzone FJ. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Conzen SD, Cole CN, Arrick BA. Characterization of functional messenger RNA splice variants of BRCA1 expressed in nonmalignant and tumor-derived breast cells. Cancer Res. 1996;56:4578–4581. [PubMed] [Google Scholar]
- 11.Orban TI, Olah E. Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem Biophys Res Commun. 2001;280:32–38. doi: 10.1006/bbrc.2000.4068. [DOI] [PubMed] [Google Scholar]
- 12.Maniccia AW, Lewis C, Begum N, Xu J, Cui J, Chipitsyna G, Aysola K, Reddy V, Bhat G, Fujimura Y, Henderson B, Reddy ES, Rao VN. Mitochondrial localization, ELK-1 transcriptional regulation and growth inhibitory functions of BRCA1, BRCA1a, and BRCA1b proteins. J Cell Physiol. 2009;219:634–641. doi: 10.1002/jcp.21708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao N, Chai YL, Shyam E, Reddy P, Rao VN. Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene. 1996;13:1–7. [PubMed] [Google Scholar]
- 14.Yuli C, Shao N, Rao R, Aysola P, Reddy V, Oprea-llies G, Lee L, Okoli J, Partridge E, Reddy ES, Rao VN. BRCA1a has antitumor activity in TN breast, ovarian and prostate cancers. Oncogene. 2007;26:6031–6037. doi: 10.1038/sj.onc.1210420. [DOI] [PubMed] [Google Scholar]
- 15.Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, Shaw MK, Van Oostveldt PM, Vaux DJ. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell. 2005;16:997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen EM, Fan S, Ma Y. BRCA1 regulation of transcription. Cancer Lett. 2006;236:175–185. doi: 10.1016/j.canlet.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 17.Fabbro M, Rodriguez JA, Baer R, Henderson BR. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- 18.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 19.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci USA. 2007;104:5794–5799. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci USA. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, Pestell RG, Tomita YA, Rosen EM. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Kim KI, Baek SH, Chung CH. Versatile protein tag, SUMO: its enzymology and biological function. J Cell Physiol. 2002;191:257–268. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- 24.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 25.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 26.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 27.Park MA, Seok YJ, Jeong G, Lee JS. SUMO1 negatively regulates BRCA1-mediated transcription, via modulation of promoter occupancy. Nucleic Acids Res. 2008;36:263–283. doi: 10.1093/nar/gkm969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Watkins T, Reddy A, Reddy ES, Rao VN. A novel mechanism whereby BRCA1/1a/1b fine tunes the dynamic complex interplay between SUMO-dependent/independent activities of Ubc9 on E2-induced ERalpha activation/repression and degradation in breast cancer cells. Int J Oncol. 2009;34:939–949. doi: 10.3892/ijo_00000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 31.Qin Y, Xu J, Aysola K, Begum N, Reddy V, Chai Y, Grizzle WE, Partridge EE, Reddy ES, Rao VN. Ubc9 mediates nuclear localization and growth suppression of BRCA1 and BRCA1a proteins. J Cell Physiol. 2011;226:3355–3367. doi: 10.1002/jcp.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronen O, Malone JP, Kay P, Bivens C, Hall K, Paruchuri LP, Mo YY, Robbins KT, Ran S. Expression of a novel marker, Ubc9, in squamous cell carcinoma of the head and neck. Head Neck. 2009;31:845–855. doi: 10.1002/hed.21048. [DOI] [PubMed] [Google Scholar]
- 33.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, Brauch H, Baisch C, Gilbert M, Harth V, Spickenheuer A, Rabstein S, Pesch B, Bruning T, Ko YD, Hamann U. Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int J Cancer. 2009;125:596–602. doi: 10.1002/ijc.24286. [DOI] [PubMed] [Google Scholar]
- 34.Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD Jr, Annunziata CM, Munshi NC. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115:2827–2834. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo YY, Yu Y, Ee PL, Beck WT. Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res. 2004;64:2793–2798. doi: 10.1158/0008-5472.can-03-2410. [DOI] [PubMed] [Google Scholar]
- 36.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, Kuan SF, Fayewicz SL, Galambos C, Acquafondata M, Dhir R, Becker D. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Lu J, Prochownik EV. Dual role for SUMO E2 conjugase Ubc9 in modulating the transforming and growth-promoting properties of the HMGA1b architectural transcription factor. J Biol Chem. 2007;282:13363–13371. doi: 10.1074/jbc.M610919200. [DOI] [PubMed] [Google Scholar]
- 38.DelloRusso C, Welcsh PL, Wang W, Garcia RL, King MC, Swisher EM. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res. 2007;5:35–45. doi: 10.1158/1541-7786.MCR-06-0234. [DOI] [PubMed] [Google Scholar]
- 39.Bogdani M, Teugels E, De Greve J, Bourgain C, Neyns B, Pipeleers-Marichal M. Loss of nuclear BRCA1 localization in breast carcinoma is age dependent. Virchows Arch. 2002;440:274–279. doi: 10.1007/s004280100526. [DOI] [PubMed] [Google Scholar]
- 40.Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008;39:857–865. doi: 10.1016/j.humpath.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, Jensen RA. Growth retardation and tumour inhibition by BRCA1. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 42.Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET. BRCA1-associated growth arrest is RB-dependent. Proc Natl Acad Sci USA. 1999;96:11866–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tait DL, Obermiller PS, Holt JT. Preclinical studies of a new generation retroviral vector for ovarian cancer BRCA1 gene therapy. Gynecol Oncol. 2000;79:471–476. doi: 10.1006/gyno.2000.5969. [DOI] [PubMed] [Google Scholar]
- 44.Randrianarison V, Marot D, Foray N, Cabannes J, Meret V, Connault E, Vitrat N, Opolon P, Perricaudet M, Feunteun J. BRCA1 carries tumor suppressor activity distinct from that of p53 and p21. Cancer Gene Ther. 2001;8:759–770. doi: 10.1038/sj.cgt.7700366. [DOI] [PubMed] [Google Scholar]
- 45.Marot D, Opolon P, Brailly-Tabard S, Elie N, Randrianarison V, Connault E, Foray N, Feunteun J, Perricaudet M. The tumor suppressor activity induced by adenovirus-mediated BRCA1 overexpression is not restricted to breast cancers. Gene Ther. 2006;13:235–244. doi: 10.1038/sj.gt.3302637. [DOI] [PubMed] [Google Scholar]
- 46.Chang YL, Huang CJ, Chan JY, Liu PY, Chang HP, Huang SM. Regulation of nuclear receptor and coactivator functions by the carboxyl terminus of ubiquitin-conjugating enzyme 9. Int J Biochem Cell Biol. 2007;39:1035–1046. doi: 10.1016/j.biocel.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi S, Shibata H, Yokota K, Suda N, Murai A, Kurihara I, Saito I, Saruta T. FHL2, UBC9, and PIAS1 are novel estrogen receptor alpha-interacting proteins. Endocr Res. 2004;30:617–621. doi: 10.1081/erc-200043789. [DOI] [PubMed] [Google Scholar]
- 48.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson ME. BRCA1 16 years later: nuclear import and export processes. FEBS J. 2010;277:3072–3078. doi: 10.1111/j.1742-4658.2010.07733.x. [DOI] [PubMed] [Google Scholar]
- 50.Mingot JM, Kostka S, Kraft R, Hartmann E, Gorlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]