Abstract
RUNX1 is a transcription factor that regulates critical processes in many aspects of hematopoiesis. RUNX1 is also integral in defining the definitive hematopoietic stem cell. In addition, many hematological diseases like myelodysplastic syndrome and myeloproliferative neoplasms have been associated with mutations in RUNX1. Located on chromosomal 21, the RUNX1 gene is involved in many forms of chromosomal translocations in leukemia. t(8;21) is one of the most common chromosomal translocations found in acute myeloid leukemia (AML), where it results in a fusion protein between RUNX1 and ETO. The RUNX1-ETO fusion protein is found in approximately 12% of all AML patients. In this review, we detail the structural features, functions, and models used to study both RUNX1 and RUNX1-ETO in hematopoiesis over the past two decades.
Keywords: RUNX1, AML1, CBFA2, RUNX1-ETO, AML1-ETO, RUNX1-RUNX1T1, Hematopoiesis, Hematopoietic Stem Cells, Leukemia, AML, Leukemogenesis, Mouse Models, Review
2. INTRODUCTION
RUNX1, also known as AML1, CBFalpha2, and PEBP2alphaB, belongs to the family of Runt-related transcription factors (RUNXs) (1). The Runt protein is encoded by the Drosophila runt gene, which is required for normal segmentation, sex determination, and neurogenesis during Drosophila embryogenesis (2–4). Other RUNX family members include RUNX2 and RUNX3. This family of proteins was first described as a component of Moloney murine leukemia virus enhancer core binding factor (CBF) and Polyomavirus enhancer binding protein 2 (PEBP2) (5–7). RUNX1 is also known as acute myeloid leukemia 1 due to the discovery of its gene sequence from human patient with acute myeloid leukemia (8). Over the past 20 years, studies have elucidated many important functions of RUNX1 in hematopoietic development, hematopoietic stem cell homeostasis, and various blood malignancies. In this review, we will focus on the role that RUNX1 plays in these various biological processes. In addition, we will discuss RUNX1-ETO, a fusion protein resulting from a translocation between chromosomes 8 and 21. The t(8;21)(q22;q22) translocation is one of the most common chromosomal translocations found in patients with AML and especially in those with the French-American-British M2 subtype of AML (9–11). Although RUNX1-ETO cannot by itself induce leukemia in mouse models, the fusion protein provides a critical hit toward leukemogenesis (12;13). The major roles that RUNX1 and RUNX1-ETO play in hematopoiesis and leukemogenesis, respectively, make them highly interesting subjects for further research.
3. RUNX1 STRUCTURE AND REGULATION
3.1. RUNX1 promoters (proximal and distal) and RUNX1 isoforms
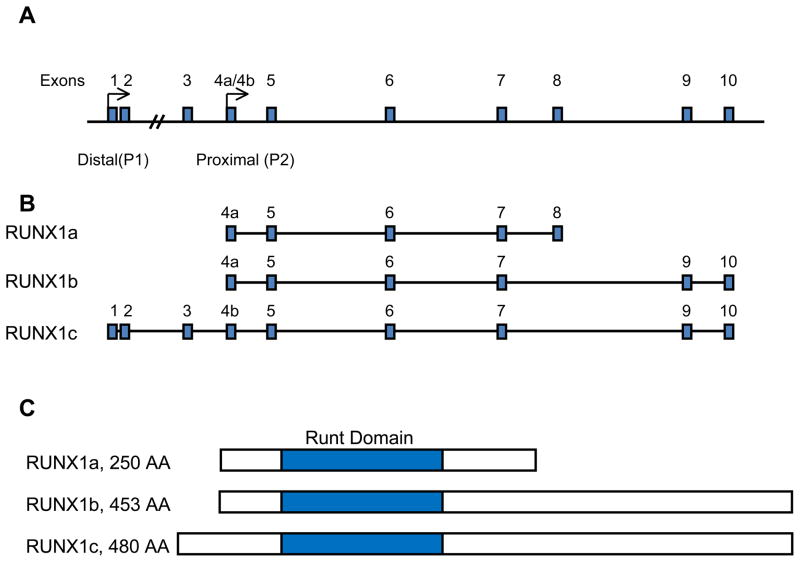
RUNX1 was first cloned from DNA obtained from an AML patient with t(8;21)-positive leukemia (8). Although there may be at least 12 different RUNX1 mRNA isoforms, three main protein isoforms of RUNX1 are primarily discussed (14). These are known as RUNX1a, RUNX1b, and RUNX1c (Figure 1a). These three major isoforms all contain the Runt domain located in the N-terminal region. RUNX1a, consisting of 250 amino acids, and RUNX1b, consisting of 453 amino acids, share the same N-terminal region and are the result of alternative splicing (15). RUNX1a lacks the transcriptional regulatory domains present in the C-terminal region common in the other two RUNX1 isoforms (16). RUNX1c, consisting of 480 amino acids, is the longest of the RUNX1 isoforms and its transcript is transcribed from a distal promoter in the RUNX1 locus, while RUNX1a and RUNX1b are transcribed from the proximal promoter (Figure 1b) (15). RUNX1b and RUNX1c have the same C-terminal region.
Figure 1.
RUNX1 isoforms and genomic locus. (A) The RUNX1 genomic locus on chromosome 21 is shown with the location of the proximal and distal promotors and exons based on the National Center for Biotechnology Information Nucleotide database. (B) The three main transcriptional isoform of RUNX1 are shown. RUNX1a is consists of exons 4a through 8. RUNX1b consists of exons 4a through 10, but excludes exon 8. RUNX1c includes exons 1 through 3 and exons 4b through 10, but also excludes exon 8. (C) The three main RUNX1 isoforms are shown with the Runt homology domain shaded.
Interestingly, the various RUNX1 isoforms play specific roles in specifying the hematopoietic stem cell (HSC) and regulating embryonic hematopoiesis. A study done by Tsuzuki et al. demonstrated that the RUNX1a isoform is found relatively more abundantly in the CD34+ progenitor population in human cord blood and that over-expression of RUNX1a compared with RUNX1b in mouse bone marrow progenitor cells can potentiate engraftment ability upon competitive transplantation (17). Hence, manipulating the levels of RUNX1a may be used to drive proliferation of human bone marrow cells for use in transplantation therapy. Another study, however, showed that over-expression of RUNX1a may also lead to the development of leukemia in a mouse transplantation model (18). These studies suggest that RUNX1a, because it includes the Runt domain but lacks the C-terminal regulatory domains, may act as a dominant-negative regulator of the other RUNX1 isoforms (16;18). As discussed in subsequent sections, the C-terminal domains are necessary for normal RUNX1 function and over-expression of a RUNX1 isoform lacking these domains may lead to abnormal hematopoiesis.
The ability of the RUNX1a isoform to direct a program of self-renewal reflects its importance in embryonic development. Early work using oligonucleotide PCR primers specific for either the proximal or distal transcriptional forms of RUNX1 in T cells showed that the distal form is more prevalent in developing T cells (19). However, when the proximal form, which in this case is RUNX1b, was retrovirally over-expressed in the 32Dcl.3 myeloid progenitor line, significantly more proliferation and neutrophil differentiation was observed when compared to over-expression of the distal RUNX1c isoform (19). In zebrafish, where the transcriptional regulation of RUNX1 using two promoters is conserved, transgenic lines that express fluorescently labeled RUNX1 isoforms specific for each of the two promoters show that the distal isoform is expressed in areas where erythromyeloid progenitors arise while the proximal isoform originates where definitive HSCs develop (20). More recent studies using mouse knock-in models to label the expression patterns of the RUNX1 distal and proximal promoters have also shown that the proximal promoter may be important for the initial development of definitive hematopoietic cells from hemogenic endothelium while the distal promoter is active in more mature progenitors (21). Furthermore, mice hypomorphic for the proximal promoter make it term but die perinatally, while mice null for the distal promoter show no overt phenotype (22;23). The aforementioned studies indicate that the two RUNX1 promoters may have varying yet overlapping functions and interestingly, the proximal RUNX1 isoform has a more involved role in defining the HSC.
The two promoters of RUNX1 rely on a cis-regulatory element located approximately 23.5 kilobases downstream of the transcriptional start site of the distal promoter (24;25). This promoter contains sites for various essential hematopoietic transcription factors like Gata2, Ets family members, and Lmo2 (24). Furthermore, this element can drive specific expression of genetic markers like lacZ or green fluorescent protein (GFP) in HSCs and the hemogenic endothelium in transgenic mice (25;26). More studies regarding the expression patterns, transcriptional control, and dosage levels of the various RUNX1 isoforms in HSCs and during embryonic hematopoiesis will be needed to further elucidate their ability to regulate HSCs and to potentiate leukemia development.
3.2. RUNX1 protein structure, domains and functions
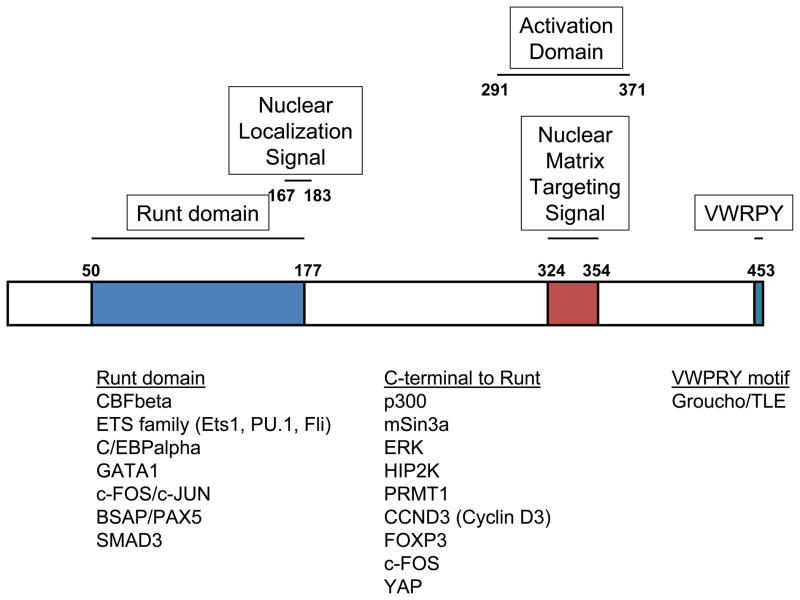
RUNX1 is defined by its 128 amino acid Runt domain found in the N-terminal region (Figure 2). The Runt domain mediates binding to DNA and interaction with its heterodimerization partner, core-binding factor beta (CBFbeta), which itself does not bind to DNA. Heterodimerization of RUNX1 and CBFbeta increases the DNA-binding affinity of RUNX1 (27;28). The complex consisting of RUNX1, CBFbeta, and DNA was one of the first gene regulatory complexes where detailed structural investigations have been conducted. NMR spectroscopy studies of the RUNX1/CBFbeta/DNA complex have revealed that it resembles an immunoglobulin fold similar to the DNA-binding domains of NF-kappaB, NFAT1, p53, and the STAT proteins (29;30). Crystal structure studies of this complex have demonstrated that CBFbeta interacts with RUNX1 allosterically to stabilize its ability to bind DNA and that diseases associated with mutations in RUNX1 correspond to sites in its DNA-binding domain (31–33).
Figure 2.
RUNX1 protein domains and interaction partners. The RUNX1b isoform is shown with the major domains listed above and interaction partners listed below.
The other protein domains of RUNX1 also help to regulate its ability to control transcription of its target genes. Various deletion studies of full-length RUNX1 have shown that the N-terminal and C-terminal regions directly adjacent to the Runt domain inhibit DNA binding (34;35). Binding to CBFbeta relieves this inhibition and allows RUNX1 to bind to DNA at its full potential (34). Furthermore, RUNX1 contains a nuclear matrix targeting signal (NMTS), a 31 amino acid region in the C-terminal region, which aids in transcriptional activation (36). At the very C-terminal end of RUNX1 is a VWRPY motif that is conserved among all Runt family members (37). This motif mediates the Groucho/TLE-dependent transcriptional repressor activities of RUNX1 (37;38). The NMTS and the VWRPY motif have roles in mediating T cell development. By itself, the C-terminal VWPRY motif is not required for developing thymocytes to properly repress CD4 expression (39). When the region containing both the nuclear matrix targeting signal and the VWRPY motif are deleted, however, the thymocytes can no longer repress CD4 (39).
In addition to interacting with CBFbeta, RUNX1 is very versatile in interacting with various other transcription factors and transcriptional co-regulators. For example, RUNX1 and Ets1 interact to coordinate transcriptional activity of the T cell receptor via the Runt domain of RUNX1, including regions just adjacent to the domain (35;40;41). RUNX1 has also been shown to interact with PU.1, C/EBPalpha, p300, mSin3a, GATA1, and Fli1 among many other factors that will not be further discussed in this review (42–47). These studies indicate that RUNX1 function relies heavily on its interaction partners and that these interaction partners may help to regulate its target genes in a tissue-specific manner.
3.3. Post-translational modifications of RUNX1
Aside from being transcriptionally regulated, RUNX1 is regulated by various post-translational mechanisms. RUNX1 was first reported to be phosphorylated by Extracellular Signal-Regulated Kinase (ERK) and phosphorylated forms of RUNX1 have been detected in CD34+ hematopoietic progenitor cells (48;49). Serines 249 and 266 of RUNX1b are ERK-associated phosphorylation sites and these sites are important in regulating the transactivation ability of RUNX1 (48). Further studies have shown that serine 273 and threonine 276 are also phosphorylated by the ERK pathway (50). Serines 249 and 266 are additionally phosphorylated by cyclin-dependent kinases (CDKs), which aids in regulating RUNX1 degradation by the anaphase-promoting complex (APC) (51). In addition, phosphorylation of RUNX1 by CDKs affects not only stability of RUNX1, but the DNA-binding affinity of RUNX1 as demonstrated by mutating three serine residues to aspartic acid which led to decreased DNA-binding affinity (52).
The effect of phosphorylation of RUNX1 is further revealed by studies looking into some of its interaction partners. For example, homeodomain-interacting protein kinase-2 (HIPK2) phosphorylates RUNX1 and subsequently triggers phosphorylation of p300, a transcriptional co-activator that interacts with RUNX1, which may explain how RUNX1 activates transcription of its target genes (53). Furthermore, CBFbeta has been shown to help recruitment of HIPK2 to phosphorylate RUNX1 (54). Phosphorylation of RUNX1 reduces its interaction with histone deacetylases and the transcriptional co-repressor mSin3A (45;55). Hence, phosphorylation of RUNX1 complements its ability to activate transcription by reducing its interactions with transcriptional repressors.
The ubiquitin-proteasome system has been well characterized and is known to regulate transcription (56). RUNX1 is a target of this system as evidenced by its enhanced stability in cells that have been treated with proteasome-specific inhibitors and when the lysine residues of RUNX1 are mutated to arginines (57). Ubiquitin-mediated degradation of RUNX1 is regulated both by heterodimerzation with CBFbeta, which enhances its stability, and by CDK phosphorylation, which promotes degradation by the APC (51;57).
Another post-translational modification that was recently described is methylation of RUNX1. PRMT1, an arginine methyltransferase, was shown to methylate RUNX1 at an area just C-terminal to the Runt domain (58). Methylation of RUNX1 inhibited its interaction with the co-repressor SIN3A which enhanced its transcriptional activity on known target genes like CD41 and PU.1 (58).
As evidenced by the aforementioned studies, RUNX1 is a protein that is heavily regulated by post-translational measures. However, the majority of these studies utilized in vitro analyses to demonstrate these post-translational effects. A more recent study involved the use of mice carrying knock-in alleles of Runx1 that had either both serine 249/serine 266 or both serine 249/serine 276 mutated to alanines, resulting in elimination of these sites for phosphorylation (59). Despite previous studies showing the importance of these serines for RUNX1 regulation, these mice exhibited no overt hematopoietic phenotype (59). Although this study serves as an example of how in vitro analyses may not necessarily predict in vivo functions, the role of phosphorylation and other post-translational modifications of RUNX1 may still be important during specific stages of hematopoiesis. Furthermore, other potential sites of phosphorylation that have yet to be examined may play more essential roles or allow for functional redundancy with serines 249, 266, and/or 276. Additional investigation will be needed to elucidate where these sites are located and whether these sites participate in RUNX1 regulation.
4. RUNX1 AS A MASTER REGULATOR OF HEMATOPOIESIS
4.1. The role of RUNX1 in the specification and development of the definitive hematopoietic stem cell using mouse models
RUNX1 plays an essential role in specifying the definitive hematopoietic stem cell (HSC). Two waves of hematopoiesis occur during embryonic development. The first wave is known as primitive hematopoiesis, which describes the differentiation of primitive macrophages and early erythrocytes from progenitors in the yolk sac to aid in the rapid development of the embryo (60). After this initial wave, definitive hematopoiesis, which describes the process of generating the various lineages of mature blood cell types from a common definitive HSC, takes place. One of the first sites that HSCs are detected in mammals is the aorta-gonad-mesonephros region at 10.5 days post conception (dpc) in the mouse embryo, where they bud off from the ventral aspect of the dorsal aorta and eventually colonize the fetal liver (61;62). Runx1 was detected in both locations during embryogenesis, indicating that expression of Runx1 marks the earliest hematopoietic precursor cells (63). One of the most important pieces of evidence implicating the role of RUNX1 in specifying the HSC was the generation of Runx1-null mice (64;65). While heterozygous mice are healthy and fertile, the homozygous knockout mice die between 12.5 to 13.5 dpc with severe hemorrhaging along the central nervous system. Such extensive hemorrhage is most likely due to defects in angiogenesis caused by a lack of angiopoietin-1 expression in these knockout mice (66). Furthermore, although these mice have nucleated primitive erythrocytes, indicating that there was no major defect in primitive hematopoiesis, they lacked definitive hematopoiesis. Cells from embryonic hematopoietic tissues, such as the yolk sac and liver, do not show colony forming units when cultured in vitro, and chimeric mice made from Runx1-deficient embryonic stem (ES) cells and wild type mouse blastocysts do not show any Runx1-deficient ES cell contribution to adult hematopoietic cells. More recent studies have further established the essential role that RUNX1 plays in derivation of HSCs from the hemogenic endothelium (67;68). Interestingly, however, once the HSC is defined, RUNX1 is no longer essential for hematopoiesis (68). These results indicate that RUNX1 plays a fundamental role in the establishment of definitive HSCs.
Although the role of RUNX1 in programming the HSC from embryonic development is well established from the investigations discussed above, one remaining question is whether RUNX1 is important in adult HSC function. Sun et al. described how mice that were haploinsufficient for Runx1 displayed a higher number of HSCs as defined by the cell surface marker phenotype, lineage− sca1+ckit+ (LSK), but contrastingly has lower number of functional long-term HSCs as assayed by limiting dilution analyses (69). To delve into this issue further, several groups have utilized a conditional Runx1 knockout model with the Mx1-Cre transgenic mouse line (70–72). The Mx1 gene is an interferon-inducible gene and its promoter allows expression of Cre to occur in HSCs when exposed to interferon (IFN) or other IFN-inducing agents like the synthetic double-stranded RNA polyinosinic/polycytidylic acid (polyIC) (73). The most striking phenotypes of conditional Runx1 knockout mice are a significant expansion of the putative HSC population (LSK cells) and myeloid progenitors, thrombocytopenia, and lymphopenia (70;71). Analysis of the spleen and thymus revealed a myeloproliferative phenotype (71;72). Further study of the stem cell compartment in these mice also revealed an expansion not only in LSK cells but in the long-term HSCs as indicated by additional labeling with CD34−Flt3− (74;75). Although Runx1-deficient cells showed enhanced proliferative ability, they are functionally impaired in their ability to engraft upon competitive transplantation with wild type hematopoietic cells into an irradiated host (71;75). Jacob at el. go further and state that lack of Runx1 causes stem cell exhaustion and use a retroviral insertional mutagenesis screen to identify Evi5 as a gene that can ameliorate this exhaustion phenotype when over-expressed. Moreover, they attribute the phenotype to a defect in the interaction between HSCs and the niche. Cxcr4, a gene important in stem cell homing and niche interactions, is a direct target of Runx1. This transcript is down-regulated in the absence of Runx1 but levels are rescued when Evi5 is over-expressed (75). Hence, absence of Runx1 may disrupt the HSC-niche interaction leading to aberrant hematopoiesis characterized as a stem cell exhaustion phenotype observed in the Runx1 knockout mice (75).
Although Jacob et al. provide one possible mechanism for the contrast in phenotype between Runx1 knockout embryos and adults, more studies will need to be conducted to describe exactly what roles and what target genes RUNX1 may additionally regulate in HSCs and hematopoiesis. Is RUNX1 only needed for specification but not maintenance of the HSC? Is RUNX1 required only to define the epigenetic environment conducive for HSC identity but is no longer needed once other hematopoietic-specific transcription factors are in place (76)? More investigation will be required to thoroughly answer these questions and further explain how RUNX1 functions as a master regulator of hematopoiesis.
4.2. RUNX1 target genes as they relate to hematopoiesis
In addition to the mouse models described earlier, numerous studies have focused on RUNX1 as a DNA-binding transcription factor and on the genes that it regulates. The Runt domain of RUNX1 mediates binding to the TGT/cGGT consensus sequence (77). In various adult blood types, target genes of RUNX1 have been fairly well characterized. In the myeloid lineage, RUNX1 directly binds and regulates the promoter activities of genes related to myeloid growth factor signaling such as IL-3, GM-CSF, the M-CSF receptor, and c-Mpl (78–81), and to the function of myeloid cells such as myeloperoxidase, neutrophil elastase, and mast cell protease 6 (82;83). In the T cell lineage, RUNX1 targets promoters and enhancers of T cell receptors and the CD11a promoter (84–86). In the B cell lineage, RUNX1 targets a B cell specific src family tyrosine kinase known as blk, Igα promoters, and the immunoglobulin antigen receptor enhancers (87–89).
Although the role of RUNX1 in regulating these genes are well described in adult blood cells, more recent studies have focused on RUNX1 target genes that are important in the regulation of hematopoiesis at the stages of stem cells and early progenitors. For example, RUNX1 has been demonstrated to play an important role in the differentiation and function of regulatory T cells by targeting and interacting with FoxP3 (90;91). RUNX1 also has an essential role in regulating PU.1, considered to be a critical transcription factor in myeloid progenitors and other mature myeloid cells (92). In this study, an upstream regulatory element located 14 kilobases upstream relative to the Pu.1 locus was found to have binding sites for Runx1, and mice harboring mutations in these binding sites exhibit a phenotype similar to the Runx1 conditional knockout phenotype described earlier (92). In addition, RUNX1 regulates miR-27a, which is involved in a feedback loop by binding to sites on the 3′ UTR of RUNX1, thereby mediating megakaryopoiesis (93). Since RUNX1 encompasses such a large role in hematopoiesis, more work is needed on the identification of its target genes, especially as they relate to HSCs or early progenitors.
4.3. Mutations in RUNX1 lead to aberrant hematopoiesis
The importance of RUNX1 in hematopoiesis is further exemplified by the mutations found in RUNX1 in patients with various hematological diseases. In one study, eight out of 160 patients with AML were found to have various mutations in the RUNX1 gene (94). Interestingly, these mutations were located in the Runt domain and further molecular analyses indicated that some of these mutations resulted in abnormal DNA binding and altered transactivation of the M-CSF promoter, a gene known to be regulated by RUNX1 (94).
In another study, six pedigrees of patients with familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML) were analyzed and found to be linked to a region of chromosome 21 encoding RUNX1 and concluded that haploinsufficiency of RUNX1 may be one cause of FPD/AML (95). While haploinsufficiency of RUNX1 may predispose patients to FPD/AML, biallelic mutations in RUNX1 resulting from a second hit may lead to full on leukemia (96).
Cases of myelodysplastic syndrome (MDS) were found to have mutations in RUNX1 (97). These mutations were found to be associated with the Runt domain of RUNX1 and the authors suggest that they result in dominant-negative forms of RUNX1 (97). Although additional studies looking into FPD/AML patients with mutations in RUNX1 have proposed a similar dominant-negative mechanism towards progression to disease, other case studies have found mutations in the C-terminal portion of RUNX1, which may suggest another mechanism toward disease progression (98–100).
Patients with myeloproliferative neoplasms (MPN) were also found to have point mutations in RUNX1 (101). Additional analyses of one of these mutated forms being transduced into human CD34+ MPN cells demonstrated that it promoted proliferation, which as the authors suggested, may allow for leukemic transformation in patients with MPN (101). Another study showed that patients with MPN have higher levels of RUNX1 mRNA transcript which up-regulate the target gene NF-E2, a gene that regulates erythropoiesis (102).
More recently patients of chronic myelomonocytic leukemia (CMML) have been found to harbor mutations in RUNX1 (103;104). Although most mutations were in the Runt homology domain, some patients had mutations in the C-terminal region after the Runt domain. Interestingly these patients progressed to full blown AML much faster when compared to patients without any mutations (104). The prevalence of RUNX1 mutations in the various hematological diseases described above highlights the important role that RUNX1 plays in normal hematopoiesis and what may happen when normal RUNX1 function goes awry.
5. RUNX1 IS A COMMONLY FOUND CONSTITUENT IN CHROMOSOMAL TRANSLOCATIONS ASSOCIATED WITH CANCER
As discussed in the previous section, RUNX1 is commonly found to be mutated in diseases associated with disrupted hematopoiesis. Many of the patients suffering from these diseases are one step away from developing full blown leukemia. Hence it is not surprising that RUNX1 is the most common target of chromosomal translocations found in acute leukemia. The three most common chromosomal translocations involving RUNX1 are: t(8;21), t(12;21), and t(3;21) (105). RUNX1-ETO (also known as AML1-ETO, RUNX1-MTG8, and RUNX1-RUNX1T1) is a result from t(8;21) and is found in about 12% of AML and 40% of the M2 subtype of AML (11). TEL-RUNX1 is a result of t(12;21) and was originally cloned from two patients with pediatric precursor B-cell acute lymphoblastic leukemia (ALL) (106). The translocation is present in about 25% of patients with childhood pre-B cell ALL and produces a fusion with the N-terminal HLH domain of the TEL protein and almost the entire RUNX1 protein, including its Runt and transactivation domains (106;107). The third most common translocation involving RUNX1 is t(3;21), which was first discovered in patients in the blast crisis phase of chronic myelogenous leukemia and later in approximately 3% of therapy-related MDS and AML (108;109). Cloning of fusion transcripts from patient tissue samples revealed that the N-terminal portion of RUNX1, including its Runt domain, was fused with either one of three genes on chromosome three including EVI, MDS1, or EAP (110). Less common chromosomal translocations involving RUNX1 have also been described in patients with a variety of other leukemias and hematological neoplasms (111–113). The prevalence of RUNX1 in these diseases obviously substantiates its role in normal blood development. Moreover, the variety of leukemias found in these patients, which have shown to include those of both myeloid and lymphoid origin, suggests that RUNX1 acts relatively upstream in the hematopoietic lineage tree to regulate the activity of HSCs.
6. RUNX1-ETO STRUCTURE AND REGULATION
6.1. RUNX1-ETO protein domains and interactions
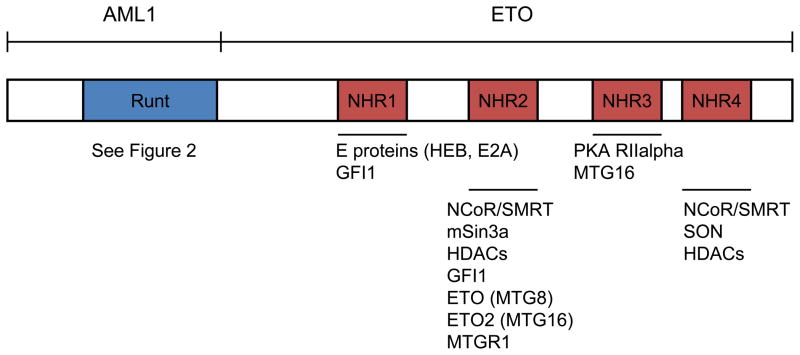
As mentioned earlier, t(8;21) is one of the most prevalent chromosomal translocations in AML, and will be the primary topic for the remainder of this review. The translocation places together the RUNX1 locus on chromosome 21 and ETO locus on chromosome 8. Both the fusion transcript and the ETO gene were first cloned in the 1990s (114;115). The breakpoints occur in intron 5 of the RUNX1 locus and in either intron 1a or 1b of the ETO locus (116–118). The RUNX1-ETO protein consists of 752 amino acids involving almost the entire ETO protein and the N-terminal portion of the RUNX1 protein containing the Runt homology domain (Figure 3) (115).
Figure 3.
RUNX1-ETO protein domains and interaction partners. The full length RUNX1-ETO is shown with the major domains listed above and interaction partners listed below.
As discussed earlier, RUNX1 has various roles in the hematopoietic system, but the Runt domain in particular functions to bind DNA and interact with other transcription factors. ETO, on the other hand, has mostly been discussed in the context of the RUNX1-ETO fusion protein in hematopoiesis. The ETO-RUNX1 transcript is not detected in t(8;21) patients, suggesting that ETO gene expression is relatively low in hematopoietic tissues (119). Mice with Eto knocked out displayed no hematopoietic abnormalities and was not found to be expressed in the hematopoietic compartment (120). Mice homozygous for the deletion die postnatally due to improper development of the gut, where it is found to be highly expressed (120). The high expression of ETO in the brain also suggests that it may play an important role in the central nervous system (121;122).
The ETO locus consists of 13 exons and spans over 87 kilobases (123). ETO was thought to be a putative transcription factor when it was first cloned based on the fact that in contained two zinc chelating domains (124). However, a screen using ETO protein revealed that it was not able to bind specific DNA sequences and hence does not seem to have any DNA-binding ability (125). ETO is mainly characterized by its four Nervy homology regions (NHRs), which are numbered one through four and share homology with the Drosophila Nervy protein (126). In Drosophila, nervy is expressed primarily in central nervous system regions and plays a role in regulating axon growth (126;127). Briefly, NHR1 has sequence similarities with TAF110 and other TAF proteins (124). NHR2 can mediate both alpha-helical tetramer formation, and homo- and hetero-oligomerization of ETO and its other family members, including ETO2 (MTG16) and MTGR1. NHR3 contains a coiled-coil motif, which shares structural homology with PKA anchoring proteins and binds to a PKA regulatory subunit (PKA RIIalpha) (128). A NHR3 point mutation that disrupts its interaction with PKA RIIalpha does not disrupt RUNX1-ETO’s ability to transform primary mouse bone marrow cells in vitro (129). NHR4 is also known as a myeloid-Nevry-DEAF1 (MYND) domain and has a zinc chelating motif (130).
These NHRs define the domains of ETO that mediate interactions with other proteins. For example, NHR1 mediates interactions with E proteins and inhibits the ability for E proteins to recruit co-activator molecules like p300/CBP (131). NHR2 can interact with co-repressors like NCoR/SMRT, mSin3a and HDACs (132–134). NHR4 also interacts with co-repressors NCoR/SMRT (133;135;136). Hence, earlier works on RUNX1-ETO have focused primarily on its function as a transcriptional repressor based on ETO’s interaction with these co-repressors (133;135;136). The results suggest that RUNX1-ETO binds to DNA using the Runt homology domain contained in its N-terminus to repress the transcription of RUNX1 target genes. Another recent study discovered SON as an RUNX1-ETO-interacting protein through the NHR4 domain. A point mutation in the NHR4 domain that disrupts RUNX1-ETO’s ability to interact with SON and NCoR/SMRT induces leukemia in a mouse transplantation model (137).
6.2. RUNX1-ETO splice isoforms and effects on leukemogenesis
The discussion of RUNX1-ETO’s various conserved domains and structure provides a natural transition to a discussion on its transcriptional isoforms. Several early reports revealed that multiple isoforms of RUNX1-ETO transcripts are detectable in t(8;21) AML patient samples (116;138). However, most RUNX1-ETO related studies have focused on the full length 752 amino acid form of RUNX1-ETO. Several models used to study the effect of RUNX1-ETO expression in mice have shown that without the presence of major additional mutations, RUNX1-ETO cannot induce leukemia on its own (12;139;140). Interestingly in one study, one mouse transplanted with RUNX1-ETO-transduced bone marrow cells was found to have developed AML rapidly (141). The leukemia cells in this mouse harbored a mutated RUNX1-ETO cDNA, which coded for a C-terminal truncated form of RUNX1-ETO that only had 552 amino acids (141). One extra exon, exon 9a, of ETO was previously reported (123). An RUNX1-ETO fusion transcript that included this exon 9a has been widely detected in t(8;21) AML. Such a transcript leads to the production of a C-terminal truncated RUNX1-ETO protein, termed RUNX1-ETO9a, which has 572 amino acids, and was almost identical to the leukemogenic 552 amino acid RUNX1-ETO protein just mentioned (142). Indeed, this RUNX1-ETO9a splice isoform rapidly induces leukemia in this mouse model (142).
The truncated version of RUNX1-ETO and RUNX1-ETO9a both lack the NHR3 and NHR4 domains. Notably, the NHR3 and NHR4 domains have been described to interact with transcriptional repressors like NCoR and SMRT (133;135;136). The ability of these truncated versions of RUNX1-ETO to rapidly induce leukemia have called into question how important these C-terminal NHR domains really are in this process. Deletion of the entire NHR4 domain or one amino acid point mutation that disrupts the zinc-chelating structure of RUNX1-ETO is sufficient to make RUNX1-ETO leukemogenic (137). This discovery demonstrates that the C-terminal portion of RUNX1-ETO does not participate in promoting leukemia. On the contrary, the C-terminal portion of full length RUNX1-ETO acts as an inhibitor to leukemogenesis. Therefore, any conditions or mutations that interfere with the function of NHR4 or its downstream signaling pathways may enhance t(8;21)-involved leukemia development. Currently, no NHR4 DNA sequence mutation has been identified in t(8;21)-positive AML (143). Further study on the biochemical function of this domain is important since it may provide valuable insights into the mechanisms of leukemogenic transformation.
6.3. Post translational modifications of RUNX1-ETO
Post translational modifications of RUNX1-ETO or ETO have not been as well described as those for RUNX1. Phosphorylated forms of ETO protein have been reported in human CD34+ hematopoietic cells (49). One study has looked into the role of the ubiquitin-proteasome pathway in the degradation of RUNX1-ETO, which demonstrated that RUNX1-ETO was ubiquitinylated and interacted with the E2-conjugase, UbcH8, and the E3-ligase, SIAH-1 (144). In addition to ubiquitin-mediated degradation, two non-classical aspartate residues (amino acids 188 and 368) of RUNX1-ETO are target sites of caspase-3 (145). They are responsible for caspase-3-mediated cleavage of RUNX1-ETO during apoptotic conditions (145).
Since RUNX1-ETO still retains the Runt homology domain, it is considered to share some of the same modifications as observed for RUNX1 and lack of the regulatory elements on the C-terminal portion of RUNX1 may contribute to RUNX1-ETO’s characteristics. For example, the arginine residues that are present just C-terminal to the Runt domain in RUNX1 are absent in the fusion protein. The lack of this particular region has been suggested to play a role in why RUNX1-ETO may have a stronger interaction with the co-repressor SIN3A than RUNX1 and therefore acts as a dominant-negative inhibitor of RUNX1 (58). Overall, however, post translational modifications of RUNX1-ETO have not been as well characterized as that of RUNX1 and offers an intriguing opportunity for further investigation.
7. RUNX1-ETO AND INDUCTION OF LEUKEMIA
7.1. Effects on hematopoiesis through the use of animal models
With t(8;21) being so prevalent in AML, there have been numerous models used to study the effects of RUNX1-ETO on hematopoiesis (Table 1). For example in Drosophila, expression of RUNX1-ETO causes an expansion of blood cell lineage progenitors (146). This model was further used to elucidate calpainB, which is a calcium-dependent protease conserved in Drosophila, as a modulator of RUNX1-ETO function (146). In zebrafish, which have a similar blood system to mammals, inducible expression of RUNX1-ETO recapitulates some of the phenotypes seen in AML patients like disruption of normal hematopoiesis and accumulation of immature blasts (147). Some of the unique qualities of zebrafish like its relatively short generation time and the large numbers of offspring per mating have allowed this model to become a useful method to screen for chemical modulators of RUNX1-ETO activity (148). The majority of models, however, that have been used to study RUNX1-ETO function center around the mouse.
Table 1.
Various animal models used to study effect of RUNX1-ETO expression
| Species | Descritpion | Hematopoietic phenotype | References |
|---|---|---|---|
| Drosophila melanogaster | Expression of RUNX1-ETO in eye | n/a | (179) |
| Drosophila melanogaster | Expression of RUNX1-ETO in blood cells | Increased number of blood progenitors | (146;180) |
| Danio rerio | Inducible expression of RUNX1-ETO by Hsp70 promoter | Increased number of immature blasts | (147;148) |
| Mus musculus | RUNX1-ETO knockin at Runx1 locus | Embryonic lethality due to hemorrhaging and block in hematopoiesis | (149;150) |
| Mus musculus | Inducible transgenic expression of RUNX1-ETO | Normal hematopoiesis | (12) |
| Mus musculus | Inducible Cre/loxP-mediated translocation of Runx1-Eto by Nestin Cre | Normal hematopoiesis | (139) |
| Mus musculus | Transgenic expression of RUNX1-ETO under MRP8 promoter | Normal hematopoiesis, treatment with ENU causes leukemia | (13) |
| Mus musculus | Inducible Cre/loxP RUNX1-ETO knockin expression by Mx1 Cre | Normal hematopoiesis, treatment with ENU causes leukemia | (140) |
| Mus musculus | Bone marrow transduction/transplantation with RUNX1-ETO | Abnormal myelopoiesis, no leukemia | (153) |
| Mus musculus | Bone marrow transduction/transplantation of ICSBP-deficient cells with RUNX1-ETO | Myeloid leukemia | (154) |
| Mus musculus | Bone marrow transduction/transplantation with RUNX1-ETO and TEL-PDGFbetaR | Myeloid leukemia | (155) |
| Mus musculus | RUNX1-ETO knockin at Sca1 locus | Myeloproliferative disease | (152) |
| Mus musculus | Bone marrow transduction/transplantation with RUNX1-ETO and FLT3 length mutation | Myeloid leukemia | (156) |
| Mus musculus | Bone marrow transduction/transplantation of human CD34+ cells with RUNX1-ETO in NOD/SCID mice | Abnormal myelopoiesis, no leukemia | (158) |
| Mus musculus | Bone marrow transduction/transplantation with WT1-overexpressing cells transduced with RUNX1-ETO | Myeloid leukemia | (181) |
| Mus musculus | Bone marrow transduction/transplantation with RUNX1-ETO9a | Myeloid leukemia | (142) |
| Mus musculus | Inducible Cre/loxP RUNX1-ETO knockin and HIP1-PDGFbetaR knockin expression by Mx1 Cre | Myeloid leukemia | (151) |
The first mouse models utilized a knock-in strategy to insert the RUNX1-ETO fusion gene into the Runx1 locus and revealed that heterozygous RUNX1-ETO mice die during embryogenesis (149;150). The embryonic lethality is similar to that exhibited by Runx1 knockout mice, but slight differences were observed. Populations of yolk sac and fetal liver cells from the heterozygous embryos were able to differentiate into macrophages and dysplastic myeloid cells, respectively, whereas the homozygous knockout lacked definitive hematopoiesis entirely (149;150). These initial mouse studies demonstrated that although RUNX1-ETO primarily acted as a dominant-negative regulator of RUNX1, it may have other properties that may be important in its ability to promote leukemogenesis.
The next wave of models consisted of transgenic mice with RUNX1-ETO under the control of various promoters like a tetracycline-responsive element and MRP8, a calcium binding protein expressed in myeloid progenitors and mature neutrophils/monocytes (12;13). Although heterozygous RUNX1-ETO mice die embryonically, these transgenic mouse appear to display normal hematopoiesis and have a healthy lifespan (12;13). Only in the presence of additional mutations induced by treating these mice with N-ethyl-N-nitrosourea do they develop overt AML (13). Another group that used the Mx1-Cre inducible system and conditional RUNX1-ETO knock-in mice observed similar results (140). The presence of another oncogene like HIP1-PDFGbetaR together with RUNX1-ETO is also sufficient to create a myeloproliferative phenotype in mice (151). Interestingly, a knock-in mouse model with RUNX1-ETO being expressed from the Sca1 gene locus, which is active in HSCs, displayed myeloproliferative disease although spontaneous AML was still not observed (152).
The general lack of a leukemia phenotype from the models above led to additional studies utilizing cell transduction with RUNX1-ETO-expressing retrovirus followed by bone marrow transplantation of these cells into lethally irradiated hosts. This model system allowed for the selection of HSCs that expressed RUNX1-ETO because they were labeled with GFP or other markers (153;154). Interestingly, these models showed a slightly more severe phenotype in the hematopoietic system as illustrated by the appearance of immature blasts and inhibition of myeloid differentiation (153;154). Survival times, however, remained relatively normal and spontaneous AML was not observed (153;154). Furthermore, in agreement with previous results, the presence of additional perturbations such as deficiency of ICSBP, presence of Flt3 length mutation, or presence of TEL-PDGFbetaR fusion protein cooperated with RUNX1-ETO to induce AML (154–156). Based on the models described so far, RUNX1-ETO has the ability to induce changes in adult hematopoiesis, especially if expressed in the HSC compartment, and establishes an environment that is conducive for leukemogenesis with the presence of additional mutations.
In contrast to the mouse systems described so far, one recent discovery has identified that truncated isoforms of RUNX1-ETO transcript are capable to inducing full blown leukemia by themselves. The RUNX1-ETO9a is one such splice isoform, that when transduced and transplanted into mice, rapidly induces leukemia (142). Further studies are warranted into why full length RUNX1-ETO requires additional mutations to generate leukemia while shorter isoforms which are missing pieces of the C-terminal domain are capable of generating leukemia on their own.
A great deal of knowledge has been elucidated using the mouse models above. Xenografts using human CD34+ cells transduced with RUNX1-ETO and transplanted into NOD/SCID mice have revealed similar results—that RUNX1-ETO alone cannot induce leukemia development (157;158). What remains to be seen is whether this xenograph model utilizing the shorter RUNX1-ETO9a isoform can induce leukemia on its own.
7.2. RUNX1-ETO target genes
Since both RUNX1 and RUNX1-ETO contain the Runt homology domain, which is responsible for their DNA-binding properties, RUNX1-ETO has generally been assumed to share the same target genes as RUNX1. However, these two DNA-binding proteins may have varying preferences for target genes because RUNX1-ETO has an affinity for binding DNA segments that contain duplicated RUNX1 consensus sites (125). Hence, presence of the fusion protein may involve a gain-of-function in some circumstances.
Most of the target gene studies conducted on RUNX1-ETO have specifically focused on its role as an inducer of leukemia. One of the first target gene studies demonstrated that RUNX1-ETO directly regulated repression of the p14Arf tumor suppressor gene, which is a mediator of p53 (159). Another study delved into the role that nerve growth factor (NGF) plays in t(8;21)-mediated leukemogenesis by observing that expression of RUNX1-ETO in human CD34+ hematopoietic cells up-regulates TRKA mRNA, which subsequently allows these cells to respond to NGF (160). Yet another study implicated the negative cell cycle regulator p21WAF1 as a transcriptional target of RUNX1-ETO and that in the absence of p21WAF1, expression of RUNX1-ETO was able to induce leukemia (161). These works have given clues to which single genes, when mutated or differentially expressed, may help RUNX1-ETO to promote leukemia development.
With the advent of more high-throughput methods like differential gene expression microarrays, a variety of studies have been conducted on a genome-wide scale to characterize the transcriptional environment that RUNX1-ETO may promote. In one study using cell lines that expressed RUNX1-ETO, the authors discovered that DNA repair genes were generally inhibited while genes that promote stem cell self-renewal, like those in the Notch pathway, were generally up-regulated (162). Another study looked at 285 samples from patients with various types of AML and were able to place t(8;21)-positive patients into a unique cluster based on their gene profiles alone (163). A similar investigation using samples from cases of pediatric AML also concluded that t(8;21)-positive patients carry a unique but common transcriptional signature (164). This transcriptional signature also includes those that are part of the endogenous microRNA (miRNA) system. Genome-wide analyses of 52 AML patient samples revealed that patients with t(8;21)-positive leukemia have a distinct miRNA expression pattern from AML patients with other chromosomal translocations (165). Recent work has already characterized specific miRNAs regulated by RUNX1-ETO like miR-126 and miR-223 (165;166).
The genome-wide microarray studies have undoubtedly revealed significant differences in gene expression with the presence of RUNX1-ETO. Advances in coupling chromatin immunoprecipitation with microarrays have provided yet another approach to analyze target genes for DNA-binding proteins like RUNX1-ETO. In a recent study utilizing this technology, Gardini et al. were able to describe thousands of potential target genes and an important interaction between RUNX1-ETO, RUNX1, and HEB in potentially regulating these target genes (167). The common theme among these single target gene and genome-wide studies reflects what has been observed in the various mouse models described in the previous section—that although RUNX1-ETO cannot induce leukemia by itself, it does promote an environment for additional mutations to occur which may eventually lead to leukemia.
7.3. Regulation of chromatin structure
RUNX1-ETO has primarily been recognized as a dominant-negative regulator of RUNX1 due to the similarities between heterozygous RUNX1-ETO knock-in mice and Runx1 knockout mice. Many reports have shown that RUNX1-ETO is a repressor of transcription. ETO and RUNX1-ETO both interact with transcriptional co-repressors and HDACs through yeast two-hybrid assays and co-immunoprecipitation studies (133;135;136). Specifically, ETO was shown to interact with NCoR and mSin3a, which themselves were shown to interact with HDACs (168). Additional studies by the same group demonstrated that RUNX1-ETO interacts with HDAC-1, HDAC-2, and HDAC-3 and that treatment with trichostatin A, an HDAC inhibitor, blocks the ability of RUNX1-ETO to suppress myeloid differentiation (169). A recent example of this mechanism was described by Fazi et al. where they detailed how RUNX1-ETO was able to recruit HDACs to the miR-223 gene locus, a myelopoiesis regulator, to silence its transcriptional activity (166). Valproic acid, another HDAC inhibitor, has also been shown to promote the differentiation of Kasumi cells, a t(8;21)-positive AML cell line (170). Valproic acid may disrupt the interaction between RUNX1-ETO and HDAC-1 and is able to reactivate transcription of RUNX1 target genes like IL-3 (171). These studies point toward the ability of RUNX1-ETO to recruit HDACs as a major mechanism that may give it its leukemogenic properties, and suggest that the use of HDAC inhibitors may be a potentially useful therapy for patients suffering from t(8;21)-positive AML. It is also worth to note that HDAC inhibitors directly promote the degradation of RUNX1-ETO (144;172).
In addition to recruiting HDACs, RUNX1-ETO has been shown to promote methylation of DNA by recruitment of DNA methyltransferases (DNMTs). The first clue that led to further characterization into whether RUNX1-ETO may be involved in recruiting DNMTs was that the combination of HDAC and DNMT inhibitors were shown to have synergistic effects on histone acetylation and release of target gene repression (173). In a follow-up study, RUNX1-ETO was shown to interact with DNMT1 by co-immunoprecipitation and that both were able to synergistically repress transcription of IL-3 when transfected together into 293T cells (174). Subsequent investigations have characterized numerous gene loci where RUNX1-ETO may promote hypermethylation (175;176). DNA methylation is not specific for RUNX1-ETO, as other cancers have also been associated with DNA hypermethylation. Interestingly, however, a large scale study of the DNA methylation patterns of patients with various AMLs revealed that the t(8;21) AML-specific gene loci may offer a unique oncologic signature that distinguishes it from other types of AML (177). Further characterization of these specific loci may offer new genes for drug targeting and additional insights into the mechanisms of t(8;21)-positive leukemogenesis.
8. PERSPECTIVES
RUNX1 is undoubtedly a master regulator of hematopoiesis. Its attribute as a critical factor in defining the identity of HSCs make RUNX1 a very attractive molecule to study for potential applications in bone marrow therapy. At the same time, the prevalence of RUNX1-ETO in AML and other types of leukemia have made the fusion protein very highly studied. Although many target gene studies have been conducted on RUNX1, a paucity of data exists pertaining to the AML target genes in HSCs or immature progenitors. A recent study has tried to tackle this problem by utilizing massively parallel sequencing coupled with chromatin-immunoprecipitation (ChIP-seq) on a murine HSC-like cell line and has presented a genome-wide map of RUNX1 chromatin occupancy along with maps of other HSC-specific transcription factors (178). Though no analyses on specific genes were conducted, the study does provide a valuable resource of the RUNX1-targeted genes that may have essential functions in HSC identification, maintenance, or differentiation.
At the time of writing this review, no ChIP-seq studies have been conducted on RUNX1-ETO. Such a study on the genome-wide occupancy status of RUNX1-ETO will be a valuable resource, especially if it could be reasonably compared to RUNX1 occupancy, which would describe how RUNX1-ETO perturbs endogenous RUNX1 function on a more global scale. As technology continues to advance, we may even see ChIP-seq done on RUNX1 and/or RUNX1-ETO occupancy using sorted HSC or progenitor populations.
Another interesting avenue for further work arises from the alternatively spliced isoform of the fusion transcript, RUNX1-ETO9a, that is capable of inducing leukemia on its own. The lack of leukemia-inducing ability of full length RUNX1-ETO suggests that the C-terminal portion of the fusion protein may not be important or may even inhibit leukemia development. The mechanisms of how RUNX1-ETO9a induces leukemia and the role of the C-terminal portion that is missing in RUNX1-ETO9a will further elucidate the pathogenesis of this very common fusion protein.
RUNX1 and RUNX1-ETO have been fixtures in hematological and leukemia research for over two decades. Despite the numerous studies on these two molecules, much remains to be discovered. The critical nature of RUNX1 in hematopoiesis and the prevalence of RUNX1-ETO in leukemia continue to make these proteins highly studied in biomedical research, and breakthroughs in this field will offer much benefit to patients suffering from RUNX1- and RUNX1-ETO-related diseases and neoplasms.
Acknowledgments
This work was supported by National Institutes of Health grants HL103106 to K.L. and grants CA096735, CA10450, and DK080665 to D.E.Z. We would also like to apologize to those whose work was not cited due to space limitations.
Abbreviations
- AML
acute myeloid leukemia
- RUNX
Runt-related transcription factor
- PCR
polyermase chain reaction
- GFP
green fluorescent protein
- CBF
core binding factor
- NMTS
nuclear matrix targeting signal
- ERK
extracellular signal-regulated kinase
- CDK
cyclin-dependent kinase
- APC
anaphase-promoting complex
- HIPK2
homeodomain-interacting protein kinase-2
- HSC
hematopoietic stem cell
- dpc
days post conception
- ES
embryonic stem
- LSK
lineage-negative, sca1-positive, ckit-positive
- IFN
interferon
- polyIC
polyinosinic/polycytidylic acid
- FDP/AML
familial platelet disorder with predisposition to acute myeloid leukemia
- MDS
myelodysplastic syndrome
- MPN
myeloproliferative neoplasm
- CMML
chronic myelomonocytic leukemia
- ALL
acute lymphoblastic leukemia
- HLH
helix-loop-helix
- NHR
Nervy homology domain
- PKA
protein kinase A
- MYND
myeloid-Nervy-DEAF1
- HDAC
histone deacetylase
- NCoR
nuclear corepressor
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
- DNMT
DNA methyltransferase
Footnotes
“This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience”. Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.”
References
- 1.van Wijnen AJ, Stein GS, Gergen JP, Groner Y, Hiebert SW, Ito Y, Liu P, Neil JC, Ohki M, Speck N. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23:4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- 2.Nussleinvolhard C, Wieschaus E. Mutations Affecting Segment Number and Polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila Segmentation Gene Runt Encodes A Novel Nuclear Regulatory Protein That Is Also Expressed in the Developing Nervous-System. Genes & Development. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 4.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 5.Speck NA, Baltimore D. 6 Distinct Nuclear Factors Interact with the 75-Base-Pair Repeat of the Moloney Murine Leukemia-Virus Enhancer. Molecular and Cellular Biology. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satake M, Ibaraki T, Ito Y. Modulation of Polyomavirus Enhancer Binding-Proteins by Ha-Ras Oncogene. Oncogene. 1988;3:69–78. [Google Scholar]
- 7.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowley JD. Identification of A Translocation with Quinacrine Fluorescence in A Patient with Acute Leukemia. Annales de Genetique. 1973;16:109–112. [PubMed] [Google Scholar]
- 10.Rowley JD. The clinical usefulness of chromosome studies in patients with leukemia. Compr Ther. 1980;6:57–64. [PubMed] [Google Scholar]
- 11.Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 12.Rhoades KL, Hetherington CJ, Harakawa N, Yergeau DA, Zhou L, Liu LQ, Little MT, Tenen DG, Zhang DE. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96:2108–2115. [PubMed] [Google Scholar]
- 13.Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, Burel SA, Lagasse E, Weissman IL, Akashi K, Zhang DE. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, Bangsow C, Hattori M, Taylor TD, Taudien S, Blechschmidt K, Shimizu N, Rosenthal A, Sakaki Y, Lancet D, Groner Y. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative Splicing and Genomic Structure of the Aml1 Gene Involved in Acute Myeloid-Leukemia. Nucleic Acids Research. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Tanaka K, Ogawa S, Kurokawa M, Mitani K, Nishida J, Shibata Y, Yazaki Y, Hirai H. An Acute Myeloid-Leukemia Gene, Aml1, Regulates Hematopoietic Myeloid Cell-Differentiation and Transcriptional Activation Antagonistically by 2 Alternative Spliced Forms. Embo Journal. 1995;14:341–350. doi: 10.1002/j.1460-2075.1995.tb07008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuzuki S, Hong DL, Gupta R, Matsuo K, Seto M, Enver T. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Medicine. 2007;4:880–896. doi: 10.1371/journal.pmed.0040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Zhang Q, Zhang DE, Zhou C, Xing H, Tian Z, Rao Q, Wang M, Wang J. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia. 2009;23:739–745. doi: 10.1038/leu.2008.350. [DOI] [PubMed] [Google Scholar]
- 19.Telfer JC, Rothenberg EV. Expression and function of a stem cell promoter for the murine CBF alpha 2 gene: Distinct roles and regulation in natural killer and T cell development. Developmental Biology. 2001;229:363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 20.Lam EY, Chau JY, Kalev-Zylinska ML, Fountaine TM, Mead RS, Hall CJ, Crosier PS, Crosier KE, Flores MV. Zebrafish runx1 promoter-EGFP transgenics mark discrete sites of definitive blood progenitors. Blood. 2009;113:1241–1249. doi: 10.1182/blood-2008-04-149898. [DOI] [PubMed] [Google Scholar]
- 21.Sroczynska P, Lancrin C, Kouskoff V, Lacaud G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009;114:5279–5289. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- 22.Pozner A, Lotem J, Xiao C, Goldenberg D, Brenner O, Negreanu V, Levanon D, Groner Y. Developmentally regulated promoter-switch transcriptionally controls Runx I function during embryonic hematopoiesis. BMC Developmental Biology. 2007;7 doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bee T, Swiers G, Muroi S, Pozner A, Nottingham W, Santos AC, Li PS, Taniuchi I, de Bruijn MFTR. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- 24.Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong AS, de Bruijn MFTR. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng CEL, Yokomizo T, Yamashita N, Cirovic B, Jin H, Wen ZL, Ito Y, Osato M. A Runx1 Intronic Enhancer Marks Hemogenic Endothelial Cells and Hematopoietic Stem Cells. Stem Cells. 2010;28:1869–1881. doi: 10.1002/stem.507. [DOI] [PubMed] [Google Scholar]
- 26.Bee T, Ashley ELK, Bickley SRB, Jarratt A, Li PS, Sloane-Stanley J, Gottgens B, de Bruijn MFTR. The mouse Runx1+23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood. 2009;113:5121–5124. doi: 10.1182/blood-2008-12-193003. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berardi MJ, Sun CH, Zehr M, Abildgaard F, Peng J, Speck NA, Bushweller JH. The Ig fold of the core binding factor alpha Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains. Structure. 1999;7:1247–1256. doi: 10.1016/s0969-2126(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 30.Nagata T, Gupta V, Sorce D, Kim WY, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nature Structural Biology. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- 31.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, Ishii S, Ogata K. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 33.Bartfeld D, Shimon L, Couture GC, Rabinovich D, Frolow F, Levanon D, Groner Y, Shakked Z. DNA recognition by the RUNX1 transcription factor is mediated by an allosteric transition in the Runt domain and by DNA bending. Structure. 2002;10:1395–1407. doi: 10.1016/s0969-2126(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 34.Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1) Mol Cell Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB, Penman S, Hiebert S, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci U S A. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–4370. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- 40.Gunther CV, Graves BJ. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994;14:7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang DE, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen HM, Hiebert SW, Tenen DG. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai Y, Kurokawa M, Yamaguchi Y, Izutsu K, Nitta E, Mitani K, Satake M, Noda T, Ito Y, Hirai H. The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol Cell Biol. 2004;24:1033–1043. doi: 10.1128/MCB.24.3.1033-1043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu G, Kanezaki R, Toki T, Watanabe S, Takahashi Y, Terui K, Kitabayashi I, Ito E. Physical association of the patient-specific GATA1 mutants with RUNX1 in acute megakaryoblastic leukemia accompanying Down syndrome. Leukemia. 2006;20:1002–1008. doi: 10.1038/sj.leu.2404223. [DOI] [PubMed] [Google Scholar]
- 47.Huang H, Yu M, Akie TE, Moran TB, Woo AJ, Tu N, Waldon Z, Lin YY, Steen H, Cantor AB. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Mol Cell Biol. 2009;29:4103–4115. doi: 10.1128/MCB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, Kadowaki T, Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erickson PF, Dessev G, Lasher RS, Philips G, Robinson M, Drabkin HA. ETO and AML1 phosphoproteins are expressed in CD34+ hematopoietic progenitors: implications for t(8;21) leukemogenesis and monitoring residual disease. Blood. 1996;88:1813–1823. [PubMed] [Google Scholar]
- 50.Zhang Y, Biggs JR, Kraft AS. Phorbol ester treatment of K562 cells regulates the transcriptional activity of AML1c through phosphorylation. J Biol Chem. 2004;279:53116–53125. doi: 10.1074/jbc.M405502200. [DOI] [PubMed] [Google Scholar]
- 51.Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol. 2006;26:7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Fried FB, Guo H, Friedman AD. Cyclin-dependent kinase phosphorylation of RUNX1/AML1 on 3 sites increases transactivation potency and stimulates cell proliferation. Blood. 2008;111:1193–1200. doi: 10.1182/blood-2007-08-109702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wee HJ, Voon DCC, Bae SC, Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo H, Friedman AD. Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. J Biol Chem. 2011;286:208–215. doi: 10.1074/jbc.M110.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 57.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, Allis CD, Tempst P, Nimer SD. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tachibana M, Tezuka C, Muroi S, Nishimoto S, Katsumoto T, Nakajima A, Kitabayashi I, Taniuchi I. Phosphorylation of Runx1 at Ser249, Ser266, and Ser276 is dispensable for bone marrow hematopoiesis and thymocyte differentiation. Biochem Biophys Res Commun. 2008;368:536–542. doi: 10.1016/j.bbrc.2008.01.124. [DOI] [PubMed] [Google Scholar]
- 60.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 61.Medvinsky AL, Samoylina NL, Muller AM, Dzierzak EA. An Early Pre-Liver Intra-Embryonic Source of Cfu-S in the Developing Mouse. Nature. 1993;364:64–67. doi: 10.1038/364064a0. [DOI] [PubMed] [Google Scholar]
- 62.Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 63.North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 64.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 66.Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, Satake M, Suda T. A role for hematopoietic stem cells in promoting angiogenesis. Cell. 2000;102:199–209. doi: 10.1016/s0092-8674(00)00025-8. [DOI] [PubMed] [Google Scholar]
- 67.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 70.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 71.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- 73.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 74.Ichikawa M, Goyama S, Asai T, Kawazu M, Nakagawa M, Takeshita M, Chiba S, Ogawa S, Kurokawa M. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. Journal of Immunology. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- 75.Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR, Asou N, Ito Y. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood. 2010;115:1610–1620. doi: 10.1182/blood-2009-07-232249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cameron S, Taylor DS, TePas EC, Speck NA, Mathey-Prevot B. Identification of a critical regulatory site in the human interleukin- 3 promoter by in vivo footprinting. Blood. 1994;83:2851–2859. [PubMed] [Google Scholar]
- 79.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae SC, Lu J, Maruyama M, Zhang YW, Oka H, Arai N, Arai K. Positive and negative regulation of granulocyte-macrophage colony- stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 80.Zhang DE, Fujioka K, Hetherington CJ, Shapiro LH, Chen HM, Look AT, Tenen DG. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1) Mol Cell Biol. 1994;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satoh Y, Matsumura I, Tanaka H, Ezoe S, Fukushima K, Tokunaga M, Yasumi M, Shibayama H, Mizuki M, Era T, Okuda T, Kanakura Y. AML1/RUNX1 Works as a Negative Regulator of c-Mpl in Hematopoietic Stem Cells. Journal of Biological Chemistry. 2008;283:30045–30056. doi: 10.1074/jbc.M804768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nuchprayoon I, Meyers S, Scott LM, Suzow J, Hiebert S, Friedman AD. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2 beta/CBF beta proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogihara H, Kanno T, Morii E, Kim DK, Lee YM, Sato M, Kim WY, Nomura S, Ito Y, Kitamura Y. Synergy of PEBP2/CBF with mi transcription factor (MITF) for transactivation of mouse mast cell protease 6 gene. Oncogene. 1999;18:4632–4639. doi: 10.1038/sj.onc.1202844. [DOI] [PubMed] [Google Scholar]
- 84.Hsiang YH, Spencer D, Wang S, Speck NA, Raulet DH. The role of viral enhancer “core” motif-related sequences in regulating T cell receptor-gamma and -delta gene expression. J Immunol. 1993;150:3905–3916. [PubMed] [Google Scholar]
- 85.Sun W, Graves BJ, Speck NA. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puig-Kroger A, Lopez-Rodriguez C, Relloso M, Sanchez-Elsner T, Nueda A, Munoz E, Bernabeu C, Corbi AL. Polyomavirus enhancer-binding protein 2/core binding factor/acute myeloid leukemia factors contribute to the cell type-specific activity of the CD11a integrin gene promoter. J Biol Chem. 2000;275:28507–28512. doi: 10.1074/jbc.M004323200. [DOI] [PubMed] [Google Scholar]
- 87.Libermann TA, Pan Z, Akbarali Y, Hetherington CJ, Boltax J, Yergeau DA, Zhang DE. AML1 (CBFalpha2) cooperates with B cell-specific activating protein (BSAP/PAX5) in activation of the B cell-specific BLK gene promoter. J Biol Chem. 1999;274:24671–24676. doi: 10.1074/jbc.274.35.24671. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Derynck R. Transcriptional regulation of the transforming growth factor-beta - inducible mouse germ line Ig alpha constant region gene by functional cooperation of Smad, CREB, and AML family members. J Biol Chem. 2000;275:16979–16985. doi: 10.1074/jbc.M001526200. [DOI] [PubMed] [Google Scholar]
- 89.Erman B, Cortes M, Nikolajczyk BS, Speck NA, Sen R. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol Cell Biol. 1998;18:1322–1330. doi: 10.1128/mcb.18.3.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S. Indispensable Role of the Runx1-Cbf beta Transcription Complex for In vivo-Suppressive Function of FoxP3(+) Regulatory T Cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 91.Rudra D, Egawa T, Chong MMW, Treuting P, Littman DR, Rudensky AY. Runx-CBF beta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nature Immunology. 2009;10:1170–1U53. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nature Genetics. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 93.Ben Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2aB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 95.Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 96.Preudhomme C, Renneville A, Bourdon V, Philippe N, Roche-Lestienne C, Boissel N, Dhedin N, Andre JM, Cornillet-Lefebvre P, Baruchel A, Mozziconacci MJ, Sobol H. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113:5583–5587. doi: 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- 97.Imai Y, Kurokawa M, Izutsu K, Hangaishi A, Takeuchi K, Maki K, Ogawa S, Chiba S, Mitani K, Hirai H. Mutations of the AML1 gene in myelodysplastic syndrome and their functional implications in leukemogenesis. Blood. 2000;96:3154–3160. [PubMed] [Google Scholar]
- 98.Michaud J, Wu F, Osato M, Cotties GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, Rossier C, Antonarakis SE, Israels S, McNicol A, Weiss H, Horwitz M, Scott HS. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 99.Heller PG, Glembotsky AC, Gandhi MJ, Cummings CL, Pirola CJ, Marta RF, Kornblihtt LI, Drachman JG, Molinas FC. Low Mpl receptor expression in a pedigree with familial platelet disorder with predisposition to acute myelogenous leukemia and a novel AML1 mutation. Blood. 2005;105:4664–4670. doi: 10.1182/blood-2005-01-0050. [DOI] [PubMed] [Google Scholar]
- 100.Churpek JE, Garcia JS, Madzo J, Jackson SA, Onel K, Godley LA. Identification and molecular characterization of a novel 3′ mutation in RUNX1 in a family with familial platelet disorder. Leukemia & Lymphoma. 2010;51:1931–1935. doi: 10.3109/10428194.2010.503821. [DOI] [PubMed] [Google Scholar]
- 101.Ding Y, Harada Y, Imagawa J, Kimura A, Harada H. AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood. 2009;114:5201–5205. doi: 10.1182/blood-2009-06-223982. [DOI] [PubMed] [Google Scholar]
- 102.Wang W, Schwemmers S, Hexner EO, Pahl HL. AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood. 2010;116:254–266. doi: 10.1182/blood-2009-11-254664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gelsi-Boyer V, Trouplin V, Adelaide J, Aceto N, Remy V, Pinson S, Houdayer C, Arnoulet C, Sainty D, Bentires-Alj M, Olschawang S, Vey N, Mozziconacci MJ, Birnbaum D, Chaffanet M. Genome profiling of chronic myelomonocytic leukemia; frequent alterations of RAS and RUNX1 genes. BMC Cancer. 2008;8 doi: 10.1186/1471-2407-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuo MC, Liang DC, Huang CF, Shih YS, Wu JH, Lin TL, Shih LY. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23:1426–1431. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 105.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 106.Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, Morgan E, Raimondi SC, Rowley JD, Gilliland DG. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
- 108.Rubin CM, Larson RA, Bitter MA, Carrino JJ, LeBeau MM, Diaz MO, Rowley JD. Association of A Chromosomal 3–21 Translocation with the Blast Phase of Chronic Myelogenous Leukemia. Blood. 1987;70:1338–1342. [PubMed] [Google Scholar]
- 109.Rubin CM, Larson RA, Anastasi J, Winter JN, Thangavelu M, Vardiman JW, Rowley JD, LeBeau MM. T(3–21)(Q26-Q22) - A Recurring Chromosomal Abnormality in Therapy-Related Myelodysplastic Syndrome and Acute Myeloid-Leukemia. Blood. 1990;76:2594–2598. [PubMed] [Google Scholar]
- 110.Nucifora G, Begy CR, Kobayashi H, Roulston D, Claxton D, Pedersenbjergaard J, Parganas E, Ihle JN, Rowley JD. Consistent Intergenic Splicing and Production of Multiple Transcripts Between Aml1 at 21Q22 and Unrelated Genes at 3Q26 in (321)(Q26Q22) Translocations. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4004–4008. doi: 10.1073/pnas.91.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mikhail FM, Serry KA, Hatem N, Mourad ZI, Farawela HM, El Kaffash DM, Coignet L, Nucifora G. A new translocation that rearranges the AML1 gene in a patient with T-cell acute lymphoblastic leukemia. Cancer Genetics and Cytogenetics. 2002;135:96–100. doi: 10.1016/s0165-4608(01)00633-1. [DOI] [PubMed] [Google Scholar]
- 112.Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- 113.Chan EM, Comer EM, Brown FC, Richkind KE, Holmes ML, Chong BH, Shiffman R, Zhang DE, Slovak ML, Willman CL, Noguchi CT, Li YJ, Heiber DJ, Kwan L, Chan RJ, Vance GH, Ramsey HC, Hromas RA. AML1-FOG2 fusion protein in myelodysplasia. Blood. 2005;105:4523–4526. doi: 10.1182/blood-2004-07-2762. [DOI] [PubMed] [Google Scholar]
- 114.Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 115.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tighe JE, Calabi F. Alternative, out-of-frame runt/MTG8 transcripts are encoded by the derivative (8) chromosome in the t(8;21) of acute myeloid leukemia M2. Blood. 1994;84:2115–2121. [PubMed] [Google Scholar]
- 117.Tighe JE, Calabi F. t(8;21) breakpoints are clustered between alternatively spliced exons of MTG8. Clin Sci (Lond) 1995;89:215–218. doi: 10.1042/cs0890215. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y, Strissel P, Strick R, Chen J, Nucifora G, Le Beau MM, Larson RA, Rowley JD. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc Natl Acad Sci U S A. 2002;99:3070–3075. doi: 10.1073/pnas.042702899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peterson LF, Boyapati A, Ahn EY, Biggs JR, Joo OA, Lo MC, Yan M, Zhang DE. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calabi F, Pannell R, Pavloska G. Gene targeting reveals a crucial role for MTG8 in the gut. Mol Cell Biol. 2001;21:5658–5666. doi: 10.1128/MCB.21.16.5658-5666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sacchi N, Tamanini F, Willemsen R, Denis-Donini S, Campiglio S, Hoogeveen AT. Subcellular localization of the oncoprotein MTG8 (CDR/ETO) in neural cells. Oncogene. 1998;16:2609–2615. doi: 10.1038/sj.onc.1201824. [DOI] [PubMed] [Google Scholar]
- 122.Alishahi A, Koyano-Nakagawa N, Nakagawa Y. Regional Expression of MTG Genes in the Developing Mouse Central Nervous System. Developmental Dynamics. 2009;238:2095–2102. doi: 10.1002/dvdy.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wolford JK, Prochazka M. Structure and expression of the human MTG8/ETO gene. Gene. 1998;212:103–109. doi: 10.1016/s0378-1119(98)00141-3. [DOI] [PubMed] [Google Scholar]
- 124.Erickson PF, Robinson M, Owens G, Drabkin HA. The Eto Portion of Acute Myeloid-Leukemia T(8–21) Fusion Transcript Encodes A Highly Evolutionarily Conserved, Putative Transcription Factor. Cancer Research. 1994;54:1782–1786. [PubMed] [Google Scholar]
- 125.Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112:1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feinstein PG, Kornfeld K, Hogness DS, Mann RS. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Terman JR, Kolodkin AL. Nervy links protein kinase a to plexin-mediated semaphorin repulsion. Science. 2004;303:1204–1207. doi: 10.1126/science.1092121. [DOI] [PubMed] [Google Scholar]
- 128.Fukuyama T, Sueoka E, Sugio Y, Otsuka T, Niho Y, Akagi K, Kozu T. MTG8 proto-oncoprotein interacts with the regulatory subunit of type II cyclic AMP-dependent protein kinase in lymphocytes. Oncogene. 2001;20:6225–6232. doi: 10.1038/sj.onc.1204794. [DOI] [PubMed] [Google Scholar]
- 129.Corpora T, Roudaia L, Oo ZM, Chen W, Manuylova E, Cai XW, Chen MJ, Cierpicki T, Speck NA, Bushweller JH. Structure of the AML1-ETO NHR3-KA(RII alpha) Complex and Its Contribution to AML1-ETO Activity. Journal of Molecular Biology. 2010;402:560–577. doi: 10.1016/j.jmb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, Klet RC, Hartman K, Laue TM, Speck NA, Bushweller JH. Structural Basis for Recognition of SMRT/N-CoR by the MYND Domain and Its Contribution to AML1/ETO’s Activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 132.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang J, Hug BA, Huang EY, Chen CW, Gelmetti V, Maccarana M, Minucci S, Pelicci PG, Lazar MA. Oligomerization of ETO is obligatory for corepressor interaction. Mol Cell Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, Lary J, Cole J, Dauter Z, Minor W, Speck NA, Bushweller JH. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 135.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci U S A. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van de Locht LT, Smetsers TF, Wittebol S, Raymakers RA, Mensink EJ. Molecular diversity in AML1/ETO fusion transcripts in patients with t(8;21) positive acute myeloid leukaemia. Leukemia. 1994;8:1780–1784. [PubMed] [Google Scholar]
- 139.Buchholz F, Refaeli Y, Trumpp A, Bishop JM. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Reports. 2000;1:133–139. doi: 10.1093/embo-reports/kvd027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 141.Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci U S A. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, Zhang DE. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 143.Hackanson B, Abdelkarim M, Jansen JH, Lubbert M. NHR4 domain mutations of ETO are probably very infrequent in AML1-ETO positive myeloid leukemia cells. Leukemia. 2010;24:860–861. doi: 10.1038/leu.2009.291. [DOI] [PubMed] [Google Scholar]
- 144.Kramer OH, Muller S, Buchwald M, Reichardt S, Heinzel T. Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J. 2008;22:1369–1379. doi: 10.1096/fj.06-8050com. [DOI] [PubMed] [Google Scholar]
- 145.Lu Y, Peng ZG, Yuan TT, Yin QQ, Xia L, Chen GQ. Multi-sites cleavage of leukemogenic AML1-ETO fusion protein by caspase-3 and its contribution to increased apoptotic sensitivity. Leukemia. 2008;22:378–386. doi: 10.1038/sj.leu.2405020. [DOI] [PubMed] [Google Scholar]
- 146.Osman D, Gobert V, Ponthan F, Heidenreich O, Haenlin M, Waltzer L. A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proc Natl Acad Sci U S A. 2009;106:12043–12048. doi: 10.1073/pnas.0902449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yeh JRJ, Munson KM, Chao YL, Peterson QP, Macrae CA, Peterson RT. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2008;135:401–410. doi: 10.1242/dev.008904. [DOI] [PubMed] [Google Scholar]
- 148.Yeh JRJ, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nature Chemical Biology. 2009;5:236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, Marin-Padilla M, Tenen DG, Speck NA, Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 150.Okuda T, Cai Z, Yang S, Lenny N, Lyu C, van Deursen JMA, Harada H, Downing JR. Expression of a knocked-In AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 151.Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, Gauvin AM, Lucas PC, Sitwala K, Downing JR, Morrison SJ, Ross TS. Persistence of Leukemia-Initiating Cells in a Conditional Knockin Model of an Imatinib-Responsive Myeloproliferative Disorder. Cancer Cell. 2009;16:137–148. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, Graubert TA. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci U S A. 2004;101:15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, Hiebert SW, Klug CA. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002;22:5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwieger M, Lohler J, Friel J, Scheller M, Horak I, Stocking C. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J Exp Med. 2002;196:1227–1240. doi: 10.1084/jem.20020824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grisolano JL, O’Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2003;100:9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM, Spiekermann K, Humphries RK, Schnittger S, Kern W, Hiddemann W, Quintanilla-Martinez L, Bohlander SK, Feuring-Buske M, Buske C. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mulloy JC, Cammenga J, Berguido FJ, Wu K, Zhou P, Comenzo RL, Jhanwar S, Moore MA, Nimer SD. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102:4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- 158.Basecke J, Schwieger M, Griesinger F, Schiedlmeier B, Wulf G, Trumper L, Stocking C. AML1/ETO promotes the maintenance of early hematopoietic progenitors in NOD/SCID mice but does not abrogate their lineage specific differentiation. Leuk Lymphoma. 2005;46:265–272. doi: 10.1080/10428190400010767. [DOI] [PubMed] [Google Scholar]
- 159.Linggi B, Muller-Tidow C, van de LL, Hu M, Nip J, Serve H, Berdel WE, van der RB, Quelle DE, Rowley JD, Cleveland J, Jansen JH, Pandolfi PP, Hiebert SW. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 160.Mulloy JC, Jankovic V, Wunderlich M, Delwel R, Cammenga J, Krejci O, Zhao H, Valk PJ, Lowenberg B, Nimer SD. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proc Natl Acad Sci U S A. 2005;102:4016–4021. doi: 10.1073/pnas.0404701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Peterson LF, Yan M, Zhang DE. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007;109:4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, Riganelli D, Sebastiani C, Cappelli E, Casciari C, Sciurpi MT, Mariano AR, Minardi SP, Luzi L, Muller H, Di Fiore PP, Frosina G, Pelicci PG. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Lowenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 164.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, Ribeiro RC, Rubnitz JE, Girtman K, Williams WK, Raimondi SC, Liang DC, Shih LY, Pui CH, Downing JR. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 165.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fazi F, Racanicchi S, Zardo G, Stames LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 167.Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, Venturini E, Zhang DE, Pelicci PG, Alcalay M. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4:e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 169.Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo CF, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu S, Klisovic RB, Vukosavljevic T, Yu J, Paschka P, Huynh L, Pang J, Neviani P, Liu Z, Blum W, Chan KK, Perrotti D, Marcucci G. Targeting AML1/ETO-histone deacetylase repressor complex: a novel mechanism for valproic acid-mediated gene expression and cellular differentiation in AML1/ETO-positive acute myeloid leukemia cells. J Pharmacol Exp Ther. 2007;321:953–960. doi: 10.1124/jpet.106.118406. [DOI] [PubMed] [Google Scholar]
- 172.Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007;26:91–101. doi: 10.1038/sj.onc.1209760. [DOI] [PubMed] [Google Scholar]
- 173.Klisovic MI, Maghraby EA, Parthun MR, Guimond M, Sklenar AR, Whitman SP, Chan KK, Murphy T, Anon J, Archer KJ, Rush LJ, Plass C, Grever MR, Byrd JC, Marcucci G. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003;17:350–358. doi: 10.1038/sj.leu.2402776. [DOI] [PubMed] [Google Scholar]
- 174.Liu S, Shen T, Huynh L, Klisovic MI, Rush LJ, Ford JL, Yu J, Becknell B, Li Y, Liu C, Vukosavljevic T, Whitman SP, Chang KS, Byrd JC, Perrotti D, Plass C, Marcucci G. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–1284. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 175.Lasa A, Carnicer MJ, Aventin A, Estivill C, Brunet S, Sierra J, Nomdedeu J. MEIS 1 expression is downregulated through promoter hypermethylation in AML1-ETO acute myeloid leukemias. Leukemia. 2004;18:1231–1237. doi: 10.1038/sj.leu.2403377. [DOI] [PubMed] [Google Scholar]
- 176.Berg T, Guo Y, Abdelkarim M, Fliegauf M, Lubbert M. Reversal of p15/INK4b hypermethylation in AML1/ETO-positive and -negative myeloid leukemia cell lines. Leukemia Research. 2007;31:497–506. doi: 10.1016/j.leukres.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 177.Figueroa ME, Lugthart S, Li YS, Erpelinck-Verschueren C, Deng XT, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, Campagne F, Mazumdar M, Greally JM, Valk PJM, Lowenberg B, Delwel R, Melnick A. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wilson NK, Foster SD, Wang XN, Knezevic K, Schutte J, Kainnakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn TRMF, Gottgens B. Combinatorial Transcriptional Control In Blood Stem/Progenitor Cells: Genome-wide Analysis of Ten Major Transcriptional Regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 179.Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132:2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- 180.Sinenko SA, Hung T, Moroz T, Tran QM, Sidhu S, Cheney MD, Speck NA, Banerjee U. Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood. 2010;116:4612–4620. doi: 10.1182/blood-2010-03-276998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nishida S, Hosen N, Shirakata T, Kanato K, Yanagihara M, Nakatsuka S, Hoshida Y, Nakazawa T, Harada Y, Tatsumi N, Tsuboi A, Kawakami M, Oka Y, Oji Y, Aozasa K, Kawase I, Sugiyama H. AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood. 2006;107:3303–3312. doi: 10.1182/blood-2005-04-1656. [DOI] [PubMed] [Google Scholar]