The crystal structure of C. jejuni anabolic ornithine transcarbamoylase has been determined at a resolution of 2.7 Å in an unliganded state.
Keywords: Campylobacter jejuni, anabolic ornithine transcarbamoylases, carbamoyl phosphate, l-ornithine, l-citrulline, arginine biosynthesis, biocatalysis
Abstract
Anabolic ornithine transcarbamoylase (aOTC) catalyzes the reaction between carbamoyl phosphate (CP) and l-ornithine (ORN) to form l-citrulline and phosphate in the urea cycle and l-arginine biosynthesis. The crystal structure of unliganded aOTC from Campylobacter jejuni (Cje aOTC) was determined at 2.7 Å resolution and refined to an R work of 20.3% and an R free of 24.0%. Cje aOTC is a trimer that forms a head-to-head pseudohexamer in the asymmetric unit. Each monomer is composed of an N-terminal CP-binding domain and a C-terminal ORN-binding domain joined by two interdomain helices. The Cje aOTC structure presents an open conformation of the enzyme with a relatively flexible orientation of the ORN-binding domain respective to the CP-binding domain. The conformation of the B2–H3 loop (residues 68–78), which is involved in binding CP in an adjacent subunit of the trimer, differs from that seen in homologous proteins with CP bound. The loop containing the ORN-binding motif (DxxxSMG, residues 223–230) has a conformation that is different from those observed in unliganded OTC structures from other species, but is similar to those in structures with bound ORN analogs. The major differences in tertiary structure between Cje aOTC and human aOTC are described.
1. Introduction
Campylobacter is among the most important of the food-borne pathogens; it is suspected of causing 2.4 million known cases of gastrointestinal illness each year in the United States and is a leading cause of diarrhea worldwide. In 99% of identified campylobacterosis cases the species responsible for illness is C. jejuni, which has been divided into two subspecies (Fagerquist et al., 2006 ▶). C. jejuni subsp. jejuni NCTC 11168 is a target organism of the Center of Structural Genomics for Infectious Disease (CSGID; Anderson, 2009 ▶).
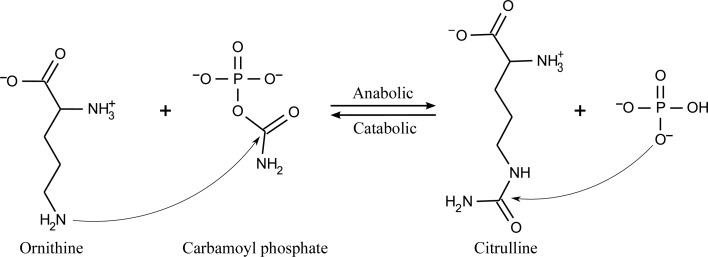
Ornithine carbamoyltransferase (OTC; EC 2.1.3.3) catalyzes the reversible transfer (Fig. 1 ▶) of the carbamoyl group from carbamoyl phosphate (CP) to the N∊ atom of l-ornithine (ORN) to produce l-citrulline (Cunin et al., 1986 ▶). There are two types of enzyme: anabolic (aOTC) and catabolic (cOTC). Anabolic OTCs catalyze the forward reaction and participate in the urea cycle and l-arginine biosynthesis. Because of its essential physiological functions, aOTC is a widespread enzyme that is found in a large variety of organisms from bacteria to mammals. In eukaryotes, the urea cycle is the main chemical pathway for the conversion of toxic ammonia to urea (Shi et al., 1998 ▶). Deleterious mutations in the human aOTC gene produce clinical hyperammonemia, with subsequent neurological symptoms or even death (Tuchman, 1993 ▶).
Figure 1.
Schematic representation of the reaction catalyzed by OTC.
In a number of microorganisms in which the arginine deiminase pathway for generating ATP from arginine is present, catabolic OTCs, which catalyze the reverse reaction, are also found (Cunin et al., 1986 ▶; Baur et al., 1990 ▶). Anabolic and catabolic OTCs function unidirectionally in vivo despite extensive sequence similarities (Nguyen et al., 1996 ▶). Anabolic OTCs catalyze the thermodynamically favored reaction and display Michaelis–Menten kinetics. The kinetic behavior of aOTCs is usually consistent with the ordered mechanism in which CP is the first substrate to bind and phosphate is the last product to be released (Cunin et al., 1986 ▶). Catabolic OTCs catalyze the thermodynamically unfavored reverse reaction yielding ORN and CP, and display allosteric inhibition by polyamines and activation by nucleoside monophosphates. The unidirectional reaction is possible owing to the poor affinity of the enzyme for CP and its high saturation cooperativity towards this substrate (Baur et al., 1990 ▶; Tricot et al., 1993 ▶, 1998 ▶; Sainz et al., 1998 ▶).
OTC belongs to the transcarbamylase family and is homologous in structure and function to the catalytic subunit of aspartate transcarbamoylase, which is the most studied enzyme of the class owing to its classical allosteric regulation and homotropic cooperativity (Lipscomb & Kantrowitz, 2011 ▶). UniProtKB (TheUniProtConsortium, 2011 ▶) contains 2591 sequences for OTC (search query EC 2.1.3.3). Currently, 21 structures of OTCs from 13 organisms, including human, Escherichia coli and Mycobacterium tuberculosis, have been deposited in the PDB (Berman et al., 2000 ▶) under the names ornithine carbamoyltransferase or ornithine transcarbamoylase. Most of the deposited OTC structures are of the anabolic enzyme, but four structures are of catabolic OTCs. Anabolic OTCs are cyclic homotrimers of about 105 kDa both in solution (Cunin et al., 1986 ▶) and as crystallized (Shi et al., 2000 ▶; Langley et al., 2000 ▶; Sankaranarayanan et al., 2008 ▶). The only exception described in the literature is the aOTC from the thermophilic archaebacterium Pyrococcus furiosus, which forms a dodecamer that has been correlated with its high thermostability (Villeret et al., 1998 ▶; Massant et al., 2003 ▶). There is one more aOTC in the PDB with a dodecameric assembly (PDB entry 1vlv), which may also be correlated with thermostability as the source organism Thermotoga maritima is hyperthermophilic. The catabolic OTCs have a diverse oligomeric composition. Both crystallographic and solution studies showed that the cOTC from Pseudomonas aeruginosa is a dodecamer and this has been correlated with its allosteric properties (Villeret et al., 1995 ▶; Tricot et al., 1998 ▶). The cOTC from Lactobacillus hilgardii is a hexamer (de Las Rivas et al., 2009 ▶) and the cOTC from the human parasite Giardia lamblia is a trimer (Galkin et al., 2009 ▶).
The mechanism of the aOTC-catalyzed reaction involves nucleophilic attack of the electron pair of the ∊-amino group of ORN on the carbonyl group of CP. In order to study the molecular mechanism of the reaction, crystal structures of Escherichia coli (Ha et al., 1997 ▶) and human (Shi et al., 1998 ▶) aOTCs liganded with the bisubstrate analogue N-(phosphonacetyl)-l-ornithine (PALO) were determined. PALO mimics the phosphate and carbonyl groups of CP and the carboxylate and α-amino groups of ORN well, but is a poor analog of the tetrahedral intermediate. Hence, the structure of the ternary complex of human aOTC with CP and l-norvaline, a competitive inhibitor of the enzyme, was determined (Shi et al., 2000 ▶). These structures indicate that binding of the bisubstrate analogue or of both substrates results in closure of the two domains and movement of the catalytic SMG loop. Two crystal structures of human aOTC complexed with just CP revealed that binding of the first substrate (CP) induces a global conformational change involving relative domain movement, whereas binding of the second substrate (ORN) brings together the flexible SMG loop (Shi et al., 2001 ▶). Comparison of the recent apo structure of M. tuberculosis aOTC (Mtu aOTC) and the structure of its complex with CP and l-norvaline clarified and confirmed the proposed catalytic mechanism of the aOTC-catalyzed reaction (Sankaranarayanan et al., 2008 ▶).
In this paper, we report the crystal structure of the anabolic ornithine carbamoyltransferase from C. jejuni subsp. jejuni NCTC 11168 (Cje aOTC; CSGID target IDP90828) at 2.7 Å resolution and describe the major differences between the Cje aOTC and human aOTC structures.
2. Materials and methods
2.1. Cloning, expression and purification
The ORF of argF was amplified by polymerase chain reaction from C. jejuni subsp. jejuni genomic DNA using the forward primer 5′-TACTTCCAATCCAATGCGATGAAACATTTTTTAACTTTAAGAG-3′ and the reverse primer 5′-TTATCCACTTCCAATGTCATTCTCTTCCTCTCTGATTA-3′. The gene was cloned into a pMCSG7 plasmid using ligation-independent cloning (Aslanidis & De Jong, 1990 ▶; Haun et al., 1992 ▶; Eschenfeldt et al., 2009 ▶). The gene was expressed in E. coli BL21-CodonPlus(DE3)-RIL. Cells were grown in selenomethionine medium at 310 K, induced by isopropyl β-d-1-thiogalactopyranoside and then grown at 293 K. Harvested cells were sonicated in lysis buffer (300 mM NaCl, 50 mM HEPES pH 7.5, 5% glycerol, 5 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine) and clarified by centrifugation; the supernatant was applied onto a nickel-chelate affinity resin (Ni–NTA, Qiagen). The resin was washed with wash buffer (300 mM NaCl, 50 mM HEPES pH 7.5, 5% glycerol, 30 mM imidazole) and the protein was eluted using elution buffer (300 mM NaCl, 50 mM HEPES pH 7.5, 5% glycerol, 250 mM imidazole). The hexahistidine tag was cleaved from the protein by the addition of recombinant His-tagged TEV protease. EDTA, TCEP and arginine were added to final concentrations of 1 mM, 0.5 mM and 0.2 M, respectively. The cleavage was performed at 277 K overnight and continued during dialysis into cleavage buffer (300 mM NaCl, 50 mM HEPES pH 7.5, 0.5 mM TCEP). Protein was separated from TEV protease by passage over nickel-chelating resin. The protein was then dialyzed into crystallization buffer (300 mM NaCl, 10 mM HEPES pH 7.5, 0.5 mM TCEP) and concentrated to a final concentration of 10 mg ml−1.
2.2. Crystallization
The crystals used for data collection were grown by the hanging-drop vapor-diffusion method. The crystallization process was monitored using the Xtaldb system (Zimmerman et al., 2005 ▶). The well solution consisted of 25% PEG 3350, 0.2 M NaCl, 0.1 M HEPES pH 8.0. Drops were formed by mixing 0.5 µl well solution and 0.5 µl of 10 mg ml−1 protein solution in crystallization buffer. Crystals were grown at room temperature and formed after one week of incubation. Immediately after harvesting, crystals were transferred into Paratone for cryoprotection and were then flash-cooled in liquid nitrogen.
2.3. Data collection and processing
Data were collected at 100 K on the 19BM beamline of the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois, USA) controlled by HKL-3000 (Minor et al., 2006 ▶). Diffraction data were reduced, scaled and merged with HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.4. Structure determination and refinement
The structure was solved by molecular replacement in HKL-3000 using MOLREP (Vagin & Teplyakov, 2010 ▶) and other CCP4 programs (Winn et al., 2011 ▶). The MR search model was a model of a Cje aOTC monomer generated by SWISS-MODEL (Arnold et al., 2006 ▶) based on the crystal structure of P. furiosus aOTC (PDB entry 1a1s; Villeret et al., 1998 ▶). There were six monomers in the asymmetric unit. The structure was also determined using SAD to locate the 78 Se atoms in the asymmetric unit, but the resulting model was less complete. Owing to difficulties in model building with 2.7 Å resolution data and a large asymmetric unit, the molecular-replacement solution was chosen as the starting point for refinement. The SAD maps were used to validate the model and verify the positions of the Se atoms. The SAD phasing and model building was performed with HKL-3000 coupled with SHELX (Sheldrick, 2008 ▶), MLPHARE (Otwinowski, 1991 ▶), Buccaneer (Cowtan, 2006 ▶), DM (Cowtan, 1994 ▶) and other CCP4 programs.
The structure was refined with REFMAC5 (Murshudov et al., 1997 ▶) in the restrained mode with isotropic B factors and H atoms in riding positions. Automatic local NCS as implemented in v.5.6 of REFMAC5 was applied throughout refinement (Murshudov et al., 2011 ▶). Six TLS groups were introduced, one group for each monomer. Water molecules were not included in the TLS groups. Coot (Emsley & Cowtan, 2004 ▶) was used for the visualization of electron-density maps and manual corrections of the atomic model. MolProbity (Chen et al., 2010 ▶) and ADIT (Yang et al., 2004 ▶) were used for structure validation. Diffraction images were deposited and can be downloaded from the CSGID webpage (http://www.csgid.org/csgid/pages/diffraction_images). Data-collection, structure-determination and refinement statistics are summarized in Table 1 ▶. The structure has been deposited in the PDB with code 3tpf.
Table 1. Summary of data-collection, structure-determination and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength (Å) | 0.9793 |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 157.2, b = 87.4, c = 141.6 |
| Resolution (Å) | 50.00–2.70 (2.77–2.70) |
| No. of unique reflections | 54348 (2709) |
| Completeness (%) | 99.9 (99.8) |
| Multiplicity | 7.8 (7.2) |
| Mean I/σ(I) | 33.4 (3.0) |
| Molecules per asymmetric unit | 6 |
| Matthews coefficient (Å3 Da−1) | 2.27 |
| Solvent content (%) | 45.8 |
| R merge † (%) | 8.9 (68.1) |
| Structure refinement | |
| Resolution range (Å) | 50.00–2.70 (2.77–2.70) |
| R work/R free (%) | 20.2/24.0 |
| No. of residues/protein atoms | 1818/13857 |
| No. of water molecules | 104 |
| No. of ligand atoms | 21 |
| Average B factors (Å2) | |
| Protein | 80.7 |
| Water | 49.4 |
| Ligands | 82.7 |
| Ramachandran plot‡ (%) | |
| Favored | 96.3 |
| Additionally favored | 3 |
| Outliers | 0.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.8 |
Bijvoet pairs were merged for calculation of R merge.
Ramachandran plot statistics were calculated by MolProbity.
2.5. Computational methods
The PISA server at the European Bioinformatics Institute (Krissinel & Henrick, 2007 ▶) was used to determine the oligomeric assembly, buried intersubunit surface areas and interacting residues for analyzed structures. Differences in the respective domain rotation between Cje OTC subunits were calculated using the DynDom server (Hayward & Berendsen, 1998 ▶). The rotation angle was calculated with the domains defined as the core β-strands of each domain. LSQKAB (Kabsch, 1976 ▶) as implemented in Coot was used to calculate the r.m.s.d. for Cα atoms between Cje aOTC subunits. Secondary-structure elements were assigned with PROMOTIF (Hutchinson & Thornton, 1996 ▶). Structure-driven sequence alignment was performed as follows: a structural alignment was prepared using the PDBeFold service using chain A of the analyzed depositions and the corresponding sequence alignment was then inspected and corrected manually. Full sequences (accounting for disordered residues) were obtained from UniProt and were aligned with the profile resulting from the previous alignment using ClustalW (Larkin et al., 2007 ▶). The online chemical editor MarvinSketch (ChemAxon) was used for the preparation of Fig. 1, PyShade (Porebski et al., manuscript in preparation) was used for the preparation of Fig. 3 and PyMOL (Schrödinger LLC) was used to generate Figs. 2, 4 and 5.
3. Results and discussion
3.1. Structure quality
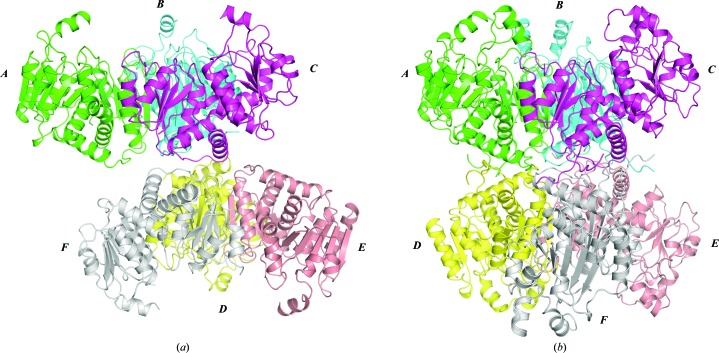
There are six monomers in the asymmetric unit of the Cje aOTC crystal structure (Fig. 2 ▶ a). According to the amino-acid sequence (Fig. 3 ▶), each Cje aOTC monomer consists of 306 amino-acid residues (UniProt accession No. Q9PNU6). With the exception of two to four C-terminal residues per monomer and the few disordered residues described below, all residues could be modeled into the electron density. The highly disordered catalytic SMG loop (residues 223–230) has poor electron density and two residues of the loop were not modeled in chains C, E and F. In chain F residues 74–76 of the catalytic B2–H3 loop also could not be modeled.
Figure 2.
(a) The pseudohexamer located in the asymmetric unit of the Cje aOTC structure. The threefold noncrystallographic axis of the ABC trimer is vertical. Colors are assigned by monomer. (b) Hexamer of the Lhi cOTC structure with the same orientation of the ABC trimer and the same color scheme as in (a).
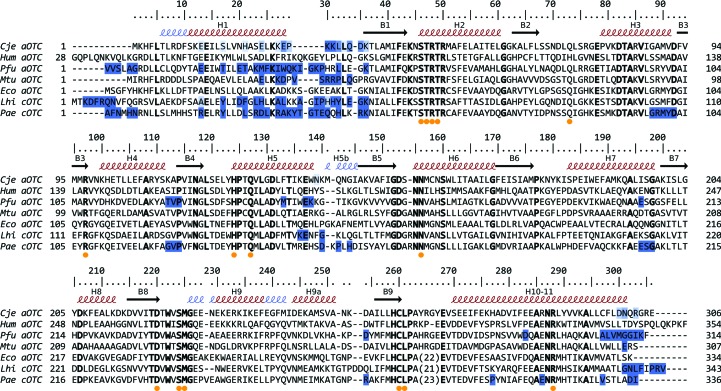
Figure 3.
Structure-driven sequence alignment of OTCases from C. jejuni (Cje aOTC), human (Hum aOTC), P. furiosus (Pfu aOTC), M. tuberculosis (Mtu aOTC), E. coli (Eco aOTC), L. hilgardii (Lhi cOTC) and P. aeruginosa (Pae cOTC). Residue numbering and secondary-structure elements are presented for Cje aOTC. β-Strands are shown as arrows; α-helices and 310-helices are shown as red and blue coils, respectively. Residues that are proposed to be involved in substrate binding are marked with an orange circle. Conserved residues are highlighted in bold. Residues that form the interface between OTC trimers in the respective hexamer, pseudohexamer or dodecamer are marked in blue. The intensity of the color corresponds to the percentage of chains in the oligomeric assembly in which the residue participates in the interface between trimers. Eco aOTC and Hum aOTC do not form this type of oligomeric assembly; therefore, they were not analysed.
The refined model has good overall geometry, with a low percentage of rotamer outliers and a low MolProbity clash score (Table 1 ▶). The structure has no Ramachandran plot outliers, with the exceptions of Leu119 and Leu261. These residues are highly conserved in OTC structures (Fig. 3 ▶) and the reasons for their unusual conformation have previously been discussed in the literature (Shi et al., 1998 ▶).
3.2. Oligomeric assembly
The oligomeric assembly of the Cje aOTC structure is a trimer (Fig. 4 ▶ a) as defined by the PISA server. The monomers in the trimer are related by a threefold noncrystallographic axis with an average of 890 Å2 buried interface area between two adjacent monomers. Two head-to-head stacked trimers form a pseudohexamer in the asymmetric unit (Fig. 2 ▶ a). The concave faces of the trimers, where the active sites are located, are exposed to large solvent cavities formed by crystal packing, while the opposite convex faces constitute the contacting interfaces between the trimers. This pseudohexameric assembly is somewhat similar to the hexameric structure of the catabolic OTC from L. hilgardii (Lhi cOTC) presented in Fig. 2 ▶(b). In the Lhi cOTC hexamer two stacked trimers are related by a twofold symmetry axis and they are rotated 60° with respect to each other along the molecular threefold symmetry axis (PDB entry 2w37; de Las Rivas et al., 2009 ▶). Distinctively, in the Cje aOTC pseudohexamer there is only a slight rotation and the trimers are not coaxial (Fig. 2 ▶). Moreover, the contact area between the Cje aOTC trimers is small. The highest value of the buried surface area at the interface between monomers from adjacent trimers is only 200 Å2, whereas in Lhi cOTC the average value of the buried surface area at the interface between contacting monomers is approximately 1260 Å2.
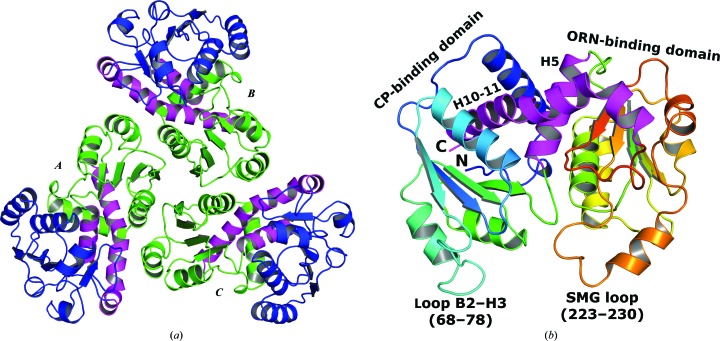
Figure 4.
(a) ABC trimer of Cje aOTC with the threefold noncrystallographic axis perpendicular to the plane of the figure. The view is from the concave face of the trimer (from the upper side of Fig. 2 ▶ a). The CP-binding domain is depicted in green, interdomain helices H5 and H10-11 are depicted in magenta and the ORN-binding domains are depicted in blue. (b) Monomer structure of Cje aOTC (subunit D). Interdomain helices are depicted in magenta. The N- and C-terminal ends, the interdomain helices and the two flexible catalytic-site loops are labeled.
In OTC structures which form a dodecameric (Pfu aOTC and Pae aOTC) or a hexameric assembly (Lhi cOTC), mainly C-terminal and N-terminal residues, together with a few residues from the middle of the polypeptide chain, participate in the interface between trimers (Fig. 3 ▶). As pointed out by de Las Rivas et al. (2009 ▶), the last eight C-terminal residues of each subunit of the Lhi cOTC hexamer constitute a key structural element for the stabilization of the hexameric assembly by forming a number of hydrophobic interactions and salt bridges. These residues are specific to Lhi cOTC and, with the exception of human aOTC, all of the OTCs with known three-dimensional structures, including Cje aOTC, have a shorter C-terminus. There are four structures of human aOTC in the PDB, but none have a pseudohexameric assembly similar to that of Lhi cOTC. One human aOTC structure complexed with CP forms a pseudohexameric assembly, but it has a head-to-tail topology (PDB entry1fvo; Shi et al., 2001 ▶). On the other hand, Mtu aOTC (PDB entries 2i6u and 2p2g; Sankaranarayanan et al., 2008 ▶) represents a further example of a bacterial OTC that forms a pseudohexamer similar to the Lhi cOTC hexamer. It can be speculated that the longer C-terminal fragment of human aOTC prevents the formation of a head-to-head pseudohexamer because it is comprised of primarily bulky hydrophilic residues (Fig. 3 ▶). In Cje aOTC and Mtu aOTC there are no additional C-terminal residues which potentially prevent the formation of a hexamer. Anyway, the interface is too small to lead to the formation of a hexamer, even if crystal formation resulted in a suitable orientation of the trimers. It can be speculated that a few mutations in the bacterial aOTC may lead to the formation of a hexamer.
3.3. Monomer structure
As in all other OTC structures, the monomer of Cje aOTC has two domains: an N-terminal CP-binding domain (residues 1–123) and a C-terminal ORN-binding domain (residues 139–269). The domains are connected by two interdomain helices H5 (residues 124–138) and H10-11 (270–301), with the C-terminal helix H10-11 penetrating the CP-binding domain (Fig. 4 ▶ b). Each domain is formed by a parallel β-sheet surrounded by α-helices and loops with α/β topology. The CP-binding domain has four β-strands, and the ORN-binding domain has five β-strands. The active site is located in the cleft between the two domains and residues from both domains are involved in catalysis. The CP-binding domains are located in the interior of the trimer and together with the interdomain helix H10-11 form the majority of the intersubunit interactions (Fig. 4 ▶ a).
The six monomers of the asymmetric unit of the Cje aOTC crystal display very similar structural organization, showing r.m.s.d. values in the range 0.2–0.4 Å for all aligned Cα atoms in pairwise alignments. It was previously shown by comparison of nonliganded and liganded OTC structures that binding of the bisubstrate analogue or both substrates results in closure of the two domains by a relative rotation of one domain versus the other by 4–9° (Ha et al., 1997 ▶; Shi et al., 2001 ▶; Sankaranarayanan et al., 2008 ▶). Accordingly, Cje aOTC has the open domain conformation, with slightly different orientations of the domains in the subunits. The largest difference corresponds to a 3.4° rotation of the ORN-binding domain to the respective CP-binding domain of subunit D in comparison with subunit F. This difference stresses the relative flexibility of the open conformation of OTCs.
Cje aOTC has 40.5% sequence identity to Lhi cOTC, 40.2% to P. furiosus aOTC, 37.9% to E. coli aOTC and 38.2% to human aOTC (Fig. 3 ▶). As in all other OTCs, Cje aOTC has the strictly conserved CP-binding motif STRTR (residues 46–50) and the ORN-binding motifs DxxxSMG (residues 220–226) and (ML)HCLP (residues 258–262). A DALI (Hasegawa & Holm, 2009 ▶) search identified P. furiosus aOTC as the closest structure (PDB entry 1a1s; Villeret et al., 1998 ▶) with a Z-score of 41.4 and an r.m.s.d. of 1.5 Å for 292 aligned Cα atoms. Cje aOTC is a potential drug target for diarrhea therapy so we will briefly describe its major differences from the structure of human aOTC (Fig. 5 ▶). There are no unliganded structures of human aOTC; thus, we are comparing the unliganded structure of Cje aOTC with the closest structure of human aOTC determined by the DALI server. This is the structure of human aOTC complexed with CP and l-norvaline (PDB entry 1c9y; Shi et al., 2000 ▶), which has a Z-score of 40.3 and an r.m.s.d. of 1.6 Å for 292 aligned Cα atoms. The main differences in the polypeptide fold are as follows.
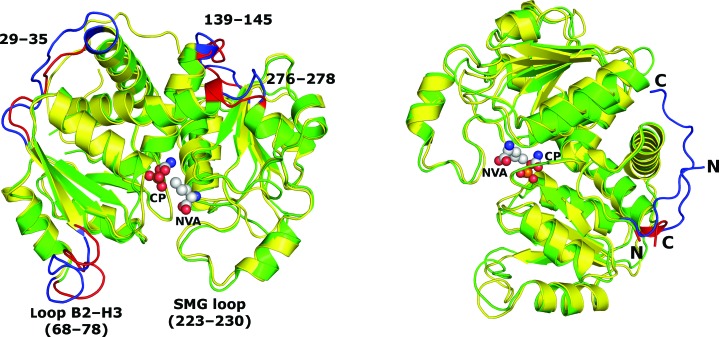
(i) Mature human aOTC has six more residues at the N-terminus (UniProt accession No. P00480), although the first residue was not modeled in the 1c9y structure.
(ii) The H1 helix is one turn shorter in Cje aOTC, which results in a slightly different conformation of the H1–B1 loop (residues 29–35).
(iii) The catalytic-site loop B2–H3 (residues 68–78) has a different conformation in Cje aOTC, which is related to CP binding in the structure of the human enzyme.
(iv) The H5–B5 loop (residues 139–145) at the interdomain bending region is one residue longer in Cje aOTC and forms one loop of a 310-helix (H5b).
(v) Residues 276–278, which constitute the second interdomain bending region in most OTC structures, is more structured in Cje aOTC, which combines helices H10 and H11 as a long, albeit kinked, C-terminal helix H10-11 (Fig. 4 ▶ b).
(vi) Human aOTC has an additional six residues at the C-terminal end and all of them have been modeled into the electron density in the 1c9y structure. In contrast, the last residue of every chain in Cje OTC could not be modeled.
Figure 5.
Superposition of Cje aOTC subunit D (green) and human aOTC (yellow; PDB entry 1c9y) in two different orientations. Carbamoyl phosphate (CP) and l-norvaline (NVA) from the human aOTC structure are shown in spherical representation. Major differences in the tertiary structures are highlighted in red for Cje aOTC and in blue for human aOTC and are labelled with Cje aOTC residue numbers. The N- and C-terminal ends of the structures are marked with the respective letters.
3.4. The active site
The active site of Cje aOTC consists of residues from both the CP-binding and the ORN-binding domains of the subunit and the B2–H3 loop (residues 68–78) from an adjacent subunit of the trimer. The active-site residues that form hydrogen bonds to the substrates in human aOTC are all conserved, with the exception of Gln73 in the B2–H3 loop, which is structurally equivalent to His117 of the human enzyme (Fig. 3 ▶). The side chain of this glutamine or histidine participates in substrate binding via a hydrogen bond to the phosphate moiety of CP, anchoring the loop in a similar conformation in all OTC structures that contain this ligand. The unliganded OTC structures demonstrate a variety of B2–H3 loop conformations, which is consistent with ordering of this loop upon CP binding (De Gregorio et al., 2003 ▶). In the Cje aOTC structure this loop is disordered and most of the side chains could not be modeled in the electron-density map. Consequently, modeled atoms in this region have high B factors. Even the main-chain atoms of residues 74–76 are not modeled in subunit F. The conformation of the B2–H3 loop is similar in all of the other Cje OTC subunits and is different from the conformation of the loop in CP-complexed OTCs (Fig. 5 ▶), with the displacement of Gln73 Cα being about 6 Å (Shi et al., 2000 ▶; De Gregorio et al., 2003 ▶).
Residues 223–230 constitute the SMG loop, so called because it contains the SMG residues of the conserved ORN-binding motif DxxxSMG. This loop has different conformations or is disordered in OTC structures without the second substrate (ORN) or its analogs bound. On the other hand, OTC binary complexes with PALO and tertiary complexes with CP and l-norvaline show identical SMG-loop conformations (Ha et al., 1997 ▶; Shi et al., 2000 ▶; Langley et al., 2000 ▶; Sankaranarayanan et al., 2008 ▶). Superposition of the binary and ternary complexes of human aOTC demonstrated that the SMG loop shifts by more than 8 Å upon binding of the second substrate analog (Shi et al., 2001 ▶). The movement of this loop seems to be essential for binding the second substrate and release of the products. In the Cje aOTC structure the SMG loop has very poor electron density, but the traced conformation is similar to those of the ternary complexes of other OTCs (Fig. 5 ▶). The loop is located on concave faces of the trimers and forms different crystallographic contacts with other subunits. We feel that the presence of these crystal contacts may be the reason that we observe a loop structure resembling the catalytically active loop structure even in the absence of bound substrate.
4. Conclusions
The crystal structure of C. jejuni anabolic ornithine transcarbamoylase displays the canonical OTC fold and has the trimeric oligomeric assembly that is found for many other anabolic OTCs. The asymmetric unit contains a head-to-head pseudohexamer that is similar to the hexameric structure of the catabolic OTC from L. hilgardii that is formed owing to specific additional C-terminal residues. The monomers in the asymmetric unit show differences in the relative orientation of the CP-binding and ORN-binding domains, reflecting the flexibility of the unliganded state of aOTCs, which has an open conformation. The catalytic B2–H3 loop, which is provided by an adjacent subunit of the trimer and participates in CP binding, has a different conformation in Cje aOTC from that in the CP-liganded state of the enzyme from other species. Thus, the Cje aOTC structure supports the previously reported observation of conformational change and ordering of the B2–H3 loop upon CP binding. The SMG loop (residues 223–230) containing the ORN-binding motif has weak electron density and presents a case where the loop is in a catalytically active conformation without ORN or its analogs bound. This fact supports the previous observations of high flexibility of the SMG loop.
Supplementary Material
PDB reference: anabolic ornithine carbamoyltransferase, 3tpf
Acknowledgments
This research was funded by Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Contract No. HHSN272200700058C. The results shown in this report are derived from work performed at Argonne National Laboratory at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by University of Chicago Argonne LLC for the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
References
- Anderson, W. F. (2009). Infect. Disord. Drug Targets, 9, 507–517. [DOI] [PMC free article] [PubMed]
- Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. (2006). Bioinformatics, 22, 195–201. [DOI] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Baur, H., Tricot, C., Stalon, V. & Haas, D. (1990). J. Biol. Chem. 265, 14728–14731. [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 31, 34–38.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Cunin, R., Glansdorff, N., Piérard, A. & Stalon, V. (1986). Microbiol. Rev. 50, 314–352. [DOI] [PMC free article] [PubMed]
- De Gregorio, A., Battistutta, R., Arena, N., Panzalorto, M., Francescato, P., Valentini, G., Bruno, G. & Zanotti, G. (2003). Org. Biomol. Chem. 1, 3178–3185. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Eschenfeldt, W. H., Lucy, S., Millard, C. S., Joachimiak, A. & Mark, I. D. (2009). Methods Mol. Biol. 498, 105–115. [DOI] [PMC free article] [PubMed]
- Fagerquist, C. K., Bates, A. H., Heath, S., King, B. C., Garbus, B. R., Harden, L. A. & Miller, W. G. (2006). J. Proteome Res. 5, 2527–2538. [DOI] [PubMed]
- Galkin, A., Kulakova, L., Wu, R., Gong, M., Dunaway-Mariano, D. & Herzberg, O. (2009). Proteins, 76, 1049–1053. [DOI] [PMC free article] [PubMed]
- Ha, Y., McCann, M. T., Tuchman, M. & Allewell, N. M. (1997). Proc. Natl Acad. Sci. USA, 94, 9550–9555. [DOI] [PMC free article] [PubMed]
- Hasegawa, H. & Holm, L. (2009). Curr. Opin. Struct. Biol. 19, 341–348. [DOI] [PubMed]
- Haun, R. S., Serventi, I. M. & Moss, J. (1992). Biotechniques, 13, 515–518. [PubMed]
- Hayward, S. & Berendsen, H. J. (1998). Proteins, 30, 144–154. [PubMed]
- Hutchinson, E. G. & Thornton, J. M. (1996). Protein Sci. 5, 212–220. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1976). Acta Cryst. A32, 922–923.
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Langley, D. B., Templeton, M. D., Fields, B. A., Mitchell, R. E. & Collyer, C. A. (2000). J. Biol. Chem. 275, 20012–20019. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Las Rivas, B. de, Fox, G. C., Angulo, I., Ripoll, M. M., Rodríguez, H., Muñoz, R. & Mancheño, J. M. (2009). J. Mol. Biol. 393, 425–434. [DOI] [PubMed]
- Lipscomb, W. N. & Kantrowitz, E. R. (2011). Acc. Chem. Res. 45, 444–453. [DOI] [PMC free article] [PubMed]
- Massant, J., Wouters, J. & Glansdorff, N. (2003). Acta Cryst. D59, 2140–2149. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nguyen, V. T., Baker, D. P., Tricot, C., Baur, H., Villeret, V., Dideberg, O., Gigot, D., Stalon, V. & Haas, D. (1996). Eur. J. Biochem. 236, 283–293. [DOI] [PubMed]
- Otwinowski, Z. (1991). Proceedings of the CCP4 Study Weekend. Isomorphous Replacement and Anomalous Scattering, edited by W. Wolf, P. R. Evans & A. G. W. Leslie, pp. 80–86. Warrington: Daresbury Laboratory.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sainz, G., Tricot, C., Foray, M. F., Marion, D., Dideberg, O. & Stalon, V. (1998). Eur. J. Biochem. 251, 528–533. [DOI] [PubMed]
- Sankaranarayanan, R., Cherney, M. M., Cherney, L. T., Garen, C. R., Moradian, F. & James, M. N. G. (2008). J. Mol. Biol. 375, 1052–1063. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, D., Morizono, H., Aoyagi, M., Tuchman, M. & Allewell, N. M. (2000). Proteins, 39, 271–277. [PubMed]
- Shi, D., Morizono, H., Ha, Y., Aoyagi, M., Tuchman, M. & Allewell, N. M. (1998). J. Biol. Chem. 273, 34247–34254. [DOI] [PubMed]
- Shi, D., Morizono, H., Yu, X., Tong, L., Allewell, N. M. & Tuchman, M. (2001). Biochem. J. 354, 501–509. [DOI] [PMC free article] [PubMed]
- The UniProt Consortium (2011). Nucleic Acids Res. 39, D214–D219. [DOI] [PMC free article] [PubMed]
- Tricot, C., Nguyen, V. T. & Stalon, V. (1993). Eur. J. Biochem. 215, 833–839. [DOI] [PubMed]
- Tricot, C., Villeret, V., Sainz, G., Dideberg, O. & Stalon, V. (1998). J. Mol. Biol. 283, 695–704. [DOI] [PubMed]
- Tuchman, M. (1993). Hum. Mutat. 2, 174–178. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Villeret, V., Clantin, B., Tricot, C., Legrain, C., Roovers, M., Stalon, V., Glansdorff, N. & Van Beeumen, J. (1998). Proc. Natl Acad. Sci. USA, 95, 2801–2806. [DOI] [PMC free article] [PubMed]
- Villeret, V., Tricot, C., Stalon, V. & Dideberg, O. (1995). Proc. Natl Acad. Sci. USA, 92, 10762–10766. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, H., Guranovic, V., Dutta, S., Feng, Z., Berman, H. M. & Westbrook, J. D. (2004). Acta Cryst. D60, 1833–1839. [DOI] [PubMed]
- Zimmerman, M., Chruszcz, M., Koclega, K., Otwinowski, Z. & Minor, W. (2005). Acta Cryst. A61, C178–C179.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: anabolic ornithine carbamoyltransferase, 3tpf