The NheA component of the B. cereus Nhe toxin was overexpressed in E. coli, purified and crystallized. Diffraction data were collected and processed to 2.05 Å resolution.
Keywords: toxins, food poisoning, nonhaemolytic enterotoxins, haemolysins
Abstract
The nonhaemolytic enterotoxin (Nhe) of Bacillus cereus plays a key role in cases of B. cereus food poisoning. The toxin is comprised of three different proteins: NheA, NheB and NheC. Here, the expression in Escherichia coli, purification and crystallization of the NheA protein are reported. The protein was crystallized by the sitting-drop vapour-diffusion method using PEG 3350 as a precipitant. The crystals of NheA diffracted to 2.05 Å resolution and belonged to space group C2, with unit-cell parameters a = 308.7, b = 58.2, c = 172.9 Å, β = 110.6°. Calculation of V M values suggests that there are approximately eight protein molecules per asymmetric unit.
1. Introduction
Bacillus cereus is a well known food-poisoning organism that causes both emetic and diarrhoeal illness (Stenfors Arnesen et al., 2008 ▶). The bacterium is responsible for the production of several cytolytic toxins, including cytolysin K (CytK), haemolysin BL (Hbl) and the nonhaemolytic enterotoxin (Nhe) (Stenfors Arnesen et al., 2008 ▶). CytK is a single-component protein toxin (Hardy et al., 2001 ▶; Lund et al., 2000 ▶), while the Hbl and Nhe toxins each consist of three different proteins (Beecher & Macmillan, 1991 ▶; Lindbäck et al., 2004 ▶). The Hbl toxin consists of a binding protein (Hbl-B; 41 kDa) and two lytic components, L1 (38 kDa) and L2 (43.5 kDa) (Ehling-Schulz et al., 2006 ▶; Madegowda et al., 2008 ▶). The Nhe toxin of B. cereus was identified from a large food-poisoning outbreak in Norway in 1995; the strain recovered, NVH 0075/95, does not contain genes producing CytK and Hbl (Ehling-Schulz et al., 2005 ▶).
B. cereus nonhaemolytic enterotoxin (Nhe) is a complex pore-forming toxin consisting of three homologous proteins, NheA (41 kDa), NheB (39 kDa) and NheC (40 kDa), and is encoded by one operon containing three genes: nheA, nheB and nheC. All three proteins are required for maximal toxicity (Lindbäck et al., 2004 ▶). Initially, Nhe was incorrectly thought to lack haemolytic activity, hence the name ‘nonhaemolytic’. However, further studies demonstrated that the toxin was able to lyse mammalian erythrocytes from various organisms, including human, horse, cat, cow, dog and pig (Fagerlund et al., 2008 ▶). In addition, Nhe has cytotoxic activity against epithelial cells, as indicated by loss of cellular adenosine 5′-triphosphate (ATP) and lactate dehydrogenase (LDH) (Haug et al., 2010 ▶). It has also been demonstrated that all three components, NheA, NheB and NheC, are required for optimal cytotoxic activity in Vero cells (Lindbäck et al., 2004 ▶). The detailed mechanism behind the pore-formation of the toxin is unknown, but is presumed to follow a pattern involving membrane binding, oligomerization and finally insertion of the transmembrane regions to form the pore. NheB has previously been shown to be a membrane-binding component (Lindbäck et al., 2004 ▶) and it has recently been shown that NheC also binds to the membrane (Lindbäck et al., 2010 ▶). NheB and NheC share 44% sequence identity, but NheA has only 22% identity to NheB and NheC and its binding is the final stage of pore formation (Lindbäck et al., 2010 ▶).
NheA, NheB and NheC also have 20–40% sequence identity to Hbl-L2, Hbl-L1 and Hbl-B (Fagerlund et al., 2008 ▶), respectively, which together comprise the second B. cereus tripartite toxin, known as Hbl, as described above. The structure solution of Hbl-B (Madegowda et al., 2008 ▶) revealed a structural resemblance to the monopartite pore-forming toxin cytolysin A (ClyA, HlyE; Fagerlund et al., 2008 ▶) from Escherichia coli. The structures of both toxins consist of an elongated four-helix bundle with two unusual elaborations, one at each end. Firstly, the ‘tail domain’ includes an extra helix, producing a five-helix bundle of unusual topology. Secondly, the ‘head domain’ consists of a hydrophobic β-hairpin and two short α-helices. The topologies of both these additional features were originally thought to be unique to ClyA (Wallace et al., 2000 ▶), but are now known to also be present in Hbl-B (Madegowda et al., 2008 ▶; Fagerlund et al., 2008 ▶). However, ClyA has a substantially different structure in its membrane-bound form, undergoing very extensive structural changes to form a dodecameric α-helical transmembrane pore in which the protomers are largely three-helix bundles with no β-sheet (Mueller et al., 2009 ▶). Similar structural changes may also occur in at least some of the components of the Hbl and Nhe toxins.
Nhe and ClyA have functional similarities in that both are cytolytic to epithelia and form large-conductance channels in planar lipid bilayers (Lai et al., 2000 ▶; Ludwig et al., 1995 ▶, 1999 ▶; Oscarsson et al., 2002 ▶). These three toxins therefore represent a novel superfamily of α-helical pore-forming toxins (Fagerlund et al., 2008 ▶). Like both ClyA (Wallace et al., 2000 ▶) and Hbl-B (Madegowda et al., 2008 ▶), the soluble form of NheA is expected to be predominantly α-helical with <10% β-sheet. However, sequence analyses show that NheA is predicted to have an amphipathic rather than a hydrophobic β-sheet (Fagerlund et al., 2008 ▶).
To date, there is limited knowledge concerning the roles of the three components of Nhe. Three-dimensional structures of these proteins would allow better understanding of the mechanism of pore formation by the Nhe toxin. This report presents the first step in the structural investigation of the Nhe toxins, namely the crystallization and initial crystallographic characterization of the 41 kDa NheA component.
2. Materials and methods
2.1. Cloning
The target gene for NheA was amplified by PCR using B. cereus NVH 0075/95 as template. PCR was carried out with DyNAzyme II DNA polymerase (supplied with 10× buffer) and deoxynucleotide triphosphate (dNTP) mixture from Finnzymes (Finland) as instructed by the manufacturer using the primers NVH1339-F, 5′-GTGAAAAAGACTTTAATTACAGG-3′, and NVH1340-R, 5′-TTAATGTACTTCAACGTTTGTAA-3′. The PCR product was cloned into pEXP5-CT/TOPO vector (Invitrogen).
2.2. Protein expression and purification
NheA expression vector was transformed into E. coli strain BL21 (DE3) cells and the cells were grown at 310 K in Luria–Bertani (LB) medium containing ampicillin (0.1 mg ml−1). When the culture reached an OD600 of 0.6–0.8, NheA expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mg ml−1. The cells were grown for a further 20 h and were harvested by centrifugation at 15 344g for 20 min at 277 K. The cell pellets were then stored at 253 K.
The following protocol was used for the purification of NheA. The cells were thawed and suspended in buffer A (50 mM Tris–HCl pH 8.0). The cells were then disrupted by ultrasonication on ice, with a short pause to prevent heating, for 3 × 29 s. Cell debris was removed by centrifugation at 70 000g for 10 min. The supernatant fraction was loaded onto a HiPrep 16/10 DEAE FF (GE Healthcare) anion-exchange column and proteins were eluted with a 200 ml 0–0.4 M NaCl gradient in buffer A. 6 ml fractions were collected and analysed using SDS–PAGE. Fractions containing NheA eluted at about 0.13 M NaCl and were combined and applied directly onto a 10 ml column packed with hydroxylapatite (Bio-Rad Laboratories; Bio-Gel HT gel) equilibrated with 10 mM NaCl. Proteins were eluted with a 70 ml gradient of 0–0.2 M sodium phosphate pH 6.8. 4 ml fractions were collected and analysed using SDS–PAGE. Fractions containing NheA eluted at approximately 0.12 M sodium phosphate and were combined and concentrated using a Vivaspin concentrator (Sartorius) with a 10 kDa molecular-weight cutoff to reduce the volume to 2 ml. Gel filtration on a HiLoad 16/60 Superdex 200 column (GE Healthcare) was used as a polishing step and was performed at a 1.5 ml min−1 flow rate in buffer A supplemented with 0.5 M NaCl. 2 ml fractions were collected and analysed by SDS–PAGE. The fractions containing the purest NheA were combined and NheA was concentrated to 10–11 mg ml−1 using a Vivaspin concentrator (as above).
2.3. Crystallization
To prepare the NheA sample for crystallization, the buffer in the sample was exchanged for 10 mM sodium phosphate pH 6.8 using a 0.5 ml Zeba Desalting Column (Thermo Scientific). The final protein concentration of the NheA sample was 9–10 mg ml−1 as estimated by the method of Bradford (1976 ▶).
Initial protein crystallization trials were set up at 290 K by a Matrix Hydra II Plus One crystallization robot using the vapour-diffusion sitting-drop method with the JSCG+ Suite and PACT crystallization screens (Qiagen). A 24-well plate (Hampton Research) was used for optimization in the sitting-drop mode. The droplets were prepared by mixing 1 or 1.5 µl of protein solution and reservoir solution in a 1:1 ratio and were equilibrated against 1 ml reservoir solution at 290 K.
2.4. X-ray diffraction, data collection and processing
X-ray diffraction data were collected from crystals flash-cooled in a stream of nitrogen gas at 100 K using an Oxford Cryosystems Cryostream device. 25% ethylene glycol (Hampton Research) was used as a cryoprotectant. X-ray diffraction data were collected on the I03 beamline at the Diamond Light Source, Oxford, England. The data-collection strategy was optimized using iMOSFLM (Battye et al., 2011 ▶). An initial data-collection run of 450 × 0.5° rotation images was interrupted twice by technical problems involving loss of the synchrotron beam and 199 images were discarded. The same 225° of data were then recollected but this time completely and without interruption. The available diffraction data were processed using XDS (Kabsch, 2010 ▶) and scaled with SCALA (Evans, 2011 ▶) from the CCP4 package (Winn et al., 2011 ▶).
3. Results and discussion
NheA was successfully expressed in E. coli BL21 (DE3) strain with a level of expression in the range of 5–7% of the total soluble protein. The protein was purified to at least 90% purity as estimated by SDS–PAGE (Fig. 1 ▶). The yield was about 3 mg from a 1 l culture. Gel-filtration studies showed that NheA is a monomer in solution.
Figure 1.
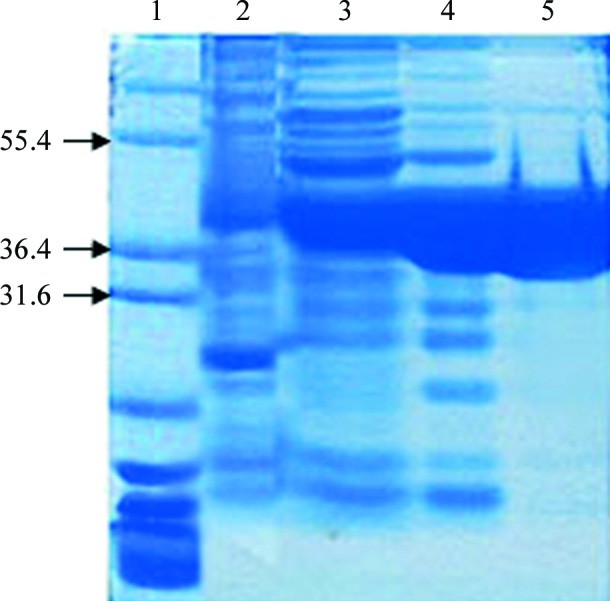
SDS–PAGE analysis of NheA purification. Lane 1, Marker12 (molecular-weights are indicated in kDa on the left); lane 2, cell extract; lane 3, after DEAE Sepharose column; lane 4, after hydroxylapatite (HA) column; lane 5, after gel-filtration column.
Initial crystallization trials yielded one prospective hit: needle-shaped crystals were observed in condition H8 of the JSCG+ Suite (0.2 M ammonium sulfate, 0.1 M bis-Tris pH 5, 25% PEG 3350). Optimization was performed by varying the PEG 3350 concentration and the buffer pH. The largest single crystal (Fig. 2 ▶), with dimensions of 0.08 × 0.05 × 0.5 mm, was obtained from 0.2 M ammonium sulfate, 0.1 M bis-Tris pH 7, 22%(w/v) PEG 3350 after 20 d.
Figure 2.

Large crystal of NheA of approximate dimensions 0.05 × 0.08 × 0.5 mm.
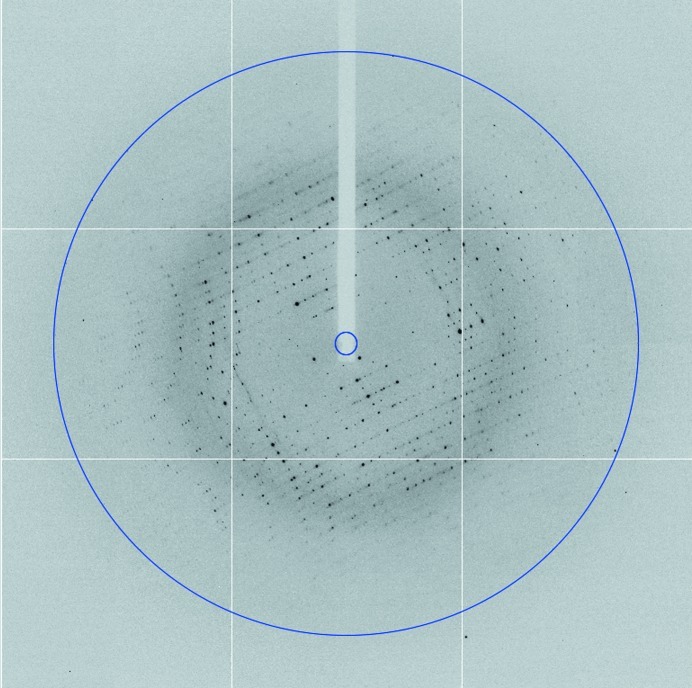
X-ray diffraction data were collected from the NheA crystals to 2.05 Å resolution (Fig. 3 ▶). The crystallographic parameters and data-collection statistics are listed in Table 1 ▶. The crystals belonged to space group C2, with unit-cell parameters a = 308.7, b = 58.2, c = 172.9 Å, β = 110.6°. A Matthews coefficient calculation (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) suggested that the number of NheA molecules in the asymmetric unit was most likely to be six, seven, eight or nine, with probabilities of 8, 24, 28 and 23%, respectively. A self-rotation function calculated at 5° intervals from 0° to 180° in κ was calculated using POLARRFN in order to investigate possible rotational pseudosymmetric relationships among the molecules in the asymmetric unit, but none were detected apart from the crystallographic twofold axis (data not shown). However, analysis of a self-Patterson function calculated with 50–6 Å resolution data showed two peaks, one with a height of 61% of the origin at (u, v, w) = (0.25, 0.25, 0.0) and another with a height of 41% of the origin at (u, v, w) = (0.5, 0.0, 0.0), indicating that the asymmetric unit contains molecules related by two different types of translational pseudo-symmetry. This suggests that the number of molecules in the asymmetric unit is a multiple of four and, given the Matthews coefficient calculation discussed above, the most probable number is therefore eight. This would give a crystal volume per protein mass (V M) of 2.2 Å3 Da−1 and a solvent content of 44.3%.
Figure 3.
X-ray diffraction pattern from an NheA crystal collected using an ADSC Q315R CCD detector at the Diamond synchrotron. The exposure time was 0.25 s, the crystal-to-detector distance was 259.0 mm and the oscillation range per frame was 0.5°. The outer edge of the image is at 1.79 Å resolution, but the data were truncated to 2.05 Å resolution (indicated by the blue circle).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 308.7, b = 58.2, c = 172.9, α = 90, β = 110.6, γ = 90 |
| Wavelength (Å) | 0.9686 |
| Resolution (Å) | 50.2–2.05 (2.16–2.05) |
| Completeness (%) | 98.9 (99.7) |
| Total No. of reflections | 504907 (74251) |
| No. of unique reflections | 179016 (26237) |
| Average I/σ(I) | 7.5 (2.5) |
| R merge † | 0.077 (0.358) |
| Solvent content‡ (%) | 44.3 |
| Matthews coefficient‡ (Å3 Da−1) | 2.21 |
R
merge = 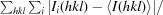
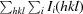 , where 〈I(hkl)〉 is the mean of the observations Ii(hkl) of reflection hkl.
, where 〈I(hkl)〉 is the mean of the observations Ii(hkl) of reflection hkl.
Assuming the presence of eight molecules of NheA in the asymmetric unit.
Attempts were made to solve the structure by molecular replacement using Phaser (McCoy, 2007 ▶) employing the soluble forms of ClyA (Wallace et al., 2000 ▶) and Hbl-B (Madegowda et al., 2008 ▶) and a protomer from the pore form of ClyA (Mueller et al., 2009 ▶) as search models, but in each case no solution was found. This is perhaps not unexpected given the low sequence similarity between NheA and Hbl-B (20% identity) and ClyA (18% identity) and the structural variability exhibited by ClyA (Wallace et al., 2000 ▶; Mueller et al., 2009 ▶) and probably by other members of the family, together with the complications that are likely to be caused by the pseudo-translational symmetry.
The availability of selenomethionine-derivative crystals should permit solution of the three-dimensional structure of NheA. This will open the way to a full structural analysis of the components of the Nhe toxin system and permit a greatly enhanced understanding of the mode of action of this important class of toxins to be developed.
Acknowledgments
MG was supported by KAU in Saudi Arabia. DP was supported by the Norwegian School of Veterinary Science, Oslo. DP is grateful for support from a FEMS Research Fellowship grant. We thank the Diamond Light Source for access to beamline I03 (BAG No. MX1218), which contributed to the results presented here, and we thank the beamline scientists for their help.
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Beecher, D. J. & Macmillan, J. D. (1991). Infect. Immun. 59, 1778–1784. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Ehling-Schulz, M., Guinebretiere, M.-H., Monthán, A., Berge, O., Fricker, M. & Svensson, B. (2006). FEMS Microbiol. Lett. 260, 232–240. [DOI] [PubMed]
- Ehling-Schulz, M., Svensson, B., Guinebretiere, M.-H., Lindbäck, T., Andersson, M., Schulz, A., Fricker, M., Christiansson, A., Granum, P. E., Märtlbauer, E., Nguyen-The, C., Salkinoja-Salonen, M. & Scherer, S. (2005). Microbiology, 151, 183–197. [DOI] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Fagerlund, A., Lindbäck, T., Storset, A. K., Granum, P. E. & Hardy, S. P. (2008). Microbiology, 154, 693–704. [DOI] [PubMed]
- Hardy, S. P., Lund, T. & Granum, P. E. (2001). FEMS Microbiol. Lett. 197, 47–51. [DOI] [PubMed]
- Haug, T. M., Sand, S. L., Sand, O., Phung, D., Granum, P. E. & Hardy, S. P. (2010). J. Membr. Biol. 237, 1–11. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Lai, X.-H., Arencibia, I., Johansson, A., Wai, S. N., Oscarsson, J., Kalfas, S., Sundqvist, K. G., Mizunoe, Y., Sjöstedt, A. & Uhlin, B. E. (2000). Infect. Immun. 68, 4363–4367. [DOI] [PMC free article] [PubMed]
- Lindbäck, T., Fagerlund, A., Rødland, M. S. & Granum, P. E. (2004). Microbiology, 150, 3959–3967. [DOI] [PubMed]
- Lindbäck, T., Hardy, S. P., Dietrich, R., Sødring, M., Didier, A., Moravek, M., Fagerlund, A., Bock, S., Nielsen, C., Casteel, M., Granum, P. E. & Märtlbauer, E. (2010). Infect. Immun. 78, 3813–3821. [DOI] [PMC free article] [PubMed]
- Ludwig, A., Bauer, S., Benz, R., Bergmann, B. & Goebel, W. (1999). Mol. Microbiol. 31, 557–567. [DOI] [PubMed]
- Ludwig, A., Tengel, C., Bauer, S., Bubert, A., Benz, R., Mollenkopf, H. J. & Goebel, W. (1995). Mol. Gen. Genet. 249, 474–486. [DOI] [PubMed]
- Lund, T., De Buyser, M. L. & Granum, P. E. (2000). Mol. Microbiol. 38, 254–261. [DOI] [PubMed]
- Madegowda, M., Eswaramoorthy, S., Burley, S. K. & Swaminathan, S. (2008). Proteins, 71, 534–540. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Mueller, M., Grauschopf, U., Maier, T., Glockshuber, R. & Ban, N. (2009). Nature (London), 459, 726–730. [DOI] [PubMed]
- Oscarsson, J., Westermark, M., Löfdahl, S., Olsen, B., Palmgren, H., Mizunoe, Y., Wai, S. N. & Uhlin, B. E. (2002). Infect. Immun. 70, 5759–5769. [DOI] [PMC free article] [PubMed]
- Stenfors Arnesen, L. P., Fagerlund, A. & Granum, P. E. (2008). FEMS Microbiol. Rev. 32, 579–606. [DOI] [PubMed]
- Wallace, A. J., Stillman, T. J., Atkins, A., Jamieson, S. J., Bullough, P. A., Green, J. & Artymiuk, P. J. (2000). Cell, 100, 265–276. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.