Rab6A′(Q72L), a constitutively active GTP-binding form of Rab6A, was purified and crystallized. The crystals were found to belong to space group P22121, with unit-cell parameters a = 36.84, b = 96.78, c = 109.99 Å. The crystals were obtained at 293 K and diffracted to a resolution of 1.9 Å.
Keywords: Rab6A, Ras superfamily, small G proteins
Abstract
Rab6A, a member of the Ras superfamily of small G proteins, is involved in the regulation of vesicle trafficking, which is critical for endocytosis, cell differentiation and cell growth. Rab6A can exist in two isoforms termed Rab6A and Rab6A′. The substitution of Gln72 by Leu (Q72L) in the Rab6A family blocks GTP-hydrolysis activity, and this mutation usually causes the Rab6A protein to be in a constitutively active form. In this study, in order to understand the functional uniqueness of Rab6A′ and the molecular mechanism of the control of activity by GTP and GDP from the crystal structure, a Rab6A′(Q72L) mutant form was overexpressed in Escherichia coli with an engineered N-terminal His tag. Rab6A′(Q72L) was then purified to homogeneity and crystallized at 293 K. X-ray diffraction data were collected to a resolution of 1.9 Å from a crystal belonging to space group P22121 with unit-cell parameters a = 36.84, b = 96.78, c = 109.99 Å. The asymmetric unit was estimated to contain two molecules.
1. Introduction
The Ras superfamily of small G proteins is a family of GTP hydrolase enzymes whose activity is regulated by the GTP-binding state, with GDP-bound inactive and GTP-bound active forms (Takai et al., 1992 ▶). Most of these enzymes are expressed ubiquitously inside cells and are key components of the molecular machinery, in which they participate in various cellular processes, including cytoskeletal organization, mitogenesis, vesicle trafficking and nuclear transport (Macara et al., 1996 ▶). The Rab GTPase family is part of the Ras superfamily of small G proteins and is responsible for vesicle trafficking, which is essential for endocytosis, biosynthesis, secretion, cell differentiation and cell growth (Bergbrede et al., 2009 ▶). The GTP-bound active form of the Rab protein can recruit specific binding partners, such as sorting adaptors, tethering factors, kinases, phosphatases and motors, and influence vesicle formation, transport and tethering (Grosshans et al., 2006 ▶). The Rab GTPase family has been extensively studied because functional loss of the Rab pathways has been implicated in a variety of diseases such as cancer, immunodeficiencies and neurological disorders (Stenmark, 2009 ▶).
Rab6A is a representative Rab GTPase family and resides in the membrane of the Golgi apparatus and the trans-Golgi network (TGN; Goud et al., 1990 ▶; Antony et al., 1992 ▶). Substitution of Gln72 by Leu (Q72L) in the Rab6A family blocks GTP-hydrolysis activity, and this mutation usually causes the Rab6 protein to be in a constitutively active form (Martinez et al., 1997 ▶). Rab6A has two isoforms termed Rab6A and Rab6A′ (Echard et al., 2000 ▶; Shan et al., 2000 ▶; Opdam et al., 2000 ▶); Rab6A′ is produced by alternative splicing of the duplicated exon within Rab6A (Echard et al., 2000 ▶). The sequence of human Rab6A′ differs from that of Rab6A by only three amino-acid residues (Val62→Ile, Thr87→Ala and Val88→Ala; Echard et al., 2000 ▶). Rab6A and Rab6A′ show similar GTP-binding activity and the active forms of these proteins inhibit the secretory pathway in vesicle transport (Echard et al., 2000 ▶). Although they appear similar, several studies have reported that the two isoforms are functionally different. For example, it is well known that Rab6A interacts with rabkinesin-6 but Rab6A′ does not (Echard et al., 1998 ▶, 2000 ▶). Although conflicting results have been reported regarding the roles of Rab6A′, it is believed that this protein plays unique roles in cellular processess (Echard et al., 2000 ▶; Young et al., 2005 ▶; Miserey-Lenkei et al., 2006 ▶; Del Nery et al., 2006 ▶; Mallard et al., 2002 ▶; Bergbrede et al., 2009 ▶).
Although several structures of Rab6A have been identified to date (Bergbrede et al., 2005 ▶), the structure of Rab6A′(Q72L) is not available. In the present study, we overexpressed, purified and crystallized Rab6A′(Q72L), which is a well known GTP-locked form (Martinez et al., 1997 ▶), as a first step towards the elucidation of the molecular structure and functional differences of Rab6A′. Details regarding the atomic structure of Rab6A′(Q72L) should enable us to understand the functional uniqueness of Rab6A′ and the molecular mechanism of control of activity by GTP and GDP.
2. Materials and methods
2.1. Expression and purification
To express the N-terminally His-tagged enzyme, the coding region for Rab6A′(Q72L) (GenBank ID NM_032144), which corresponds to residues 5–178, was cloned into pET15b. The plasmid was transformed into Escherichia coli BL21 (RIPL) competent cells and its expression in LB medium was induced by treatment with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K when the OD600 reached 0.62. Cells expressing Rab6A′(Q72L) were pelleted by centrifugation, resuspended and lysed by sonication in 50 ml lysis buffer (20 mM Tris pH 7.9, 500 mM NaCl, 20 mM imidazole). The lysate was then centrifuged at 16 000 rev min−1 for 30 min at 277 K, after which the supernatant fractions were applied onto a gravity-flow column (Bio-Rad) packed with Ni–NTA affinity resin (Qiagen). Next, the unbound bacterial proteins were removed from the column using lysis buffer (20 mM Tris pH 7.9, 500 mM NaCl, 20 mM imidazole). The N-terminally His-tagged Rab6A′(Q72L) was eluted from the column using elution buffer (20 mM Tris buffer pH 7.9, 500 mM NaCl, 250 mM imidazole). The elution fractions were collected as 0.5 ml volumes to a total volume of 2 ml. The collected Rab6A′(Q72L) was applied onto a Superdex 200 gel-filtration column (GE Healthcare) that had been pre-equilibrated with 20 mM Tris pH 8.0, 150 mM NaCl. Rab6A′(Q72L) (molecular mass 22 kDa) eluted at around 19 ml and was collected and concentrated to 10–12 mg ml−1. The protein concentration was measured using a protein-assay kit (Bio-Rad) and was determined using the Bradford method (Bradford, 1976 ▶). Purified Rab6A′(Q72L) contained the additional residues MGSSHHHHHHSSGLVPRGSHM at the N-terminus. The additional residues at the N-terminus, which included the hexahistidine tag, were not removed.
2.2. Crystallization
Crystallization conditions were initially screened at 293 K by the hanging-drop vapour-diffusion method using screening kits from Hampton Research (Crystal Screen and Crystal Screen 2). Initial crystals were grown on plates by equilibrating a mixture consisting of 1 µl protein solution (4.53 mg ml−1 protein in 20 mM Tris pH 8.0, 150 mM NaCl) and 1 µl of reservoir solution No. 46 from Crystal Screen (18% PEG 8000, 0.2 M calcium acetate, 0.1 M sodium cacodylate pH 6.5) against 0.4 ml reservoir solution. Following optimization, crystals appeared within 3 d and grew to maximum dimensions of 0.2 × 0.2 × 0.1 mm in the presence of 20% PEG 8000, 0.3 M calcium acetate, 0.1 M sodium cacodylate pH 6.7. The crystals diffracted to a resolution of 1.9 Å.
2.3. Crystallographic data collection
For data collection, the crystals were briefly soaked in a solution corresponding to the reservoir solution supplemented with 30%(v/v) glycerol and were then flash-cooled in liquid nitrogen. A 1.9 Å resolution native diffraction data set was collected from a single crystal at 110 K using a MAR CCD detector (crystal-to-detector distance 250 mm, 1° oscillation per image, total rotation angle 180°) on beamline BL-4A at the Pohang Accelerator Laboratory (PAL), Republic of Korea. The data sets were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The exact mechanism of the GTP- and GDP-mediated control of the activity of the Rab GTPase family is still unknown. To obtain a better understanding of this process, we overexpressed, purified and crystallized Rab6A′(Q72L), a well known GTP-locked form of the protein.
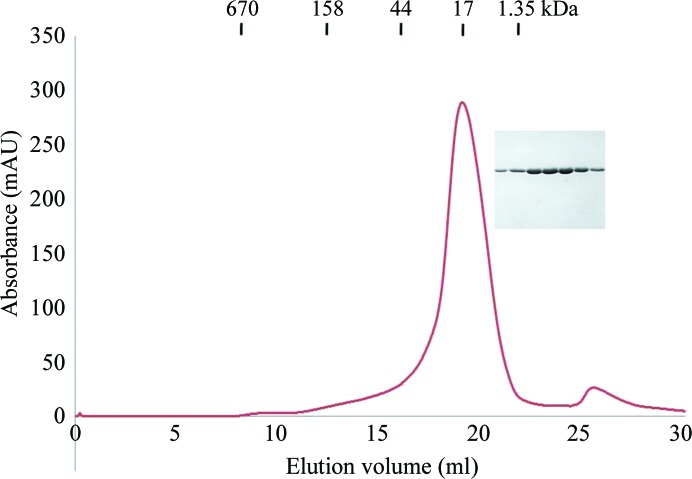
His-tag affinity chromatography followed by gel-filtration chromatography produced 90% pure Rab6A′(Q72L) and no contaminating bands were observed upon SDS–PAGE analysis (Fig. 1 ▶). The calculated monomeric molecular mass of Rab6A′(Q72L), including the N-terminal His tag, was 22 000 Da and its elution peak from size-exclusion chromatography suggests that it exists as a monomer in solution (Fig. 1 ▶). A gel-filtration standard (Bio-Rad) containing a mixture of molecular-mass markers (thyroglobulin, 670 000 Da; globulin, 158 000 Da; ovalbumin, 44 000 Da; myoglobulin, 17 000 Da; vitamin B12, 1350 Da) was used for size calibration.
Figure 1.
Gel-filtration chromatography and SDS–PAGE of Rab6A′(Q72L)
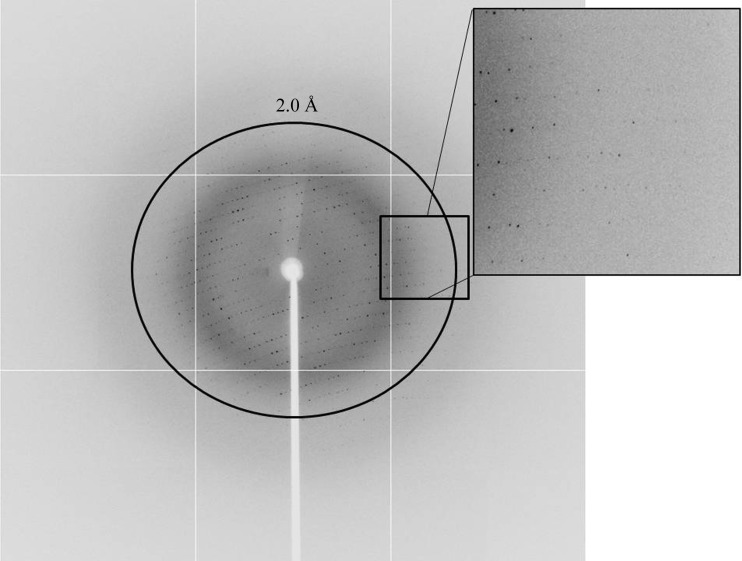
An initial plate-shaped crystal that diffracted poorly was obtained using condition No. 46 of Crystal Screen (18% PEG 8000, 0.2 M calcium acetate, 0.1 M sodium cacodylate pH 6.5). Optimization of the crystallization conditions using a range of concentrations of protein, PEG 8000 and calcium acetate and a range of pH values led to better crystals for diffraction (Fig. 2 ▶). The optimized crystals grew to dimensions of 0.2 × 0.2 × 0.1 mm in 3 d and diffracted to 1.9 Å resolution (Fig. 3 ▶). The crystals belonged to space P22121, with unit-cell parameters a = 36.84, b = 96.78, c = 109.99 Å. Diffraction data statistics are shown in Table 1 ▶.
Figure 2.
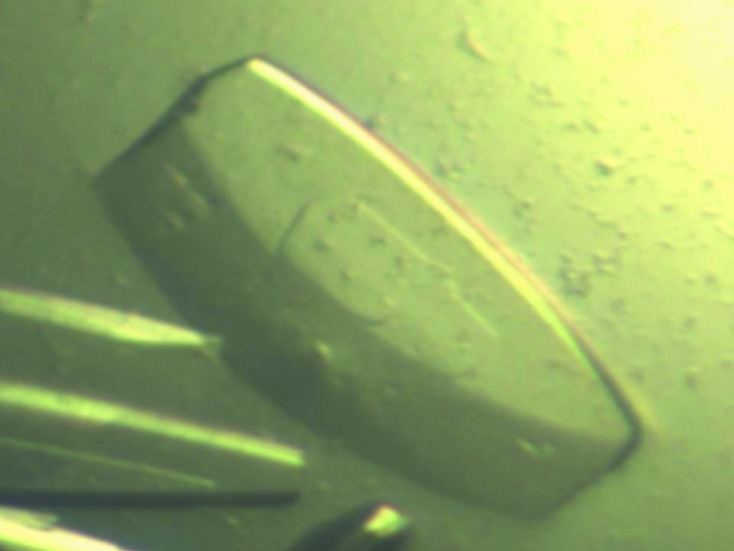
Crystal of Rab6A′(Q72L). Crystals were grown in 3 d in the presence of 20% PEG 8000, 0.3 M calcium acetate, 0.1 M sodium cacodylate pH 6.7. The approximate dimensions of the crystals were 0.2 × 0.2 × 0.1 mm.
Figure 3.
A diffraction image (1° oscillation) from a Rab6A′(Q72L) crystal with a 1.9 Å resolution limit.
Table 1. Diffraction data statistics of the Rab6A′(Q72L) crystals.
Values in parentheses are for the highest resolution shell.
| X-ray source | BL-4A, PAL |
| Wavelength (Å) | 0.9999 |
| Space group | P22121 |
| Unit-cell parameters (Å) | a = 36.84, b = 96.78, c = 109.99 |
| Resolution limits (Å) | 50–1.9 (1.95–1.90) |
| No. of observations | 222576 |
| No. of unique reflections | 31899 |
| Multiplicity | 7.0 (6.9) |
| Mean I/σ(I) | 29.1 (3.6) |
| Completeness (%) | 99.8 (99.9) |
| R merge † | 8.9 (48.7) |
R
merge = 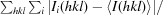
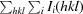 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all i observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all i observations of reflection hkl.
Assuming the presence of two molecules in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.23 Å3 Da−1, which corresponds to a solvent content of 44.8% (Matthews, 1968 ▶). The molecular-replacement phasing method was conducted using Phaser (McCoy et al., 2007 ▶); GTP-bound Rab6A (PDB entry 2gil; Bergbrede et al., 2005 ▶) was used as a search model. A clear solution with rotation-function and translation-function Z-scores of 18.2 and 22.8, respectively, was initially obtained. Initial refinement with REFMAC5 (Vagin & Teplyakov, 2010 ▶) using the initial Phaser model gave an R work of 32.8% and an R free of 38.7%. Further structural refinement is currently in progress.
Acknowledgments
This research was supported by Yeungnam University research grants in 2010.
References
- Antony, C., Cibert, C., Géraud, G., Santa Maria, A., Maro, B., Mayau, V. & Goud, B. (1992). J. Cell Sci. 103, 785–796. [DOI] [PubMed]
- Bergbrede, T., Chuky, N., Schoebel, S., Blankenfeldt, W., Geyer, M., Fuchs, E., Goody, R. S., Barr, F. & Alexandrov, K. (2009). J. Biol. Chem. 284, 2628–2635. [DOI] [PubMed]
- Bergbrede, T., Pylypenko, O., Rak, A. & Alexandrov, K. (2005). J. Struct. Biol. 152, 235–238. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Del Nery, E., Miserey-Lenkei, S., Falguières, T., Nizak, C., Johannes, L., Perez, F. & Goud, B. (2006). Traffic, 7, 394–407. [DOI] [PubMed]
- Echard, A., Jollivet, F., Martinez, O., Lacapère, J.-J., Rousselet, A., Janoueix-Lerosey, I. & Goud, B. (1998). Science, 279, 580–585. [DOI] [PubMed]
- Echard, A., Opdam, F. J. M., de Leeuw, H. J. P. C., Jollivet, F., Savelkoul, P., Hendriks, W., Voorberg, J., Goud, B. & Fransen, J. A. M. (2000). Mol. Biol. Cell, 11, 3819–3833. [DOI] [PMC free article] [PubMed]
- Goud, B., Zahraoui, A., Tavitian, A. & Saraste, J. (1990). Nature (London), 345, 553–556. [DOI] [PubMed]
- Grosshans, B. L., Ortiz, D. & Novick, P. (2006). Proc. Natl Acad. Sci. USA, 103, 11821–11827. [DOI] [PMC free article] [PubMed]
- Macara, I. G., Lounsbury, K. M., Richards, S. A., McKiernan, C. & Bar-Sagi, D. (1996). FASEB J. 10, 625–630. [DOI] [PubMed]
- Mallard, F., Tang, B. L., Galli, T., Tenza, D., Saint-Pol, A., Yue, X., Antony, C., Hong, W., Goud, B. & Johannes, L. (2002). J. Cell Biol. 156, 653–664. [DOI] [PMC free article] [PubMed]
- Martinez, O., Antony, C., Pehau-Arnaudet, G., Berger, E. G., Salamero, J. & Goud, B. (1997). Proc. Natl Acad. Sci. USA, 94, 1828–1833. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Miserey-Lenkei, S., Couëdel-Courteille, A., Del Nery, E., Bardin, S., Piel, M., Racine, V., Sibarita, J.-B., Perez, F., Bornens, M. & Goud, B. (2006). EMBO J. 25, 278–289. [DOI] [PMC free article] [PubMed]
- Opdam, F. J. M., Echard, A., Croes, H. J. E., van den Hurk, J. A. J. M., van de Vorstenbosch, R. A., Ginsel, L. A., Goud, B. & Fransen, J. A. M. (2000). J. Cell Sci. 113, 2725–2735. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Shan, J., Mason, J. M., Yuan, L., Barcia, M., Porti, D., Calabro, A., Budman, D., Vinciguerra, V. & Xu, H. (2000). Gene, 257, 67–75. [DOI] [PubMed]
- Stenmark, H. (2009). Nature Rev. Mol. Cell Biol. 10, 513–525. [DOI] [PubMed]
- Takai, Y., Kaibuchi, K., Kikuchi, A. & Kawata, M. (1992). Int. Rev. Cytol. 133, 187–230. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Young, J., Stauber, T., del Nery, E., Vernos, I., Pepperkok, R. & Nilsson, T. (2005). Mol. Biol. Cell, 16, 162–177. [DOI] [PMC free article] [PubMed]