AIM2 is an innate immune sensor of microbial double-stranded DNA. The HIN-200 domain of mouse AIM2 bound to a 15 bp and an 18 bp dsDNA were crystallized and diffract to about 4.0 Å.
Keywords: mouse AIM2, HIN-200 domain, DNA binding
Abstract
AIM2 (absent in melanoma 2) is an innate immune receptor for cytosolic double-stranded DNA (dsDNA). The engagement of dsDNA by AIM2 activates the AIM2 inflammasome, resulting in the cleavage of pro-interleukin-1β by caspase-1. The DNA-binding HIN-200 domain of mouse AIM2 bound to a 15 bp dsDNA and to an 18 bp dsDNA was purified and crystallized. The AIM2 HIN-200 domain in complex with the 15 bp DNA crystallized in the cubic space group I23 or I213, with unit-cell parameter a = 235.60 Å. The complex of the AIM2 HIN-200 domain and the 18 bp DNA crystallized in a similar unit cell. Diffraction data for the two complexes were collected to about 4.0 Å resolution. Mutagenesis and DNA-binding studies suggest that mouse AIM2 uses a similar surface to human AIM2 to recognize DNA.
1. Introduction
Nucleic acids from bacteria and viruses induce potent immune responses in infected cells (Takeuchi & Akira, 2010 ▶; Hornung & Latz, 2010 ▶; Barber, 2011 ▶). A number of pattern-recognition receptors (PRRs) mediate the sensing of microbial nucleic acids in innate immunity (Takeuchi & Akira, 2010 ▶; Hornung & Latz, 2010 ▶; Vilaysane & Muruve, 2009 ▶; Kato et al., 2011 ▶). Absent in melanoma 2 (AIM2) is a cytosolic microbial DNA sensor that regulates the activation of procaspase-1 through the AIM2 inflammasome (Schattgen & Fitzgerald, 2011 ▶; Hornung et al., 2009 ▶; Fernandes-Alnemri et al., 2009 ▶; Roberts et al., 2009 ▶). AIM2 has a PYRIN domain at its N-terminus and a HIN-200 domain at its C-terminus. The HIN-200 domain is directly involved in the recognition of microbial DNA (Hornung et al., 2009 ▶; Schattgen & Fitzgerald, 2011 ▶). The PYRIN domain interacts with the downstream adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD) through homotypic PYRIN–PYRIN interactions. ASC in turn recruits procaspase-1 through its C-terminal CARD (caspase activation and recruitment domain), forming the AIM2 inflammasome. The induced proximity of procaspase-1 at the AIM2 inflammasome facilitates the activation of caspase-1 (Schroder & Tschopp, 2010 ▶; Hornung & Latz, 2010 ▶). The cleavage of pro-interleukin-1β by caspase-1 results in the secretion of interleukin-1β (IL-1β) to mediate inflammatory responses to infection. IL-1β is a key cytokine involved in the recruitment of immune cells to sites of infection and the regulation of immune-cell proliferation, differentiation and apoptosis (Sims & Smith, 2010 ▶; Dinarello, 2009 ▶). The critical roles of AIM2 in host defense against bacterial and viral infections have been confirmed in AIM2-knockout mice (Rathinam et al., 2010 ▶; Fernandes-Alnemri et al., 2010 ▶; Schattgen & Fitzgerald, 2011 ▶). The crystal structures of human AIM2 and IFI16 (interferon-inducible protein 16) HIN-200 domains bound to DNA have recently been determined (Jin et al., 2012 ▶), providing insight into the mechanism of DNA recognition by AIM2-like receptors.
In order to understand the mechanism of DNA recognition by mouse AIM2 (mAIM2), we have crystallized the HIN-200 domain of mAIM2 bound to several different DNAs and conducted preliminary crystallographic analysis.
2. Materials and methods
2.1. Expression and purification of mAIM2 HIN-200
The HIN-200 domain of mAIM2 (residues 154–354) was cloned into the pET22b(+) plasmid vector (Novagen). Initial expression tests showed heterogeneous oligomerization, which was eliminated after the C264S mutation. The final construct including a hexahistidine tag was composed of a total of 211 residues (mAIM2 HIN-200, molecular mass 22.8 kDa). The sequences of the plasmid were confirmed by DNA sequencing.
mAIM2 HIN-200 was overexpressed in Escherichia coli BL21 (DE3) cells. The cells were grown in LB medium with 100 µg ml−1 ampicillin at 310 K until the cell density reached an OD600 of 0.8. Expression of the protein was induced by the addition of 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) after cooling the culture to 288 K. The cell culture was then incubated for an additional 16 h at 288 K. The cells were harvested by centrifugation at 4000g for 10 min at 277 K. The pellet was resuspended in 150 mM NaCl, 20 mM Tris pH 7.5 (buffer A) and lysed by sonication. The crude lysate was cleared of cell debris by centrifugation at 8000g for 10 min followed by additional centrifugation at 16 000g for 30 min at 277 K.
The protein was first purified by affinity chromatography using an Ni2+ Sepharose column (GE Healthcare). Nonspecifically bound proteins and mAIM2 HIN-200-bound bacterial DNA were removed by washing the column with 500 mM NaCl, 20 mM Tris pH 7.5 buffer plus 10 mM imidazole. The protein was eluted with 150 mM NaCl, 20 mM Tris pH 7.5 buffer plus 250 mM imidazole. After adding 5 mM DTT to the pooled fractions, the samples were concentrated and further purified by gel-filtration chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare), eluting with buffer A.
2.2. Purification of the mAIM2 HIN-200–dsDNA complex
The dsDNA used to form the complex with the mAIM2 HIN-200 was derived from a 60 bp DNA fragment from the genomic DNA of the herpes simplex virus (HSV), a fragment that is known to activate AIM2 in murine myeloid cells (Rasmussen et al., 2011 ▶). The 15 bp DNA (HSV15) forward and reverse oligonucleotides were 5′-TAAGACACGATGCGA-3′ and 5′-TCGCATCGTGTCTTA-3′, respectively. The 18 bp DNA (HSV18) forward and reverse oligonucleotides were 5′-CAAGACACGATGCGATAG-3′ and 5′-CTATCGCATCGGTCTTG-3′, respectively. All of the DNA oligonucleotides were ordered from Integrated DNA Technologies (IDT) and dissolved in buffer A to a concentration of 1 mM. The oligonucleotides were mixed in a 1:1 molar ratio and annealed by heating in a 368 K water bath and cooling to 293 K at room temperature. The annealed dsDNAs were purified by gel-filtration chromatography using a HiLoad 16/60 Superdex 75 column, eluting with buffer A.
To obtain the proper molar ratio for complex formation, small-scale gel-filtration chromatography binding assays were performed with HSV15 using a Superdex 200 10/300 GL column (GE Healthcare). The molar ratio to obtain total saturation of mAIM2 HIN-200 by dsDNA was estimated to be 1.6:1 protein:dsDNA; this ratio was also used for the complex formed using HSV18. The mAIM2 HIN-200/dsDNA mixture was concentrated and the complex was purified by gel-filtration chromatography using a HiLoad 16/60 Superdex 75 column. The eluted complex was immediately concentrated to 13 mg ml−1. The concentrations of the purified mAIM2 HIN-200 and dsDNA were determined from the UV absorbance at 280 and 260 nm, respectively. The concentration of the mAIM2 HIN-200–dsDNA complex was determined by the Bradford method (Bio-Rad).
2.3. Mutagenesis and DNA-binding studies of mAIM2 HIN-200
The mAIM2 HIN-200 domain construct containing residues 135–354 was used for mutagenesis and DNA-binding studies to determine which residues were critical for DNA binding. A structural model of the mAIM2 HIN-200 domain was generated with SWISS-MODEL using the IFI16 HIN-200 domain structure (PDB entry 3b6y; Liao et al., 2011 ▶) as a template. A total of 15 positively charged residues on the surface of mAIM2 were mutated to glutamate. To test the effect of the mutations on DNA binding, each mutant was mixed with HSV18 in a 5:1 mAIM2 HIN-200:dsDNA molar ratio and analyzed by electrophoretic mobility shift assay (EMSA) using a 10% nondenaturing polyacrylamide gel in 1× Tris–boric acid buffer. DNA was detected within the gel by ethidium bromide staining and was visualized using Gel Doc XR (Bio-Rad). DNA-binding studies were also performed by gel-filtration chromatography using a Superdex 200 10/300 GL column (GE Healthcare), but with a 3:1 molar ratio of mAIM2 HIN-200:dsDNA; wild-type mAIM2 HIN-200 and HSV18 dsDNA were used as controls.
2.4. Crystallization of mAIM2 HIN-200 in complex with DNA
The complex of mAIM2 HIN-200 with HSV15 was crystallized using 24% Jeffamine ED-2100, 0.1 M HEPES pH 7.0. The complex of mAIM2 HIN-200 with HSV18 was crystallized using 26% pentaerythritol ethoxylate (15/4 EO/OH), 0.05 M ammonium sulfate, 0.05 M bis-Tris pH 6.5. Both of the complexes were crystallized by the hanging-drop vapor-diffusion method over a 0.5 ml reservoir at 277 K.
To verify the presence of mAIM2 HIN-200 and DNA within the crystals, the crystals were harvested, washed and analyzed using 10% nondenaturing polyacrylamide gel in 1× Tris–boric acid buffer. The gel was stained with ethidium bromide and visualized using Gel Doc XR (Bio-Rad). The protein–DNA complex was observed in the dissolved crystals, confirming that the AIM2–DNA complex was present in both crystals (data not shown).
2.5. Data collection and processing
An mAIM2 HIN-200–HSV15 complex crystal measuring 0.22 × 0.22 × 0.24 mm grew over 13 d at 277 K and an mAIM2 HIN-200–HSV18 complex crystal measuring 0.25 × 0.23 × 0.23 mm grew over 8 d at 277 K. The mAIM2 HIN-200–HSV15 complex crystal was transferred into a cryobuffer consisting of 30% PEG 400, 24% Jeffamine ED-2100, 0.1 M HEPES pH 7.0. The mAIM2 HIN-200–HSV18 complex crystal was transferred into a cryobuffer consisting of 30% 15/4 EO/OH, 0.05 M ammonium sulfate, 0.05 M bis-Tris pH 6.5. Both crystals were then mounted in cryoloops and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on SSRL beamline 11.1 using a PILATUS 6M pixel-array detector. The crystals were rotated through 90° with 0.2° oscillation per frame and 5 s exposure per frame. The diffraction data were processed with the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
3.1. Mouse AIM2 uses a similar surface to human AIM2 to bind DNA
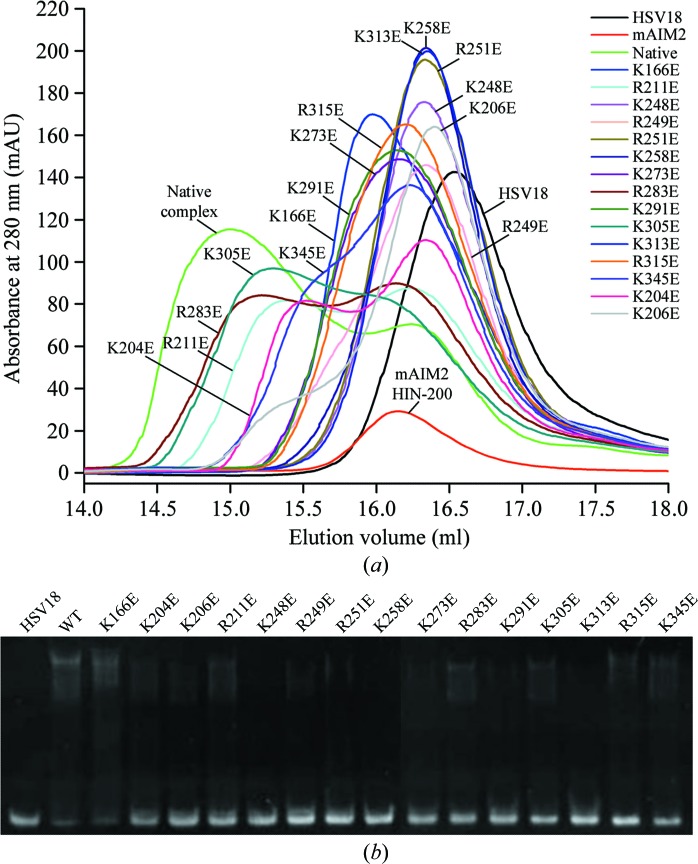
To interpret the mutagenesis results, an mAIM2 HIN-200–DNA complex model was generated by superposition of the mAIM2 HIN-200 homology model onto the human AIM2 HIN-200–DNA complex structure (PDB entry 3rn2; Jin et al., 2012 ▶). DNA-binding studies of mAIM2 mutants by gel-filtration chromatography (Fig. 1 ▶ a) showed that the mutation of Lys206, Lys248, Arg249, Arg251, Lys258 and Lys313 to glutamate almost disrupted DNA binding. Three of these six residues, Lys248, Lys251 and Lys313, are on the DNA-binding surface in the mAIM2–DNA complex model. In addition, the mutation of Lys166, Lys273, Lys291 and Arg315 to glutamate severely reduced DNA binding. Three of these four residues, Lys166, Lys291 and Arg315, are on the DNA-binding surface in the complex model. In contrast, the mutation of Lys204, Arg211 and Lys345 only slightly reduced the DNA binding. Of these three residues, Lys204 is on the DNA-binding surface in the mAIM2–DNA complex model. Consistent with these results, residues Lys166, Lys204, Lys206, Lys313 and Arg315 are conserved in human AIM2 and are involved in DNA binding. As predicted, two control mutations at residues Arg283 and Lys305, which are not on the DNA-binding surface, do not affect DNA binding significantly. Similar results were observed in DNA-binding studies by EMSA (Fig. 1 ▶ b). These results indicate that mAIM2 uses a similar binding surface to human AIM2 to bind DNA, but is likely to interact with the DNA in a different orientation.
Figure 1.
Mutagenesis and DNA-binding studies of mAIM2 HIN-200. (a) DNA-binding studies of mAIM2 HIN-200 mutants with HSV18 by gel-filtration chromatography. (b) DNA-binding studies of mAIM2 HIN-200 mutants with HSV18 by EMSA.
3.2. mAIM2 HIN-200 bound to different lengths of dsDNA crystallizes in similar unit cells
The crystals of mAIM2 HIN-200 bound to 15 bp and 18 bp DNA (Fig. 2 ▶) belonged to space group I23 or I213. The exact space group cannot be determined from the diffraction data since the two space groups have the same reflection conditions. The unit-cell parameters for the HSV15 and HSV18 complexes were similar, with a = 235.60 Å for the HSV15 complex and a = 236.06 Å for the HSV18 complex. Statistics of data collection are summarized in Table 1 ▶. The estimated molecular masses of the mAIM2 HIN-200 complexes with HSV15 and HSV18 were 35 and 36 kDa, respectively, based on their elution volumes from the gel-filtration column. The unit cell may contain 3–6 complexes, corresponding to a V M of 2.5–5.1 Å3 Da−1 and a solvent content of 52–76%. Since the crystals diffracted weakly, it is likely that they have high solvent contents. Data analysis using phenix.xtriage from the PHENIX package (Adams et al., 2010 ▶) indicated that both crystals were twinned, with twin fractions of 0.31 for the HSV15 complex and 0.29 for the HSV18 complex. Crystals of mAIM2 HIN-200 bound to a 20 bp or a 30 bp dsDNA were also obtained and diffraction data have been collected to about 4 Å resolution (data not shown). Surprisingly, these complexes crystallized with almost the same unit cell as the 15 and 18 bp DNA complexes. This suggests that dsDNAs of different lengths are likely to form similar higher order structures with mAIM2. The unit cells of the mAIM2–DNA complexes are different from those of hAIM2 complexes with similar length dsDNAs. Since AIM2 binds DNA in a sequence-independent manner, an 18 bp dsDNA with a palindromic sequence was also used in crystallization to reduce the heterogeneity of the samples. Crystals of mAIM2 bound to the palindromic 18 bp DNA were similar to the HSV18 complex crystals and diffracted to about 4 Å resolution.
Figure 2.
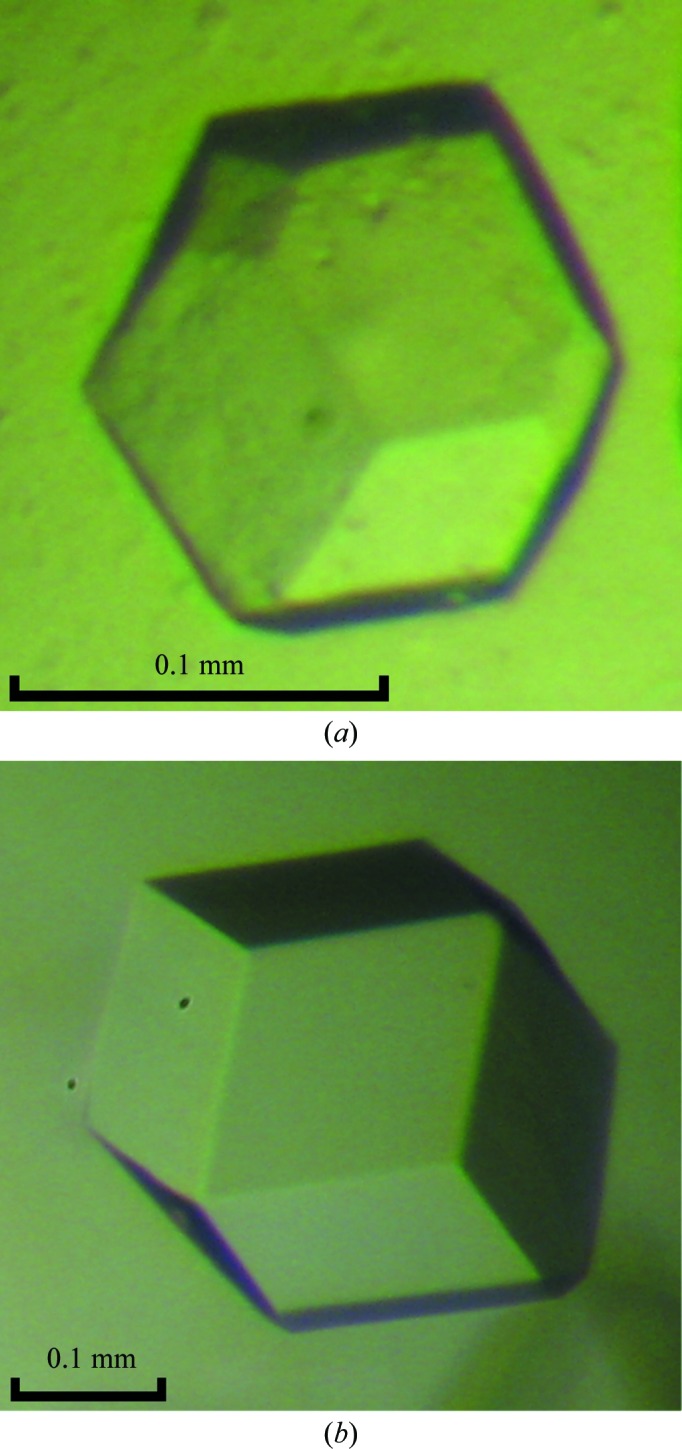
Crystals of mAIM2 HIN-200 bound to (a) 15 bp dsDNA and (b) 18 bp dsDNA.
Table 1. Statistics of diffraction data for mAIM2 HIN-200 complexes.
| HSV15 complex | HSV18 complex | |
|---|---|---|
| Wavelength (Å) | 1.0331 | 1.0331 |
| Resolution range (Å) | 48.10–4.00 (4.07–4.00) | 48.20–4.10 (4.17–4.10) |
| Space group | I23 or I213 | I23 or I213 |
| Unit-cell parameter (Å) | a = 235.60 | a = 236.06 |
| Total No. of reflections | 183453 | 171925 |
| No. of unique reflections | 18444 | 17319 |
| Multiplicity | 9.9 (10.2) | 9.9 (10.3) |
| Completeness (%) | 99.8 (100.0) | 100.0 (100.0) |
| Mean I/σ(I) | 33.8 (3.0) | 27.5 (2.6) |
| R merge † (%) | 6.2 (60.6) | 6.7 (63.0) |
R
merge = 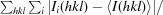
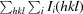 , where Ii(hkl) is the intensity measured for a given reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection.
, where Ii(hkl) is the intensity measured for a given reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection.
Attempts to solve the structure by molecular replacement using both MOLREP and Phaser (Winn et al., 2011 ▶) were unsuccessful for the mAIM2 HIN-200–dsDNA complexes. Crystal structures of the HIN-200 domains of IFI16 (PDB entries 2oq0 and 3b6y; Liao et al., 2011 ▶) and of human AIM2 HIN-200 and its complexes with DNA (PDB entry 3rn2; Jin et al., 2012 ▶) were used as search models. Space groups I213 and I23 were both tested in molecular replacement. It is likely that mouse AIM2 HIN-200 forms a special higher order complex with DNA that differs from that of human AIM2. The structures of these complexes may provide critical insights into how AIM2 forms the inflammasome upon DNA binding. Twinning of the crystals also made the molecular replacement challenging. Molecular replacement with detwinned data and a homology model of mAIM2 HIN-200 also failed to solve the structure. Crystals of SeMet derivatives of the complexes have been generated. The structures will be determined using either single-wavelength or multiple-wavelength anomalous diffraction methods.
Acknowledgments
The diffraction data were collected at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209) and the National Institute of General Medical Sciences. This work was supported in part by a grant from the National Institutes of Health (AI087741 to PL).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Barber, G. N. (2011). Curr. Opin. Immunol. 23, 10–20. [DOI] [PMC free article] [PubMed]
- Dinarello, C. A. (2009). Annu. Rev. Immunol. 27, 519–550. [DOI] [PubMed]
- Fernandes-Alnemri, T., Yu, J.-W., Datta, P., Wu, J. & Alnemri, E. S. (2009). Nature (London), 458, 509–513. [DOI] [PMC free article] [PubMed]
- Fernandes-Alnemri, T., Yu, J.-W., Juliana, C., Solorzano, L., Kang, S., Wu, J., Datta, P., McCormick, M., Huang, L., McDermott, E., Eisenlohr, L., Landel, C. P. & Alnemri, E. S. (2010). Nature Immunol. 11, 385–393. [DOI] [PMC free article] [PubMed]
- Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., Latz, E. & Fitzgerald, K. A. (2009). Nature (London), 458, 514–518. [DOI] [PMC free article] [PubMed]
- Hornung, V. & Latz, E. (2010). Nature Rev. Immunol. 10, 123–130. [DOI] [PubMed]
- Jin, T., Perry, A., Jiang, J., Smith, P., Curry, J. A., Unterholzner, L., Jiang, Z., Horvath, G., Rathinam, V. A., Johnstone, R. W., Hornung, V., Latz, E., Bowie, A. G., Fitzgerald, K. A. & Xiao, T. S. (2012). Immunity, 36, 561–571. [DOI] [PMC free article] [PubMed]
- Kato, H., Takahasi, K. & Fujita, T. (2011). Immunol. Rev. 243, 91–98. [DOI] [PubMed]
- Liao, J. C., Lam, R., Brazda, V., Duan, S., Ravichandran, M., Ma, J., Xiao, T., Tempel, W., Zuo, X., Wang, Y. X., Chirgadze, N. Y. & Arrowsmith, C. H. (2011). Structure, 19, 418–429. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Rasmussen, S. B., Horan, K. A., Holm, C. K., Stranks, A. J., Mettenleiter, T. C., Simon, A. K., Jensen, S. B., Rixon, F. J., He, B. & Paludan, S. R. (2011). J. Immunol. 187, 5268–5276. [DOI] [PMC free article] [PubMed]
- Rathinam, V. A., Jiang, Z., Waggoner, S. N., Sharma, S., Cole, L. E., Waggoner, L., Vanaja, S. K., Monks, B. G., Ganesan, S., Latz, E., Hornung, V., Vogel, S. N., Szomolanyi-Tsuda, E. & Fitzgerald, K. A. (2010). Nature Immunol. 11, 395–402. [DOI] [PMC free article] [PubMed]
- Roberts, T. L., Idris, A., Dunn, J. A., Kelly, G. M., Burnton, C. M., Hodgson, S., Hardy, L. L., Garceau, V., Sweet, M. J., Ross, I. L., Hume, D. A. & Stacey, K. J. (2009). Science, 323, 1057–1060. [DOI] [PubMed]
- Schattgen, S. A. & Fitzgerald, K. A. (2011). Immunol. Rev. 243, 109–118. [DOI] [PubMed]
- Schroder, K. & Tschopp, J. (2010). Cell, 140, 821–832. [DOI] [PubMed]
- Sims, J. E. & Smith, D. E. (2010). Nature Rev. Immunol. 10, 89–102. [DOI] [PubMed]
- Takeuchi, O. & Akira, S. (2010). Cell, 140, 805–820. [DOI] [PubMed]
- Vilaysane, A. & Muruve, D. A. (2009). Semin. Immunol. 21, 208–214. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.