The cloning, expression, purification, crystallization and preliminary X-ray characterization of a trimodular type III cohesin–dockerin complex from the R. flavefaciens cellulosome system are described.
Keywords: scaffoldins, X modules, selenomethionine, single-wavelength anomalous diffraction
Abstract
In Ruminococcus flavefaciens, a predominant fibre-degrading bacterium found in ruminants, cellulosomal proteins are anchored to the bacterial cell wall through a relatively small ScaE scaffoldin which includes a single type III cohesin. The cotton-binding protein CttA consists of two cellulose-binding modules and a C-terminal modular pair (XDoc) comprising an X-module and a contiguous dockerin, which exhibits high affinity towards the ScaE cohesin. Seleno-l-methionine-labelled derivatives of the ScaE cohesin module and the XDoc from CttA have been expressed, copurified and cocrystallized. The crystals belonged to the tetragonal space group P43212, with unit-cell parameters a = b = 78.7, c = 203.4 Å, and the unit cell contains a single cohesin–XDoc complex in the asymmetric unit. The diffraction data were phased to 2.0 Å resolution using the anomalous signal of the Se atoms.
1. Introduction
The exocellular multienzyme cellulosome complex is considered to be the most efficient cellulase system, in which a consortium of catalytic and noncatalytic modules are involved in the degradation of crystalline cellulosic substrates and associated plant cell-wall polysaccharides. The overall architecture and assembly of the cellulosome are dictated by the high-affinity species-specific interaction between two complementary modules: the cohesin (Coh), which is found on the scaffoldin protein, and the dockerin (Doc), which is borne by either a catalytic subunit or a scaffoldin protein (Bayer et al., 2004 ▶; Pagès et al., 1997 ▶). To date, hundreds of different Cohs and Docs have been sequenced from over 100 different bacterial and archaeal species (Peer et al., 2009 ▶).
One of the most important cellulose-based ecosystems is the digestive tract of herbivores, and in particular ruminants. Ruminococcus flavefaciens is an important Gram-positive, anaerobic, cellulosome-producing rumen bacterium involved in plant cell-wall degradation (Julliand et al., 1999 ▶; Krause et al., 1999 ▶; Nelson et al., 2003 ▶; Flint, 1997 ▶; Wedekind et al., 1988 ▶; Flint & Bayer, 2008 ▶). The cellulosome system of R. flavefaciens strain FD-1 has recently been discovered to bear one of the most intricate complexes in the cellulosome world, much more so than those reported previously for cellulolytic Clostridium species (Bayer et al., 2008 ▶; Flint et al., 2008 ▶; Berg Miller et al., 2009 ▶; Rincon et al., 2010 ▶).
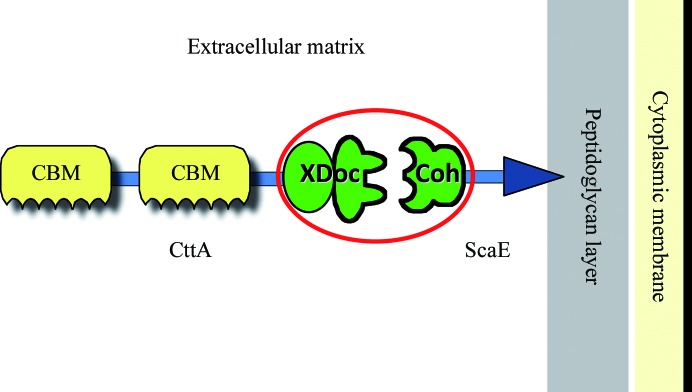
According to gene-clustering analysis of several R. flavefaciens strains, four scaffoldins constitute the cellulosomal backbone of this bacterium, ScaC, ScaA, ScaB and ScaE, all of which are encoded by the sca gene cluster (Jindou et al., 2008 ▶). In this context, several cohesin–dockerin specificities between ScaA, ScaB and ScaC dictate the assembly of various enzymatic components onto the complex, including the enzyme equivalents of Cel44A and Ce3B. The central adaptor scaffoldin ScaB has been shown to bear an X-module followed by a dockerin module at its C-terminus (XDoc). The ScaB XDoc exhibits a high level of specificity towards the single cohesin (CohE) of the bacterial cell-wall-anchored scaffoldin ScaE, thereby anchoring the entire complex to the cell surface (Jindou et al., 2006 ▶; Rincon et al., 2005 ▶). Moreover, the sca gene cluster also encodes a cotton-binding protein, CttA, which bears a couple of putative cellulose-binding modules and a similar C-terminal XDoc that also interacts with CohE (Fig. 1 ▶; Rincon et al., 2007 ▶). The monovalent (single-cohesin) ScaE has a C-terminal Gram-positive LPXTG-like motif that mediates its covalent binding to the bacterial cell wall via a sortase-mediated attachment mechanism (Navarre & Schneewind, 1994 ▶; Rincon et al., 2005 ▶). ScaE thus plays a major role in anchoring components of the cellulosomal system to the cell wall, thereby facilitating the proximity of the enzymatic assembly to the cellulose substrate via its interaction with ScaB and CttA.
Figure 1.
Schematic representation of the intermodular interaction in the R. flavefaciens FD-1 cellulosome system. ScaE fulfils a key role in the cell-attachment and substrate-binding functions of the R. flavefaciens cellulosome system via the interaction of its cohesin module with the XDoc dyad of the cotton-binding protein CttA. CttA contains two putative CBMs, which are associated with binding to cellulosic substrates (e.g. cotton). The RfCohE–XDoc complex is circled.
The cohesins of ScaA, ScaB and ScaE were found to be phylogenetically distinct from the previously described type I and type II cohesins. They were therefore designated into a new group of cohesins: the type III cohesins (Ding et al., 2001 ▶; Rincon et al., 2005 ▶). To date, the structures of several type I and type II Coh–Doc complexes have been solved, thereby providing direct insight into the mode of interaction between the two modular counterparts in Clostridium thermocellum and C. cellulolyticum (Carvalho et al., 2003 ▶, 2007 ▶; Adams et al., 2006 ▶, 2010 ▶; Pinheiro et al., 2008 ▶). A single type III cohesin structure from R. flavefaciens strain 17 has been reported to date (Alber et al., 2008 ▶, 2009 ▶), but the achievement of a structural description of a type III Coh–Doc complex has remained challenging. Here, we report the cloning, expression, co-purification, crystallization and preliminary X-ray characterization of a type III RfCohE–XDoc complex between CohE and the C-terminal XDoc dyad of CttA from R. flavefaciens strain FD-1.
2. Experimental methods
2.1. Cloning of CohE and XDoc in expression vectors
The DNA encoding the cohesin module from the scaE scaffoldin gene of R. flavefaciens strain FD-1 (gi:268610849) was cloned into the pET28a expression vector (Novagen, Madison, Wisconsin, USA) together with a sequence encoding a hexa-His tag (peptide sequence MAHHHHHHAMAL) attached to the 5′ end. Independently, the DNA encoding the XDoc dyad from the cttA scaffoldin gene of R. flavefaciens strain FD-1 (gi:268610848) was cloned into the pETDuetACYC expression vector (Novagen, Madison, Wisconsin, USA) lacking the sequence encoding a hexa-His tag. The resultant plasmids were sequenced by capillary electrophoresis using a DNA analyzer (Applied Biosystems, Foster City, California, USA) and were separately transferred into Escherichia coli strain BL21 (DE3).
2.2. Expression and copurification of the RfCohE–XDoc complex
Expression of the native and seleno-l-methionine-labelled CohE module (residues 30–230) from R. flavefaciens ScaE scaffoldin and of the XDoc dyad from CttA scaffoldin (residues 565–803) was conducted according to the method described previously (Van Duyne et al., 1993 ▶; Alber et al., 2008 ▶). CohE and XDoc were expressed in 1 and 0.75 l cell culture, respectively. The cultured cells were harvested by centrifugation (1600g for 15 min) at 277 K and were resuspended in 20 ml binding buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) adjusted to pH 7.0, 150 mM NaCl, 10 mM CaCl2, 5 mM imidazole]. The suspension was kept on ice during sonication, after which cell debris was removed by centrifugation (20 000g at 277 K for 20 min). The supernatant fluids of both expressed proteins were combined, allowing formation and purification of the complex. The supernatant fractions were applied onto a column packed with 5 ml nickel–nitrilotriacetic acid resin (Zephyr ProteomiX, Rehovot, Israel) equilibrated with binding buffer. Bound proteins were eluted with binding buffer containing 150 mM imidazole. The pooled purified fractions were applied onto a Hi-Load 16/60 Superdex 75 size-exclusion column (Amersham Pharmacia Biosciences) equilibrated with 25 mM HEPES adjusted to pH 7.0, 25 mM NaCl, 10 mM CaCl2, 5 mM dithiothreitol. The collected fractions (1 ml) were further examined for protein purity by SDS–PAGE (Fig. 2 ▶). Selected complex fractions were pooled and concentrated using Amicon Ultra centrifugal filters of 10 000 molecular-weight cutoff (Millipore, Cork, Ireland), yielding 0.5 ml purified concentrated RfCohE–XDoc complex (30 mg ml−1). Affinity and size-exclusion purification were performed at room temperature. The concentration of the complex was estimated by measuring the absorbance at 280 nm using the calculated extinction coefficient of the proteins (∊280 = 60 740).
Figure 2.
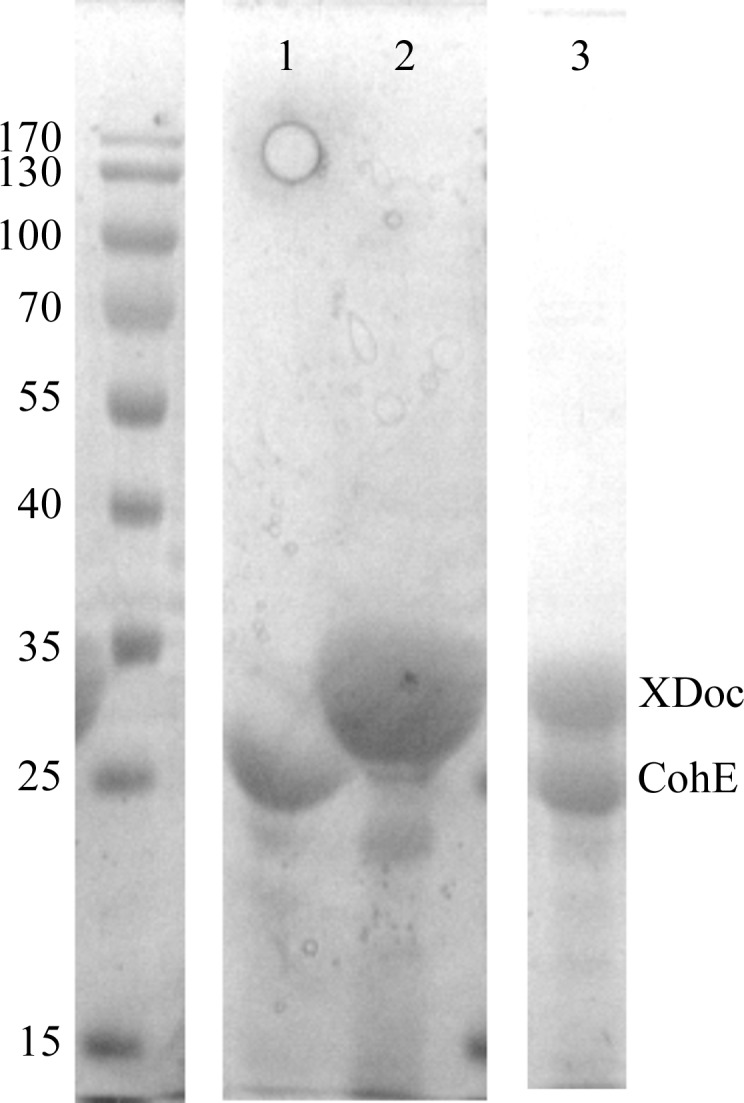
SDS–PAGE gel (12%). The left column contains molecular-weight markers (kDa). Lanes 1 and 2 contain purified CohE and XDoc, respectively. Lane 3 contains the purified RfCohE–XDoc complex.
2.3. Protein crystallization
Native and SeMet-labelled RfCohE–XDoc protein complexes were crystallized at 293 K by the hanging-drop vapour-diffusion method. Screening for crystallization conditions was performed using the Index kit (Hampton Research, California, USA). Initial screening yielded single crystals using condition No. 6 (0.1 M Tris pH 8.5, 2.0 M ammonium sulfate). Further optimization of the conditions yielded a final reservoir solution consisting of 0.1 M Tris pH 8.5, 1.8 M ammonium sulfate. The protein solution (2 µl of a 30 mg ml−1 protein solution in 25 mM HEPES adjusted to pH 7.0, 25 mM NaCl, 10 mM CaCl2, 5 mM dithiothreitol) was mixed with 2 µl reservoir solution and equilibrated against 0.5 ml reservoir solution in a 24-well VDX plate (Hampton Research). The crystals grew within 2–3 d at 293 K and reached their full size of 0.05 × 0.1 × 0.5 mm within 5 d, exhibiting a prismatic morphology (Fig. 3 ▶).
Figure 3.
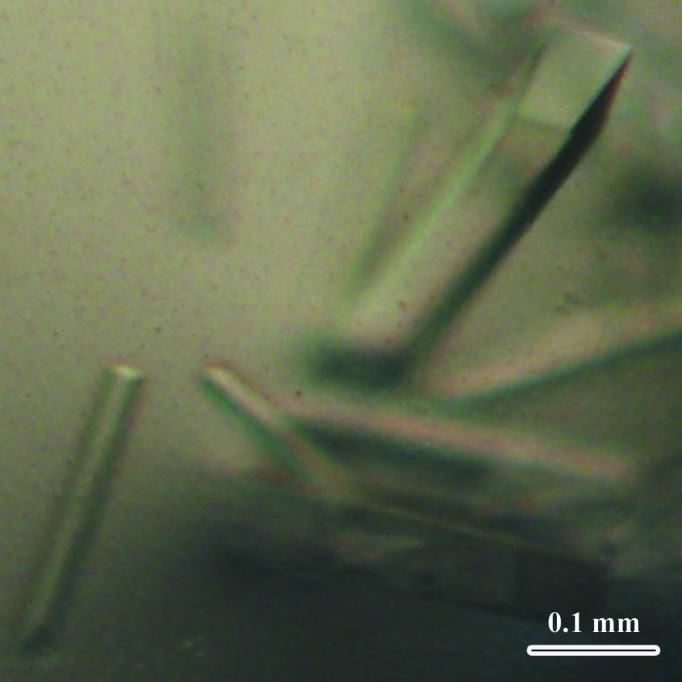
Crystals of the SeMet-labelled RfCohE–XDoc complex.
2.4. X-ray data collection and processing
The crystals were gently removed from the crystallization drop, incubated for a moment in a cryostabilization solution comprised of a mixture of equal volumes of a twofold-concentrated solution of the crystallization components and a solution consisting of 18%(w/v) sucrose, 16%(w/v) glycerol, 16%(w/v) ethylene glycol and 4%(w/v) glucose. For data collection, crystals were mounted on a MiTeGen MicroMount, plunged into liquid nitrogen and placed in pucks for mounting at the synchrotron. Diffraction data for the SeMet-labelled and native RfCohE–XDoc complex crystals were collected using synchrotron radiation on beamline BM14 at the ESRF, Grenoble, France equipped with a MAR 225 CCD detector using wavelengths of 0.97841 Å (the Se edge as determined by a fluorescence-energy scan) and 0.97833 Å, respectively. The native and SeMet-derivative crystals diffracted to resolutions of 1.97 and 2.00 Å, respectively. Diffraction images were obtained using 0.5° oscillation steps over oscillation ranges of 135° (270 frames) and 77° (154 frames) for the SeMet-labelled and native crystals, respectively. Images were integrated and scaled using DENZO and SCALEPACK as implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶).
The observed anomalous signal measurability (Zwart, 2005 ▶) of 0.092 for the SeMet-labelled structure was significantly larger than the required lower limit of 0.05. Calculations were performed using phenix.xtriage as implemented in PHENIX (Adams et al., 2011 ▶). One molecule of the RfCohE–XDoc complex per asymmetric unit and 63% solvent content corresponded to a Matthews coefficient V M of 3.33 Å3 Da−1 (Matthews, 1968 ▶). Data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics for the native and SeMet RfCohE–XDoc crystals.
Values in parentheses are for the highest resolution shell.
| Native RfCohE–XDoc | SeMet RfCohE–XDoc | |
|---|---|---|
| Data collection | ||
| X-ray source | BM14, ESRF | BM14, ESRF |
| Wavelength (Å) | 0.97833 | 0.97841 |
| Space group | P43212 | P43212 |
| Unit-cell parameters (Å, °) | a = b = 78.53, c = 202.81 | a = b = 78.73, c = 203.24 |
| Resolution (Å) | 20–1.97 (2.0–1.97) | 30.0–2.00 (2.03–2.00) |
| Mosaicity (°) | 0.4–0.41 | 0.32–0.40 |
| Matthews coefficient V M (Å3 Da−1) | 3.31 | 3.33 |
| Solvent content (%) | 63.1 | 63.0 |
| Data processing | ||
| No. of measured reflections | 284650 | 481008 |
| No. of unique reflections | 46047 | 44177 |
| Completeness (%) | 98.5 (97.6) | 100 (100) |
| R merge † (%) | 10.7 (94.8) | 13.7 (85.6) |
| Mean I/σ(I) | 16.4 (2.0) | 17.1 (2.6) |
| Wilson B factor (Å2) | 24.27 | 21.69 |
R
merge = 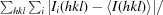
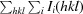 , where
, where 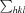 denotes the sum over all reflections and
denotes the sum over all reflections and 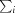 is the sum over all equivalent and symmetry-related reflections.
is the sum over all equivalent and symmetry-related reflections.
3. Results and discussion
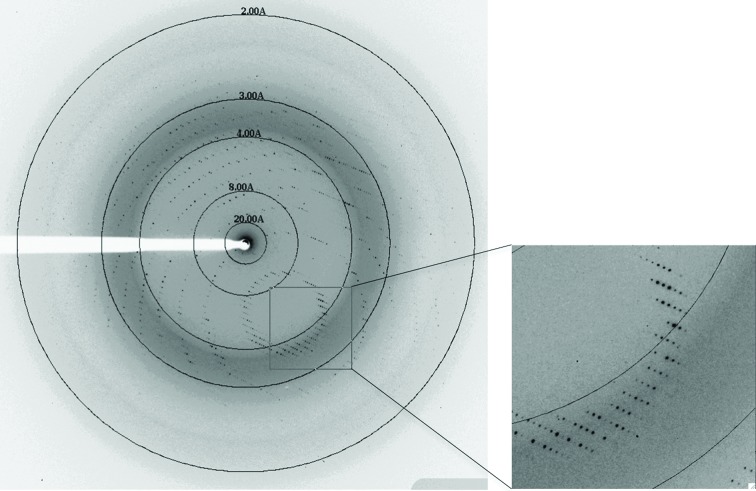
The preliminary crystal structure of the SeMet derivative of a type III RfCohE–XDoc complex was determined by single-wavelength anomalous diffraction (SAD). Attempts to obtain an ordered diffraction pattern that would allow data collection were challenging. Numerous crystals were tested. Crystals characterized by a clear sharp-edged morphology (Fig. 3 ▶) that yielded ordered X-ray diffraction pattern were infrequent, whereas crystals that exhibited amorphous edges gave a scattered diffraction pattern (Fig. 4 ▶). The diffraction quality of the crystals was a function of both the crystallization trial and the time that passed between crystal formation and harvesting. Fresh sharp-edged crystals resulted in the best diffraction. The SeMet RfCohE–XDoc crystal contained a single copy of the heterodimeric complex in the asymmetric unit, which contained five Se atoms (corresponding to four SeMet residues in the CohE module and one in the Doc module) and three calcium ions. The SHELXC/D/E pipeline (Sheldrick, 2010 ▶) and HKL2MAP graphical user interface (Pape & Schneider, 2004 ▶) were used during the data-collection session for determination of the selenium substructure followed by phasing, electron-density modification and initial tracing of the complex structure as polyalanine chains. The structure was further rebuilt using ARP/wARP (Langer et al., 2008 ▶) and Coot (Emsley et al., 2010 ▶). Model building of the remaining part of the structure and refinement are currently in progress.
Figure 4.
X-ray diffraction pattern from an ordered RfCohE–XDoc crystal.
Acknowledgments
We would like to thank the ESRF for synchrotron beam time and the staff scientists of the BM14 station for their assistance. This research was supported by the Israel Science Foundation (Grant Nos. 159/07, 293/08, 966/09 and 24/11) and by grants from the United States–Israel Binational Science Foundation (BSF), Jerusalem, Israel. EAB holds The Maynard I. and Elaine Wishner Chair of Bio-Organic Chemistry at the Weizmann Institute of Science.
References
- Adams, J. J., Currie, M. A., Ali, S., Bayer, E. A., Jia, Z. & Smith, S. P. (2010). J. Mol. Biol. 396, 833–839. [DOI] [PubMed]
- Adams, J. J., Pal, G., Jia, Z. & Smith, S. P. (2006). Proc. Natl Acad. Sci. USA, 103, 305–310. [DOI] [PMC free article] [PubMed]
- Adams, P. D. et al. (2011). Methods, 55, 94–106.
- Alber, O., Noach, I., Lamed, R., Shimon, L. J. W., Bayer, E. A. & Frolow, F. (2008). Acta Cryst. F64, 77–80. [DOI] [PMC free article] [PubMed]
- Alber, O., Noach, I., Rincon, M. T., Flint, H. J., Shimon, L. J. W., Lamed, R., Frolow, F. & Bayer, E. A. (2009). Proteins, 77, 699–709. [DOI] [PubMed]
- Bayer, E. A., Belaich, J. P., Shoham, Y. & Lamed, R. (2004). Annu. Rev. Microbiol. 58, 521–554. [DOI] [PubMed]
- Bayer, E. A., Lamed, R., White, B. A. & Flint, H. J. (2008). Chem. Rec. 8, 364–377. [DOI] [PubMed]
- Berg Miller, M. E. et al. (2009). PLoS One, 4, e6650. [DOI] [PMC free article] [PubMed]
- Carvalho, A. L., Dias, F. M., Nagy, T., Prates, J. A., Proctor, M. R., Smith, N., Bayer, E. A., Davies, G. J., Ferreira, L. M., Romão, M. J., Fontes, C. M. & Gilbert, H. J. (2007). Proc. Natl Acad. Sci. USA, 104, 3089–3094. [DOI] [PMC free article] [PubMed]
- Carvalho, A. L., Dias, F. M., Prates, J. A., Nagy, T., Gilbert, H. J., Davies, G. J., Ferreira, L. M., Romão, M. J. & Fontes, C. M. (2003). Proc. Natl Acad. Sci. USA, 100, 13809–13814. [DOI] [PMC free article] [PubMed]
- Ding, S.-Y., Rincon, M. T., Lamed, R., Martin, J. C., McCrae, S. I., Aurilia, V., Shoham, Y., Bayer, E. A. & Flint, H. J. (2001). J. Bacteriol. 183, 1945–1953. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Flint, H. J. (1997). Trends Microbiol. 5, 483–488. [DOI] [PubMed]
- Flint, H. J. & Bayer, E. A. (2008). Ann. N. Y. Acad. Sci. 1125, 280–288. [DOI] [PubMed]
- Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R. & White, B. A. (2008). Nature Rev. Microbiol. 6, 121–131. [DOI] [PubMed]
- Jindou, S., Borovok, I., Rincon, M. T., Flint, H. J., Antonopoulos, D. A., Berg, M. E., White, B. A., Bayer, E. A. & Lamed, R. (2006). J. Bacteriol. 188, 7971–7976. [DOI] [PMC free article] [PubMed]
- Jindou, S., Brulc, J. M., Levy-Assaraf, M., Rincon, M. T., Flint, H. J., Berg, M. E., Wilson, M. K., White, B. A., Bayer, E. A., Lamed, R. & Borovok, I. (2008). FEMS Microbiol. Lett. 285, 188–194. [DOI] [PubMed]
- Julliand, V., de Vaux, A., Millet, L. & Fonty, G. (1999). Appl. Environ. Microbiol. 65, 3738–3741. [DOI] [PMC free article] [PubMed]
- Krause, D. O., Dalrymple, B. P., Smith, W. J., Mackie, R. I. & McSweeney, C. S. (1999). Microbiology, 145, 1797–1807. [DOI] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Navarre, W. W. & Schneewind, O. (1994). Mol. Microbiol. 14, 115–121. [DOI] [PubMed]
- Nelson, K. E., Zinder, S. H., Hance, I., Burr, P., Odongo, D., Wasawo, D., Odenyo, A. & Bishop, R. (2003). Environ. Microbiol. 5, 1212–1220. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pagès, S., Bélaïch, A., Bélaïch, J.-P., Morag, E., Lamed, R., Shoham, Y. & Bayer, E. A. (1997). Proteins, 29, 517–527. [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst. 37, 843–844.
- Peer, A., Smith, S. P., Bayer, E. A., Lamed, R. & Borovok, I. (2009). FEMS Microbiol. Lett. 291, 1–16. [DOI] [PMC free article] [PubMed]
- Pinheiro, B. A., Proctor, M. R., Martinez-Fleites, C., Prates, J. A., Money, V. A., Davies, G. J., Bayer, E. A., Fontes, C. M., Fierobe, H. P. & Gilbert, H. J. (2008). J. Biol. Chem. 283, 18422–18430. [DOI] [PubMed]
- Rincon, M. T., Cepeljnik, T., Martin, J. C., Barak, Y., Lamed, R., Bayer, E. A. & Flint, H. J. (2007). J. Bacteriol. 189, 4774–4783. [DOI] [PMC free article] [PubMed]
- Rincon, M. T., Cepeljnik, T., Martin, J. C., Lamed, R., Barak, Y., Bayer, E. A. & Flint, H. J. (2005). J. Bacteriol. 187, 7569–7578. [DOI] [PMC free article] [PubMed]
- Rincon, M. T., Dassa, B., Flint, H. J., Travis, A. J., Jindou, S., Borovok, I., Lamed, R., Bayer, E. A., Henrissat, B., Coutinho, P. M., Antonopoulos, D. A., Berg Miller, M. E. & White, B. A. (2010). PLoS One, 5, e12476. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Wedekind, K. J., Mansfield, H. R. & Montgomery, L. (1988). Appl. Environ. Microbiol. 54, 1530–1535. [DOI] [PMC free article] [PubMed]
- Zwart, P. H. (2005). Acta Cryst. D61, 1437–1448. [DOI] [PubMed]