Abstract
Purpose
Metastatic uveal melanoma (UM) represents the most common intraocular malignancy with very poor prognosis and no effective treatments. Oncogenic mutations in the G protein alpha subunit q and 11 have been described in about 85% of uveal melanomas and confer constitutive activation. Multiple signaling pathways are induced as a consequence of GNAQ/11 activation, which include the MEK/ERK kinase cascade. We analyzed the transcriptional profile of cell lines treated with a MEK inhibitor to identify gene targets of activated GNAQ and evaluate the biological importance of these genes in UM.
Experimental Design
We performed microarray analysis of UM cell lines with GNAQ mutations treated with the MEK inhibitor selumetinib. For comparison, we used cells carrying BRAFV600E and cells without either mutation. Changes in the expression of selected genes were then confirmed by real-time qPCR and immunoblotting.
Results
We found that GNAQ mutant cells have a MEK-dependent transcriptional output and identified a unique set of genes that are down-regulated by MEK inhibition, including the RNA helicase DDX21 and the cyclin dependent kinase regulator CDK5R1, while JUN was induced. We provide evidence that these genes are involved in cell proliferation, tumor cell invasion and drug resistance, respectively. Furthermore, we show that selumetinib treatment regulates the expression of these genes in tumor tissues of patients with metastatic GNAQ/11 mutant uveal melanoma. Conclusions: Our findings define a subset of transcriptionally regulated genes by selumetinib in GNAQ mutant cells and provide new insights into understanding the biologic effect of MEK inhibition in this disease.
Keywords: selumetinib, microarray, metastasis, JUN
Introduction
The Ras/Raf/MEK/ERK pathway is often activated by genetic alterations in upstream signaling molecules, such as Ras and BRAF. Hyperactivation of this signaling cascade results in dysregulated cell proliferation and malignant transformation of a number of human tumors (1). Recently, it has been demonstrated the presence of somatic activating mutations in the heterotrimeric G protein alpha subunit q (GNAQ) and 11 (GNA11) in 46% and 30% of primary uveal melanomas, respectively (2-4). These mutations occur in codon 209 in the Ras-like domain, resulting in loss of GTPase activity and constitutive activation. Approximately 85% of ocular melanomas arise from melanocytes within the uveal tract, which consists of the iris, ciliary body, and choroid of the eye (5), and account for a significant rate of deaths. Uveal melanoma is biologically distinct from cutaneus melanoma, as recurrent chromosomal abnormalities have been detected in chromosome 3 and 8 (6), as well as a very high tendency to metastasize to the liver (7). NRAS mutations have not been detected in UM (8), and BRAF mutations are considered rare (9, 10); however, activation of the MAPK pathway by mutant GNAQ/11 appears to be critical for the development of this disease (2). G protein alpha subunits transduce external signals from G protein-coupled seven transmembrane domain receptors (GPCR) to intracellular signaling pathways. The canonical target of GNAQ/11 stimulation is phospholipase C-β, which, in turn, stimulate inositol lipid signaling (i.e. calcium and PKC), in response to mitogenic GPCR agonists (11, 12). The constitutively active mutant GNAQ and GNA11 have been reported to activate the ERK pathway (2, 4), and knockdown of mutant GNAQ in UM cells resulted in MAP-kinase inhibition, reduced growth and induced apoptosis (2). However, the transcriptional output of ERK signaling downstream of mutant G proteins is not well characterized. Targeting MEK with allosteric small molecule inhibitors has been reported to be effective in suppressing cell growth and, in some cells, inducing apoptosis. In particular, melanoma, thyroid and non-small cell lung cancer with mutant BRAFV600E have been shown to be sensitive to MEK inhibitors (13-15). In a large number of tumor types treated with the MEK inhibitor selumetinib (AZD6244), Dry et al. have reported transcriptional signatures that predict MEK addiction or drug resistance (16). In another study, using microarray analysis of cells treated with a MEK inhibitor, Pratilas et al. have found genes, including members of the dual specificity phosphatase and sprouty gene families, that were differentially regulated by MEK inhibition in BRAFV600E cells but not in receptor tyrosine kinase-driven tumor cells with similarly elevated levels of p-ERK (17). They also demonstrated that BRAFV600E cells have elevated ERK-dependent transcriptional output and disabled feedback inhibition of RAF-MEK signaling. Here, we report that UM cells with GNAQ mutations are highly sensitive to MEK inhibition with selumetinib. Expression microarray analysis identified a MEK dependent transcriptional profile that is in part similar to that of BRAFV600E melanoma cells. In addition, we identified several genes unique to GNAQ mutant cells, which are involved in proliferation and tumor cell invasion. Furthermore, pre- and post-treatment tumor biopsies from an ongoing clinical trial of selumetinib in patients with uveal melanoma indicate that these genes are transcriptionally regulated and may correlate with clinical benefit.
Materials and Methods
Cell culture
Omm1.3, Mel202, Mel270 have been kindly provided by Dr Bruce Ksander (Harvard Medical School, Boston, MA). OCM1A and 92.1 were from Dr William Harbour (Washington University, St. Louis, MO). OCM3, Mel290, C918 were from Robert Folberg (University of Illinois, Chicago, IL). UM cell lines have been sequenced for the presence of activating mutations in codons 209 (exon 5) and 183 (exon 4) of GNAQ and GNA11. Two cell lines had Q209L mutation (92.1, Mel202), while Omm1.3 and Mel270 had Q209P mutation. None had GNA11 mutations. Cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100μg/ml streptomycin, and maintained at 37°C in 5% CO2. Cells were treated with selumetinib (AZD6244, graciously supplied by AstraZeneca).
Cell viability Assays
Cells were plated in 96-well plates, and treated with the indicated concentrations of selumetinib, PD0325901, or DMSO in triplicates. Viability was assessed after four days of treatment using the Cell Counting Kit 8 (CCK8) from Dojindo Molecular Technologies according to the manufacturer’s instructions. Survival is expressed as a percentage of untreated cells. The calculations of CI values were conducted using the CompuSyn software (ComboSyn) (18).
Microarray data analysis
Cells were treated in triplicate with 250nM selumetinib, or 0.01% DMSO as control, for 8 hours. Following RNA extraction with Trizol Reagent (Invitrogen), cDNA was synthesized in the presence of oligo(dT)24-T7 from Genset Corp. cRNA was prepared using biotinylated UTP and CTP and was hybridized to Human HT-12 oligonucleotide Illumina arrays in triplicates. We used a single array slide for each cell line to minimize the effect of experimental artifact. Differential expression analysis was performed to identify genes whose expression is affected by the treatment by comparing the post-treatment expression vs. the pre-treatment expression. The array data were log2 transformed and quantile normalized. For each cell line, gene expression was compared between time points using the empirical Bayesian method and the R LIMMA package. An empirical Bayes t test was applied to each gene (19). Linear models and empirical Bayes methods were used for assessing differential expression in microarray experiments and a p-value cutoff of 0.0001 was used to select differentially expressed genes. There are ~47,000 markers on the Illumina array, and 5 genes are expected by chance to have a p-value <0.0001. Data is deposited at GEO accession no. GSE33655.
Immunoblotting
Cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail tablets (Roche Diagnostics) and 1mM Na3VO4. Equal amounts of protein were loaded on 4-12% PAGE gels (Invitrogen). PVDF membranes were blocked with 5% nonfat dried milk and probed with pERK, ERK, Cyclin D1, CDK5R1 (p35), c-Jun, α-tubulin (Cell Signaling), SPRY2, DUSP6, ETV5, DDX21 (Abcam).
Quantitative real-time PCR
Reverse transcription of 1μg of RNA was done using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative real-time PCR assays were done on the 7300 Real Time PCR System (Applied Biosystems). TaqMan gene expression assays, which include gene-specific probe primer sets (Applied Biosystems), were used to detect the indicated genes and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)/HPRT mRNA. The relative expression of each gene was calculated by the ΔΔCT method.
RNAi-mediated gene knockdown
The list of siRNA is reported in the Supplementary methods. siRNAs were transfected using Lipofectamine RNAiMAX reagent (Invitrogen). After transfections cells were counted using a Nexcelom cell counter and plated in 96 well plates for cell viability assays. The statistical significance of the experimental results was determined by the two-sided t test.
Tissue sample collection and analysis
Matched tumor biopsies were collected from patients with metastatic uveal melanoma (clinicaltrials.gov # NCT01143402) before and after 14 days of selumetinib treatment. Flash frozen specimens were then lysed in RIPA lysis buffer and analyzed by immunoblotting. The protocol was approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center, and all patients signed informed consent forms.
Results
The MEK inhibitor selumetinib inhibits cell viability of GNAQ mutant uveal melanoma
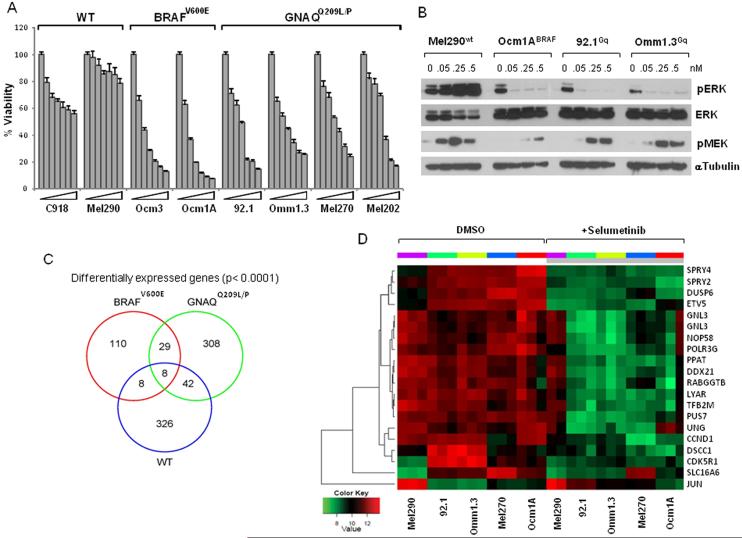
Using a panel of UM cell lines expressing GNAQQ209L/P, mutant BRAFV600E or WT for both, we investigated the effects of MEK inhibition on cell viability with selumetinib. As shown in Figure 1A, the GNAQQ209L/P cells exhibited dose dependent decrease in cell viability at nanomolar concentrations (IC50<0.1 μM). The BRAFV600E cells were the most sensitive, with IC50<0.05 μM, while cells without either BRAF or GNAQ mutations exhibited little sensitivity, with IC50 >1 μM. This corresponded to inhibition of pERK with selumetinb in the sensitive mutant cells (Fig. 1B and Supplemental Fig. 1A), while induction of pMEK is consistent with relief of negative feedback (20). The effect of MEK inhibition on GNAQ mutant UM demonstrated an accumulation in the G1 phase of the cell cycle (Supplemental Fig. 1B). GNAQ downregulation by siRNA induced a decrease in expression of pERK in the GNAQQ209L/P cells (Supplemental Fig. 2A). This corresponded to a decrease in cell viability which was not appreciably increased by selumetinib (Supplemental Fig. 2B). Suppression of GNAQ in WT and BRAFV600E cells did not inhibit pERK (Supplemental Fig. 2A) and did not affect cell viability with or without selumetinib (Supplemental Fig. 2B). In addition, overexpression of a GnaqQ209L plasmid in a WT cell line (Supplemental Fig. 2C) sensitized the cells to selumetinib, as compared to cells with an empty vector (Supplemental Fig. 2D). These results confirm that GNAQQ209L/P signals to MEK and specifically renders mutant cells susceptible to MEK inhibition, as reported by Van Raamsdonk et al (2).
Figure 1. Selumetinib inhibits cell viability of BRAFV600E and GNAQQ209L/P cell lines.
A. Cells with wild-type GNAQ/BRAF (C918, Mel290), mutant GNAQ (92.1, Omm1.3, Mel270, Mel202) and BRAFV600E (OCM3, OCM1A) were treated with increasing concentrations of selumetinib (0, 25, 50, 100, 250, 500, 1000nM), and analyzed for cell viability on day 5, calculated as percent of untreated controls. Data are representative of three independent experiments. Bars, mean s.d. B. Immunoblots of pERK and pMEK in response to increasing concentration of selumetinib in cells with different mutational status. Microarray results: C, Venn-diagram summarizing differentially expressed genes in selumetinib-treated GNAQQ209L/P cell lines (green circle), BRAFV600E (red circle) and WT cells (blue circle), with corresponding overlapping genes as indicated. D, Heat map representation of top 19 genes identified as meeting the statistical threshold (see methods) for significant change in expression in all the cell lines and with fold-change > 2 in GNAQQ209L/P cell lines, in response to 250nM selumetinib (grey bar), or DMSO as control for 8 hours, in triplicate. One replicate of the Mel290 cell line was excluded as it did not cluster with the other two replicates. Cell lines with individual replicates are columns, genes are rows.
Expression profile of GNAQQ209L/P cells compared to BRAFV600E MEK signature
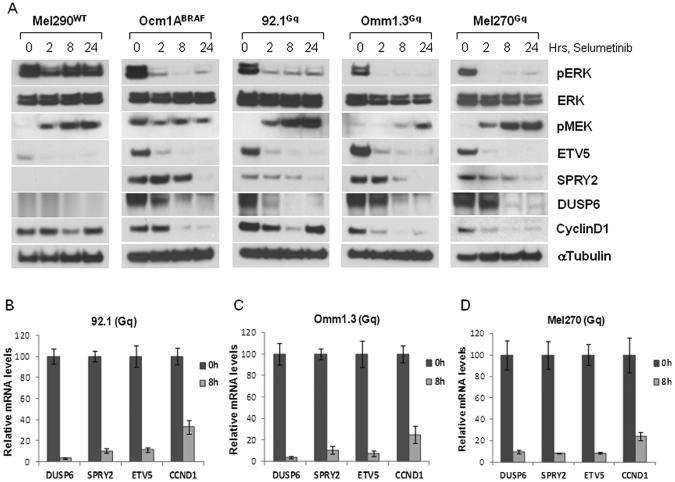
In order to identify the transcriptional profile of MEK inhibition in GNAQQ209L/P cells and potential novel targets of GNAQQ209L/P signaling, we utilized microarray gene expression analysis. Cells from each genetically defined subgroup of UM (three cell lines with GNAQQ209L/P, one line with BRAFV600E and one cell line WT for both) were treated with 250nM selumetinib or vehicle for eight hours. Genes whose change upon MEK inhibition exceeded defined statistical thresholds for all three GNAQQ209L/P cell lines were considered significant and defined the ERK-dependent transcriptional output of GNAQQ209L/P UM cells. A total number of 387 genes met the significance level of p ≤0.0001 in the GNAQQ209L/P cells. Identical parameters were used to define the set of genes regulated differentially by MEK inhibitor in the BRAFV600E and WT cell lines, and a direct comparison was performed to evaluate common genes among the genetic subgroups. Of the 387 genes determined to have changed significantly in response to selumetinib, 308 were differentially expressed only in GNAQQ209L/P cells, while 29 overlapped with BRAFV600E MEK-dependent genes, 42 overlapped with genes meeting significance in WT cells, and 8 genes were common to all three groups (Venn diagram, Fig. 1B). The top 19 genes with highest significance and fold change common to all GNAQQ209L/P cells before and after treatment with selumetinib are displayed in a heat map (Fig. 1D). A list of p-values and fold change of representative genes for each group (GNAQQ209L/P only and overlap groups), are shown in Table 1. Of note, several of the genes that were shared between GNAQQ209L/P and BRAFV600E cells were previously described ERK targets, such as CCND1, transcription factors ETV5, MYC, and genes involved in the feedback inhibition of MEK/ERK signaling, i.e. dual specificity phosphatase 6 (DUSP6), and sprouty family members SPRY2, SPRY4. These genes are key components of “MEK signatures” described in other tumor cells (16, 17), and we confirmed their decline in expression (mRNA and protein levels) after selumetinib treatment, using immunoblotting and quantitative RT-PCR (Fig. 2). Following exposure to selumetinib for up to 24 hours, pERK levels were determined by immunoblotting (Fig. 2A). In the less sensitive WT cell line, pERK was slightly downregulated at 2 hours and quickly rebounded at later time points, while its inhibition was more complete in cells with BRAFV600E and GNAQQ209L/P mutations. Basal expression levels of ETV5, DUSP6 and SPRY2 proteins were almost undetectable in the WT cell line, while they were highly expressed in the mutant cells (both BRAFV600E and GNAQQ209L/P), confirming elevated transcriptional output of MEK signaling in these cells (17). Cyclin D1 was durably downregulated in BRAFV600E cells, while its expression rebounded in GNAQQ209L/P and WT cells at later time points. The induction of pMEK did not occur in BRAFV600E cells, in accordance with the reported abrogation of negative feedback between ERK and RAF proteins by BRAFV600E (17, 20). The microarray results of these MEK-dependent genes were also confirmed by quantitative RT-PCR in three GNAQQ209L/P cell lines, where these genes were suppressed (Fig. 2B, C, D). The high basal expression of these transcripts and their downregulation by selumetinib confirmed that the MEK/ERK pathway is active in GNAQQ209L/P cells, and that the transcriptional events described in GNAQQ209L/P are at least, in part, similar to reported MEK functional activation signatures. For example, Pratilas et al. reported a signature of 52 MEK-dependent genes in BRAFV600E cells (17), of which 19 are represented among our 345 genes (5.5%) and are significantly over-enriched (p<0.0001).
Table 1.
List of differentially expressed genes in response to selumetinib
| Genotype | Probe ID | Gene Name | p-value | log2 FC |
|---|---|---|---|---|
|
| ||||
| GNAQ Q209L/P | ilmn_ 1730928 | cdk5r1 | < 10−11 | −1.96 |
| ilmn_ 1735461 | ddx21 | < 10−7 – 10−11 | −1.73 | |
| ilmn_ 1806023 | jun | < 10−3 – 10−12 | +1.64 | |
| ilmn_ 1748476 | nop58 | < 10−8 – 10−9 | −1.49 | |
| ilmn_ 2330267 | abce1 | < 10−6 – 10−10 | −1.51 | |
| ilmn_ 1806106 | gnl3 | < 10−8 – 10−10 | −1.50 | |
| ilmn_ 1776577 | dscc1 | < 10−5 – 10−9 | −1.41 | |
| ilmn_ 1674302 | ppat | < 10−9 – 10−11 | −1.37 | |
| ilmn_ 2221564 | lyar | < 10−9 – 10−10 | −1.36 | |
| ilmn_ 2067708 | tfb2m | < 10−7 – 10−10 | −1.35 | |
| ilmn_ 1683120 | ung | < 10−8 – 10−10 | −1.34 | |
|
|
||||
|
GNAQQ209L/P- BrafV600E |
ilmn_ 1739222 | etv5 | < 10−13 – 10−14 | −2.34 |
| ilmn_ 2089329 | spry2 | < 10−12 – 10−15 | −2.33 | |
| ilmn_ 2396020 | dusp6 | < 10−9 – 10−12 | −1.94 | |
| ilmn_ 1688480 | ccnd1 | < 10−11 – 10−12 | −1.81 | |
| ilmn_ 1729691 | slc16a6 | < 10−8 – 10−12 | −1.80 | |
| ilmn_ 2086105 | sprv4 | < 10−10 – 10−13 | −1.61 | |
| ilmn_ 2329914 | spry1 | < 10−6 – 10−10 | −1.50 | |
| ilmn_ 1737184 | cdca7 | < 10−6 – 10−10 | −1.20 | |
| ilmn_ 1677765 | lrp8 | < 10−7 – 10−10 | −1.20 | |
| ilmn_ 1680618 | myc | < 10−6 – 10−10 | −1.10 | |
|
|
||||
|
GNAQQ209L/P- WT |
ilmn_ 1744147 | osbpl8 | < 10−7 – 10−10 | −1.56 |
| ilmn_ 1782459 | cebpz | < 10−7 – 10−8 | −1.35 | |
| ilmn_ 2131523 | sacs | <10−6 – 10−9 | −1.34 | |
| ilmn_ 1687978 | phlda1 | < 10−5 – 10−10 | −1.33 | |
| ilmn_ 1733356 | prei3 | <10−5 – 10−10 | −1.11 | |
log2 FC = average fold-change for three GNAQQ209L/Pcell lines
Figure 2. Validation of MEK-regulated genes in GNAQQ209L/P cells.
These genes were representative of those shared between GNAQQ209L/P and BRAFV600E MEK-dependent signature. A, Immunoblot analysis of cells treated with 250nM selumetinib over the time, using antibodies for the indicated proteins. Each blot is representative of at least two experiments showing same results. B, C, D, Relative mRNA levels of selected ERK output genes before and after selumetinib in three GNAQQ209L/P cell lines. Cells were treated with selumetinib for 8h and RT-PCR was performed using gene-specific primers for DUSP6, ETV5, SPRY2 and cyclin D1. Values were normalized with GADPH and HPRT as housekeeping genes using the ΔΔCT method. Each experiment was performed in triplicates. Bars, ± sd.
MEK inhibition regulates unique genes in GNAQQ209L/P cells
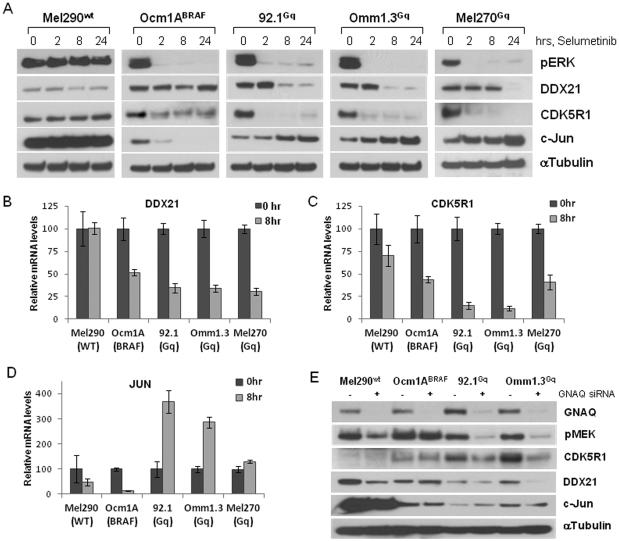
The gene profile of MEK inhibition in GNAQQ209L/P cell lines also comprised uniquely regulated genes (Table 1). From this list, we selected three genes, two down-regulated (DDX21, CDK5R1) and one up-regulated (JUN) for validation by immunoblotting and quantitative RT-PCR (Fig. 3). Immunoblot detection showed that DDX21 and CDK5R1 protein levels were downregulated in GNAQQ209L/Pcells after MEK inhibition over time, but not in the BRAFV600E and WT cell lines (Fig. 3A). JUN increased exclusively in the GNAQQ209L/P cells, while it was quickly downregulated in BRAFV600E cells. In contrast, JUN protein expression was high in WT cells and did not change upon treatment (Fig. 3A). Selumetinib also reduced DDX21 and CDK5R1 mRNA levels in GNAQQ209L/P cells, while they did not significantly change in WT cells (Fig. 3B & C). In the BRAFV600E cells there was a decrease in mRNA levels, but generally to a lesser extent than the GNAQQ209L/P cells and this was associated with relatively minor changes in the protein levels of DDX21 and CDK5R1 (Fig. 3B & C, and Fig 3A). Also, JUN was induced upon treatment in GNAQQ209L/P cells, while it was profoundly decreased in BRAFV600E (Fig. 3D), in accordance with the protein levels. There was also a slight decrease in JUN mRNA in WT cells that did not affect the protein levels (Fig. 3D).
Figure 3. Confirmation of MEK-dependent genes differentially regulated by selumetinib in GNAQQ209L/P cells.
A, Immunoblot analysis of DDX21, CDK5R1 and JUN expression after treatment with 250nM selumetinib over the time in cell lines with different mutational status. Total RNA was extracted from cells after 8h of treatment and qRT-PCR was performed using gene-specific primers for DDX21 (B), CDK5R1 (C) and JUN (D). Values are relative to mRNA levels of untreated cells. Each experiment was performed two or three times in triplicates. E, Western blot analysis of UM cells after GNAQ knockdown by siRNA using the indicated antibodies.
To prove specificity of MEK-dependent signaling downstream of GNAQ and to exclude drug unrelated effects, the expression levels of these genes were analyzed in cells after GNAQ knock down. Phospho-MEK was markedly downregulated in the GNAQQ209L/P cells after GNAQ depletion (Fig. 3E). A slight decrease was seen in WT cell line possibly due to the disruption of wild type GNAQ signaling. Consistent with the effects of selumetinib, CDK5R1 and DDX21 were downregulated in the GNAQQ209L/P cells, while c-Jun was increased, though less than was observed with drug alone (Fig. 3E). In order to determine whether the effects on GNAQQ209L/P cells were restricted to selumetinib, we treated UM cells with a different MEK inhibitor, PD0325901. Both the GNAQQ209L/P and BRAFV600E cells were sensitive to increasing concentrations of the drug (Supplemental Fig. 3B). In terms of protein expression, the change in c-jun (induction in GNAQQ209L/P, suppression in BRAFV600E, and no change in WT) was similar to selumetinib, but suppression of CDK5R1 and DDX21 was observed in both GNAQQ209L/P and BRAFV600E cell lines (Supplemental Fig. 3A). Interestingly, PD0325901 inhibited pERK in the WT cell line but this resulted in essentially no reduction in cell viability, nor a decrease in cyclin D1 expression. Overall, these results suggest that all the effects of selumetinib on transcriptional expression in UM cells may not be shared by other MEK inhibitors. This will require further investigation.
MEK-dependent GNAQQ209L/P specific genes mediate cell proliferation and cell migration
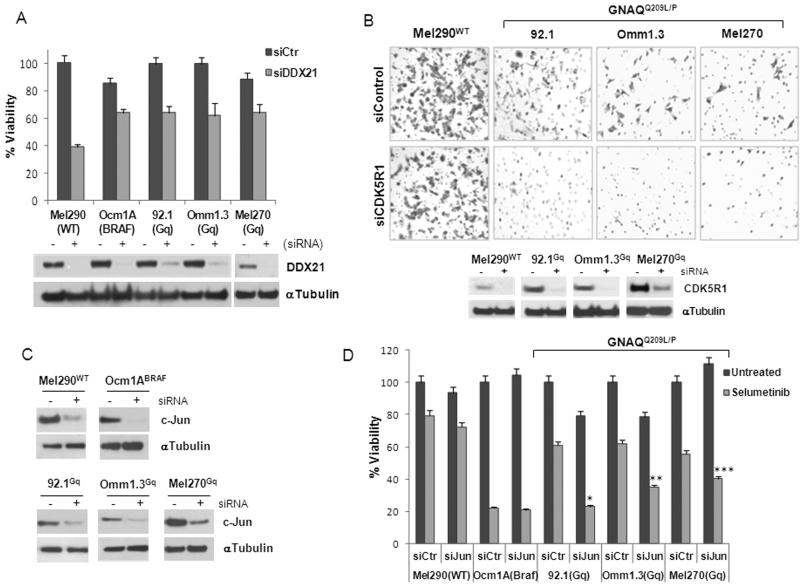
Of the genes uniquely down-regulated by selumetinib in GNAQQ209L/P cells, we focused on DDX21 and CDKR51. DDX21 is a member of RNA helicase family, which contains a motif (DExD/H) highly conserved from bacteria to humans (21). They are multifunctional proteins involved in RNA unwinding and play a role in transcription regulation (22). To test whether DDX21 is involved in the anti-proliferative effects of selumetinib, we carried out gene knockdown experiments. Three GNAQQ209L/P mutant cell lines, as well as BRAFV600E and WT cells, were transiently transfected with two different DDX21-specific or control siRNAs (Fig. 4A, lower panel, and Supplemental Fig. 4A). Depletion of DDX21 reduced cell viability of all the cell lines, indicating a role for DDX21 in cell proliferation that is independent of the GNAQ status. In fact, the most affected was the WT cell line with 60% inhibition of proliferation. The BRAFV600E cells were the least sensitive, with 20% inhibition, whereas GNAQQ209L/P cells showed ~40% inhibition relative to control siRNA-transfected cells (Fig. 4A, upper panel, Supplemental Fig. 4B). The addition of selumetinib decreased viability of the GNAQQ209L/P cell lines by only an additional 10-20% (Supplemental Fig. 5C, D, E), suggesting that DDX21 downregulation contributes to the anti-proliferative effects of selumetinib in these cells. Furthermore, DDX21 downregulation did not affect migration of the cell lines (data not shown).
Figure 4. GNAQQ209L/P-MEK-dependent genes play a role in cell proliferation and metastasis.
A, siRNA mediated knockdown of DDX21 (+) in five cell lines (lower panel); (-), control siRNA. Cell viability was measured after 4 days (upper panel). B, Migration assays of WT and GNAQQ209L/P cell lines transfected with CDK5R1 or control siRNA (lower panels). Cells migrated through the membrane pores were stained and counted under the microscope. Representative fields of x200 magnification are shown. C, UM cell lines were trasnfected with control or c-Jun siRNA and tested in proliferation assays (D) after 4 days of 250nM selumetinib treatment. Experiments were repeated three times in triplicates. * p<0.0001; ** p<0.001, *** p<0.005 for comparison of c-Jun versus control siRNA with selumetinib treatment.
CDK5R1 encodes p35, a specific activator of the serine/threonine kinase CDK5, which plays a crucial role in CNS development and maintenance (23). A role for CDK5 and p35 in cell migration has also been reported (24, 25). Seeking to establish a possible functional role underlying the observed differences in CDK5R1 expression, we assessed the impact of knockdown of this protein in UM cells using two different siRNA. CDK5R1 siRNA did not significantly inhibit cell viability (Supplemental Fig. 4C). All three GNAQQ209L/P cell lines depleted of CDK5R1 (Fig. 4B, lower panels) showed a decrease in cell migration (Fig. 4B, upper panels) compared to cells with control siRNA. The WT cells (Fig. 4B), as well as the BRAFV600E (Supplemental Fig. 6) showed no inhibition of migration with siRNA to CDK5R1, whereas GNAQ siRNA inhibited migration of the GNAQQ209L/P cell line Omm1.3 (Supplemental Fig. 6). Interestingly, selumetinib inhibited migration of both GNAQQ209L/P and BRAFV600E, but not the WT cells (Supplemental Fig. 6), suggesting that selumetinib may have effects on the cell migration of BRAFV600E cells, but this effect is independent of CDK5R1.
JUN expression is related to sensitivity to MEK inhibition
c-Jun is a component of the AP-1 transcription complex regulated by JNK, and it is involved in a number of cell responses, like cell proliferation and cell death (26). c-Jun was up-regulated in GNAQQ209L/P cells after selumetinib treatment, showing a differential regulation compared to cells with BRAFV600E. Knock down of c-Jun by two different siRNAs (Fig. 4C and Supplemental Fig. 7A) significantly increased the antiproliferative effects of selumetinib in GNAQQ209L/P cell lines (Fig. 4D and Supplemental Fig. 7B), suggesting that c-Jun induction may be involved in mechanisms of resistance to MEK inhibition. In contrast, c-Jun did not seem to play a role in WT and BRAFV600E cells as its knockdown did not alter the sensitivity to the selumetinib when compared to control siRNA (Fig. 4D and Supplemental Fig. 7B).
Validation of GNAQQ209L/P-specific ERK transcriptional output in tumor tissues
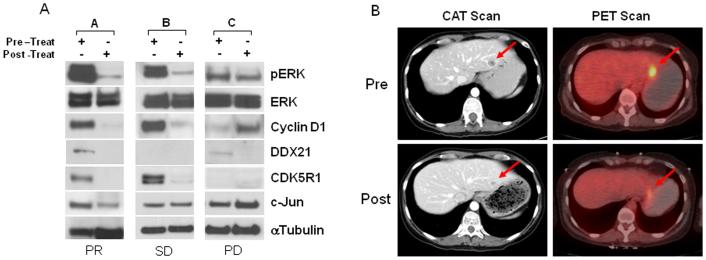
In order to assess the clinical efficacy of MEK inhibition on uveal melanoma, we are conducting a phase II clinical trial of selumetinib versus temozolomide in patients with metastatic uveal melanoma (clinicaltrials.gov # NCT01143402). Matched tumor biopsies are collected to examine target modulation between baseline and day 14 from patients with GNAQ/11 mutations receiving selumetinib. Results from three representative biopsy pairs are shown (Fig. 5A). The results of all the patients in the trial will be reported separately. Sustained inhibition of pERK and suppression of cyclin D1 were observed in patients A and B on day 14, but not in patient C. This correlated to best clinical response by RECIST criteria (27): partial response in liver metastases in patient A (Fig 5B), stable disease in patient B and progression of disease in patient C. Furthermore, DDX21 was downregulated by selumetinib in patients A and C, CDK5R1 decreased in patient A and B, whereas they could not be detected in patient B and C, respectively. c-Jun expression increased in patient C (Fig 5A). A similar trend has been seen in other patients treated on this clinical trial. These preliminary results are consistent with our in vitro studies indicating the existence of unique subset of genes in GNAQQ209L/P cells that are regulated by selumetinib and the expression of which could have an impact on clinical outcome.
Figure 5. Validation of MEK inhibition and expression of ERK-dependent genes in tumor tissues.
A, Baseline and day 14 liver tumor biopsies were collected in patients with mutant GNAQ/11 metastatic uveal melanoma receiving selumetinib in a phase II clinical trial. Matched-pair biopsies were analyzed by immunoblotting for expression levels of pERK, ERK, Cyclin D1, CDK5R1, DDX21, c-Jun and α-tubulin. Clinical efficacy is also shown: PR= partial response, SD= stable disease, PD= progressive disease. B, Liver metastases of patient A with uveal melanoma (pre- and post-treatment with selumetinib). A partial response in the liver lesion is assessed by CAT scan (left, arrows) and PET scan (right, arrows).
Discussion
Uveal melanoma represents the most common intraocular malignancy. However, there are no effective treatments for this aggressive disease. Selumetinib is the only MEK inhibitor in clinical trials currently in the United States for patients with uveal melanoma. Here we report that cells with GNAQQ209L/P mutations are sensitive to MEK inhibition by selumetinib, and sensitization was associated with a MEK-dependent gene expression profile. Some features of this profile are overlapping with that elicited in BRAFV600E UM cells and other cell types (16), which supports the MEK dependence of GNAQQ209L/P cells. This gene profile includes the dual-specificity phosphatases (DUSP4/6), the sprouty homologues (SPRY1/2/4), which are known transcriptional targets of the ERK pathway involved in negative feedback regulation of ERK. The Ets variant transcription factor ETV5 was also regulated by MEK inhibition, along with cell division cycle associated protein 7 (CDCA7), the proto-oncogene MYC, and the solute carrier family 16, member 6 (SLC16A6).
Additional features of the MEK profile were identified as specific for GNAQQ209L/P cells. A number of genes suppressed in GNAQQ209L/P cells by selumetinib, like LYAR, NOP58, GNL3 and PPAT were reported as nuclear proteins involved in cell growth and tumorigenesis (28-31). DDX21 was recently identified as a novel biomarker for colorectal cancer (32), while CDK5R1 was involved in metastasis (24, 33) and associated with meningioma progression (34). Interestingly, it has been reported that mutant K-RAS regulates expression/stability of CDK5 and CDK5R1 (p35) to increase malignant progression and invasion of pancreatic cancer cells (35). It is plausible that mutant GNAQ acts similarly to mutant K-RAS, as CDK5R1 expression was in fact elevated in the GNAQQ209L/P cells compared to cells with other genetic backgrounds. Furthermore, it has been reported that CDK5 negatively regulates c-Jun N-terminal kinase 3 activity and its target c-Jun, to prevent apoptosis in developing neurons (36). This implicates a possible interaction between these proteins in promoting survival of UM cells. CDK5R1 and DDX21 were also downregulated by selumetinib in vivo in tissues of patients enrolled in a Phase II clinical trial we are conducting.
JUN was upregulated after selumetinib treatment in the GNAQQ209L/P cells only. Depending on the cell type and drug treatment, c-Jun and Jun kinase have been implicated in both pro- and anti-apoptotic responses (37). In cutaneous melanoma cells, active ERK induces c-Jun expression (38). In contrast, c-Jun was induced by MEK inhibition in GNAQQ209L/P UM cells, suggesting a differential regulation of the ERK/JNK pathway. G protein-mediated signaling is complex and involves multiple downstream binding partners and various regulatory scaffolding/adaptor and effector proteins (12). For example, PKC is a target of GNAQ activation, and it might be involved in feedback regulation of c-Jun when ERK is inhibited. The upregulation of c-Jun could represent an alternative route to cell proliferation, which would explain the relative lower sensitivity to selumetinib of GNAQQ209L/P cells as compared to BRAFV600E cells. Interestingly, increased expression of c-Jun has also been reported in colorectal cancer cells with KRAS or BRAF mutations after acquired resistance to selumetinib (39). We demonstrated that the anti-proliferative effect of selumetinib can be enhanced by suppressing c-Jun in the GNAQQ209L/P cells. This would suggest that targeting c-Jun in the presence of MEK inhibition would result in enhanced anti-tumor effects and may prevent selumetinib resistance. In conclusion, our findings define a unique molecular profile of MEK inhibition by selumetinib in UM cells with mutant GNAQ, and point to a set of transcriptionally modified genes that could have an impact on the activity of this agent in this disease.
Supplementary Material
Translational Relevance.
Uveal melanoma is the most common primary intraocular malignancy in adults. Metastasis occurs frequently and there are no effective therapies. Recently, it has been demonstrated that 85% of uveal melanomas have oncogenic mutations in the GNAQ/11, which activate the MAPK pathway. Here, we analyzed the transcriptional profile of GNAQ mutant cell lines treated with selumetinib, a MEK inhibitor, currently in clinical trial for uveal melanoma. We found that these cells have a MEK functional activation signature and a unique set of MEK-dependent genes involved in proliferation, cell invasion and drug resistance. These genes are also regulated in pre- and post-treatment tumor biopsies obtained from patients with uveal melanoma treated with selumetinib, and their expression may correlate to clinical activity. Our findings provide new insights into the biologic effect of selumetinib in this disease and may have profound implications for the clinical development of MEK inhibitors in uveal melanoma.
Acknowledgments
Grant Support: This work was supported by Melanoma Research Alliance and Cycle for Survival funds. C.A. Pratilas was supported by the NIH grant # K08-127350.
References
- 1.Zebisch A, Czernilofsky AP, Keri G, Smigelskaite J, Sill H, Troppmair J. Signaling through RAS-RAF-MEK-ERK: from basics to bedside. Curr Med Chem. 2007;14:601–23. doi: 10.2174/092986707780059670. [DOI] [PubMed] [Google Scholar]
- 2.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strickland D, Lee JA. Melanomas of eye: stability of rates. Am J Epidemiol. 1981;113:700–2. doi: 10.1093/oxfordjournals.aje.a113150. [DOI] [PubMed] [Google Scholar]
- 6.Horsman DE, White VA. Cytogenetic analysis of uveal melanoma. Consistent occurrence of monosomy 3 and trisomy 8q. Cancer. 1993;71:811–9. doi: 10.1002/1097-0142(19930201)71:3<811::aid-cncr2820710325>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 7.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18:75–84. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Mooy CM, Van der Helm MJ, Van der Kwast TH, De Jong PT, Ruiter DJ, Zwarthoff EC. No N-ras mutations in human uveal melanoma: the role of ultraviolet light revisited. Br J Cancer. 1991;64:411–3. doi: 10.1038/bjc.1991.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuidervaart W, van Nieuwpoort F, Stark M, Dijkman R, Packer L, Borgstein AM, et al. Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer. 2005;92:2032–8. doi: 10.1038/sj.bjc.6602598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malaponte G, Libra M, Gangemi P, Bevelacqua V, Mangano K, D’Amico F, et al. Detection of BRAF gene mutation in primary choroidal melanoma tissue. Cancer Biol Ther. 2006;5:225–7. doi: 10.4161/cbt.5.2.2429. [DOI] [PubMed] [Google Scholar]
- 11.Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213:589–602. doi: 10.1002/jcp.21246. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard KB, Hepler JR. Cell signalling diversity of the Gqalpha family of heterotrimeric G proteins. Cell Signal. 2006;18:135–50. doi: 10.1016/j.cellsig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leboeuf R, Baumgartner JE. BRAFV600E mutation is associated with preferential sensitivity to mitogen-activated protein kinase kinase inhibition in thyroid cancer cell lines. J Clin Endocrinol Metab. 2008;93:2194–201. doi: 10.1210/jc.2007-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–83. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–73. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68:6145–53. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 21.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 23.Lopes JP, Agostinho P. Cdk5: multitasking between physiological and pathological conditions. Prog Neurobiol. 2011;94:49–63. doi: 10.1016/j.pneurobio.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Strock CJ, Park JI, Nakakura EK, Bova GS, Isaacs JT, Ball DW, et al. Cyclin-dependent kinase 5 activity controls cell motility and metastatic potential of prostate cancer cells. Cancer Res. 2006;66:7509–15. doi: 10.1158/0008-5472.CAN-05-3048. [DOI] [PubMed] [Google Scholar]
- 25.Contreras-Vallejos E, Utreras E, Gonzalez-Billault C. Going out of the brain: Non-nervous system physiological and pathological functions of Cdk5. Cell Signal. 2011;24:44–52. doi: 10.1016/j.cellsig.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–77. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 27.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 28.Su L, Hershberger RJ, Weissman IL. LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes & Dev. 1993;7:735–48. doi: 10.1101/gad.7.5.735. [DOI] [PubMed] [Google Scholar]
- 29.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–64. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida R, Fujimoto T, Kudoh S, Nagata M, Nakayama H, Shinohara M, et al. Nucleostemin affects the proliferation but not differentiation of oral squamous cell carcinoma cells. Cancer Sci. 2011;102:1418–23. doi: 10.1111/j.1349-7006.2011.01935.x. [DOI] [PubMed] [Google Scholar]
- 31.Gassmann MG, Stanzel A, Werner S. Growth factor-regulated expression of enzymes involved in nucleotide biosynthesis: a novel mechanism of growth factor action. Oncogene. 1999;18:6667–76. doi: 10.1038/sj.onc.1203120. [DOI] [PubMed] [Google Scholar]
- 32.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–9. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 33.Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, et al. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PloS One. 2011;6:e20038. doi: 10.1371/journal.pone.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrobel G, Roerig P, Kokocinski F, Neben K, Hahn M, Reifenberger G, et al. Microarray-based gene expression profiling of benign, atypical and anaplastic meningiomas identifies novel genes associated with meningioma progression. Int J Cancer. 2005;114:249–56. doi: 10.1002/ijc.20733. [DOI] [PubMed] [Google Scholar]
- 35.Eggers JP, Grandgenett PM, Collisson EC, Lewallen ME, Tremayne J, Singh PK, et al. Cyclin-Dependent Kinase 5 Is Amplified and Overexpressed in Pancreatic Cancer and Activated by Mutant K-Ras. Clin Cancer Res. 2011;17:6140–50. doi: 10.1158/1078-0432.CCR-10-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li BS, Zhang L, Takahashi S, Ma W, Jaffe H, Kulkarni AB, et al. Cyclin-dependent kinase 5 prevents neuronal apoptosis by negative regulation of c-Jun N-terminal kinase 3. EMBO J. 2002;21:324–33. doi: 10.1093/emboj/21.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasilevskaya I, O’Dwyer PJ. Role of Jun and Jun kinase in resistance of cancer cells to therapy. Drug Resist Updat. 2003;6:147–56. doi: 10.1016/s1368-7646(03)00043-8. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–60. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.