Abstract
Phosphorylation of CARMA1 is a crucial event initiating the assembly of IKK and JNK signaling complexes downstream of activated antigen receptors. We previously mapped three PKC target sites in murine CARMA1 in vitro, and demonstrated that mutation of two of these serines (S564 and S657) resulted in reduced NF-κB activation, while mutation of the third serine (S649) had no clear effect. Here we report that, when low concentrations of antigen receptor activators are used, loss of S649 (by mutation to alanine) promotes enhanced IKK and JNK activation in both B and T cell lines. Reconstitution of CARMA1−/− DT40 B-cells with CARMA1 S649A leads to increased cell death and reduced cell growth in comparison to WT CARMA1, likely a result of enhanced JNK activation. To directly determine if S649 is modified in vivo, we generated phospho-specific antibodies recognizing phospho-S649, and phospho-S657 as a positive control. While phospho-S657 peaked and declined rapidly after antigen receptor stimulation, phospho-S649 occurred later and was maintained for a significantly longer period post-stimulation in both B and T cells. Interestingly, phospho-S657 was completely abolished in PKCβ-deficient B cells, whereas delayed phosphorylation at S649 was partially intact and depended, in part, upon novel PKC activity. Thus, distinct PKC-mediated CARMA1 phosphorylation events exert opposing effects on the activation status of CARMA1. We propose that early phosphorylation events at S657 and S564 promote the initial assembly of the CARMA1 signalosome, while later phosphorylation at S649 triggers CARMA1 down-regulation.
Keywords: CARMA1, lymphocytes, Serine phosphorylation, NF-κB, JNK
Introduction
The transcription factor NF-κB and the MAPK JNK are both activated downstream of antigen receptors (AR), and together regulate the activation, proliferation, apoptosis, survival and differentiation of lymphocytes (1, 2). In AR signaling, both NF-κB and JNK share requirements for molecules involved in the proximal tyrosine phosphorylation cascade leading to PLCγ activation. They also both require PKC-dependent activation of CARMA1, as well as the downstream adaptors BCL10 and MALT1 (3). Downstream of MALT1, many of the molecular players required for activation of the classical NF-κB pathway have been described, and are shared with other families of membrane-bound receptors. Activation of JNK is less fully understood, but is known to require phosphorylation mediated by MEK4, MEK7 and MEK6 in lymphocytes (1, 4). Once activated, JNK phosphorylates various downstream substrates, including c-Jun, to promote activation of the AP-1 transcription factor (5).
The adaptor protein caspase-recruitment domain (CARD)-MAGUK containing protein 1 (CARMA1) is required for activation of both of these pathways. CARMA1-deficient cellular and animal models have demonstrated the importance of this protein in immunity (6–11). CARMA1-deficient lymphocytes exhibit reduced proliferation, differentiation and survival to AR engagement or AR agonists (P/I; chemical activators of PKCs). At the molecular level, BCR or TCR stimulation of CARMA1-deficient lymphocytes induces normal phosphotyrosine signals, Ca2+ flux, and ERK phosphorylation; however NF-κB and JNK activities remain inactive. Structurally, CARMA1 contains several domains important for the recruitment of signaling proteins as well as for its oligomerization and plasma membrane recruitment (12, 13). The N-terminal portion of CARMA1 includes both a CARD and a coiled-coil domain, required for the recruitment of the adaptor proteins BCL-10 (10, 12–15) and CARMA1 oligomerization (16), respectively. These domains are linked to a C-terminal region highly related to the MAGUK family of proteins (17). The MAGUK region contains a postsynaptic density 95/disc large/zona occludens 1 (PDZ), a SRC-homology 3 (SH3), and a guanylate kinase-like (GUK) domain. The CARD, CC, SH3 and GUK domains are each required for CARMA1 activity (12, 18) and interact with distinct proteins to coordinate CARMA1 signaling (e.g., CARD with BCL10, CC for homodimerization, etc)(3)
We and others have demonstrated that CARMA1 activation is controlled by PKCβ in B cells and PKCθ in T cells (19, 20). Activated PKCβ or PKCθ phosphorylate a flexible region of CARMA1 linking the coiled-coil and PDZ domains termed the PKC-Regulated Domain (PRD) (3, 21). We demonstrated that specific serine residues within the murine PRD (S564, S649 and S657) were phosphorylated by recombinant PKCβ/θ isoenzymes in vitro, and mutation of these residues to alanine revealed that both S564 and S657 were required for AR-induced NF-κB activation in Jurkat T cells. In contrast, the S649 residue was apparently not required for the activation of a NF-κB reporter gene (20). Similar results were recently reported for the analogous serine residue (S660) of chicken CARMA1 (22).
These findings suggested two alternative possibilities with regard to the role for S649: that this residue is not a functionally relevant in vivo phosphorylation site in lymphocytes; or that S649 phosphorylation occurs in vivo, but has a separate role from that of activating CARMA1 signaling. In this report we demonstrate that, while S657 phosphorylation delivers a positive signal for NF-κB and JNK activation, S649 phosphorylation negatively impacts these molecular events in lymphocytes. Functionally, S649 mutation results in a decrease in the survival and growth of reconstituted CARMA1−/− DT40 B-cells, compared to WT CARMA1. Using antibodies directed to phosphorylated S649 and S657 (p-S649 and p-S657), we show that CARMA1 is phosphorylated in vivo at both S657 and S649 in response to AR engagement, yet with markedly different kinetics. Thus, specific phosphorylation of serine residues in the PRD domain differentially regulates the downstream signaling activity of CARMA1.
Materials and Methods
Cells lines, reagents and antibodies
Chicken B cell lines with genetic deletions of PKCβ or CARMA1 (PKCβ−/− DT40 and CARMA1−/− DT40) were gifts from Tomohiro Kurosaki (23); human Ramos B cells, Jurkat T cells and HEK-293 fibroblasts were obtained from ATCC. Cells were cultured as described (20). Rottlerin, Ro318425, PMA and Ionomycin were from EMD Biosciences. Antibodies utilized were against: chicken IgM (Bethyl Labs), myc epitope (9E10; Sigma-Aldrich), human CD28 and IKK-γ (BD Biosciences), pERK, ERK, actin, IκBα and PKCβ-II (Santa Cruz Biotechnology), p-IκBα and p-JNK (Cell Signaling Technology) and human CD3 (OKT3; a gift from Dr. Steve Ziegler). Polyclonal antibodies against p-S649 and p-S657 of murine CARMA1 were generated in rabbits by Open Biosystems using the following phosphorylated peptides: p-S649, LRKF(pS)LERPFR; p-S657, ERPFRP(pS)VTSG.
Generation of retroviral supernatants and stable cells lines
HEK 293 cells (5×106/plate) were transfected using calcium phosphate with 10 μg of the retroviral packing plasmid 10A1 and each of the following cDNAs: myc-CARMA1-WT, -S649A, -S657A, or -S564A sub-cloned into MSCV-IRES-GFP retroviral vector. Culture supernatants were harvested, filtered using 0.22 μm filters, and incubated with the lymphoid cells lines in the presence of 8 μg/ml of polybrene. Cells were spinfected for 20 min at 800×g RPM. Depending on the experiment, cells were counted and analyzed for cell survival at 0 and 24 hours post-transduction, or cultured and expanded for cell sorting based on GFP expression using a FACSAria cell sorter (BD Biosciences).
Cell stimulation, immunoprecipitation and immunoblotting
DT40, Ramos and Jurkat cells (2×106 cell/condition) were stimulated with PMA (at doses ranging from 3.9 nM to 1 μM) and Ionomycin at 1 μg/mL; anti-IgM 5, 2.5, 1.25 μg/mL), or anti-CD3 (10 μg/mL)/anti-CD28 (1 μg/ml) for selected time points at 37°C. Cells were lysed in RIPA buffer (250 mM NaCl, 10 mM Tris-HCl, pH 7.4, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1 mM Na3VO4, 1 mM NaF) containing a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich). For immunoprecipitations, 10–30×106 cells/condition were lysed in RIPA buffer without Na-deoxycholate and SDS. Lysates were incubated with 1.5 μg of anti-myc for 15 min followed by 2 h incubation with protein-G beads (GE Healthcare) at 4°C. For testing phospho-specific antibodies, GST-PRD, GST-PRD-S649A, and GST-PRD-S657A recombinant proteins were produced and phosphorylated in vitro as described, using non-radioactive ATP (20). Protein complexes were resolved by SDS-PAGE and transferred to PVDF membranes. Primary antibodies were incubated overnight followed by 1–2 h incubation with anti-rabbit-IgG IRDye800 (Rockland Inc), and/or anti-mouse IgG Alexa Fluor 680 (Molecular Probes). Proteins were detected and quantified by an Odyssey Infrared Imaging System from Li-Cor Biosciences.
IKK in vitro kinase assay
DT40 cells (20 × 106/condition) were serum-starved 1 h at 37°C then either left unstimulated or stimulated with P/I for 10 min at 37°C. Cells were lysed in ice-cold lysis buffer, and an IKK in vitro kinase assay (Cell Signaling) was carried out as described by the manufacturer.
NF-κ B reporter gene assays
HEK 293 cells (5 × 105/sample) or Jurkat Tcells (1 ×106/sample) were transfected for 24 h with Fugene 6 (Roche) at a DNA/Fugene-6 ratio of 1:6 with 250 ng of an NF-κB reporter plasmid (Igk2-IFN-Luciferase, a gift from Joel Pomerantz (12)) and pRenilla-TK (Promega; transfection control) reporter vector, and several doses of myc-CARMA1 or myc-CARMA1-S649A mutant cDNA in the pIRES-PURO backbone (Clontech). Six h before harvesting, the cells were stimulated with P/I (1 μM/1 μg/mL) for HEK 293 cells, or CD3 (5 μg/mL)/CD28 (1 μg/mL) for Jurkat T cells. NF-κB activation was analyzed measuring the firefly and Renilla luciferase activities according to the Dual-Luciferase Reporter assay from Promega.
Results
IKK and JNK activation DT40 cells expressing CARMA1 phosphorylation site mutants
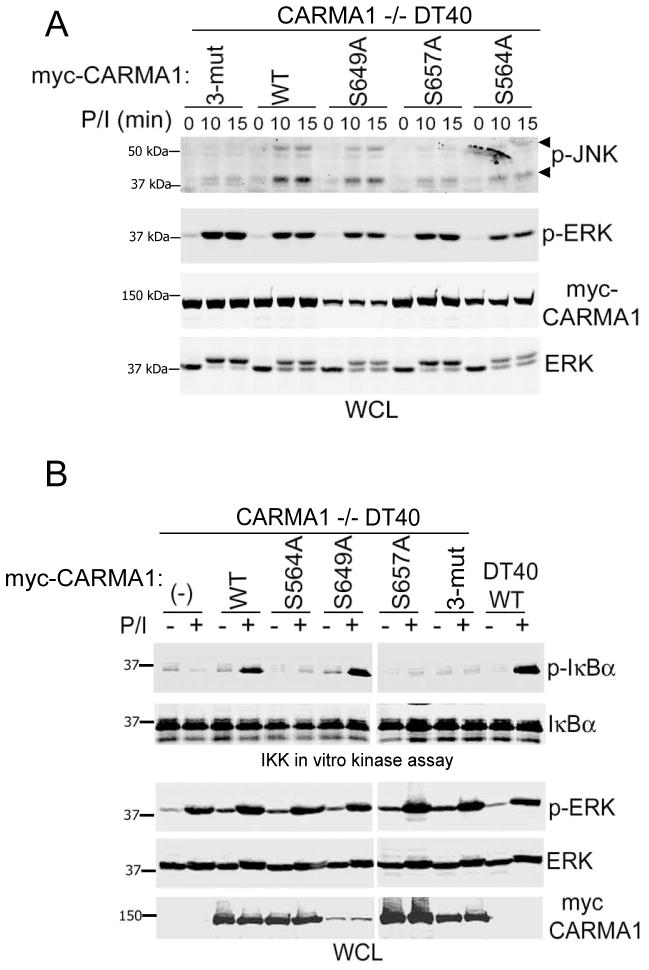
Using CARMA1-deficient chicken D40 cell lines reconstituted with mutant chicken CARMA1 coding sequences, Shinohara et al. (22) were able to validate various potential PKC target sites in CARMA1 downstream of AR stimulation. We performed an identical set of experiments using P/I stimulation to confirm that the murine CARMA1 PKC target sites that we have previously identified (S564, 649, and 657; (20)), have the same effect on CARMA1 function as the correlating chicken CARMA1 residues. CARMA1−/− DT40 cells were reconstituted with either CARMA1-WT or CARMA1 constructs with S564A, S649A, or S657A mutations. IKK and JNK activation was analyzed after treatment with a high dose of the AR chemical mimetics P/I (1 μM/1 μg/ml, respectively; Fig. 1). JNK activity was assessed using p-JNK specific antibodies, and IKK activity assessed using an in vitro kinase assay.
FIGURE 1.
Mutation of serines 564 and 657, but not 649, interferes with CARMA1-dependent IKK and JNK activation in DT40 B cells. A and B, WT or CARMA1−/− DT40 cells stably expressing myc-CARMA1-WT, myc-CARMA1 (S564,649,657A) (3-mut) or the specified single serine-to-alanine mutants (2×106 cells/condition) were activated with P/I (1 μM and 1 μg/ml, respectively) for the indicated time (A), or for 0 (−) and 10 min (+; B). JNK activation was assessed by immunoblotting using anti-p-JNK antibodies (A), and IKK activity (measured as IκBα phosphorylation) was analyzed as described in material and methods (B). p-ERK immunoblot was used as an activation control, and anti- myc (CARMA1), and ERK blots were used as loading controls. WCL, whole cell lysates.
Mutation of all three murine serine residues that we have reported to be PKC-specific substrates (CARMA1-3-mut) produced a CARMA1 molecule unable to reconstitute IKK or JNK activation, indicating that some of these residues are required to initiate CARMA1 signaling. To determine the impact of the individual residues, CARMA1−/− DT40 cells expressing CARMA1 single serine mutants were activated and analyzed as described above. Consistent with previous reports (19, 20, 22), while S564A and S657A mutations substantially decreased both JNK (Fig. 1A) and IKK (Fig. 1B) activation, S649A mutation did not show any functional defect under these activation conditions.
Phosphorylation of CARMA1 at S649 negatively regulates IKK activation
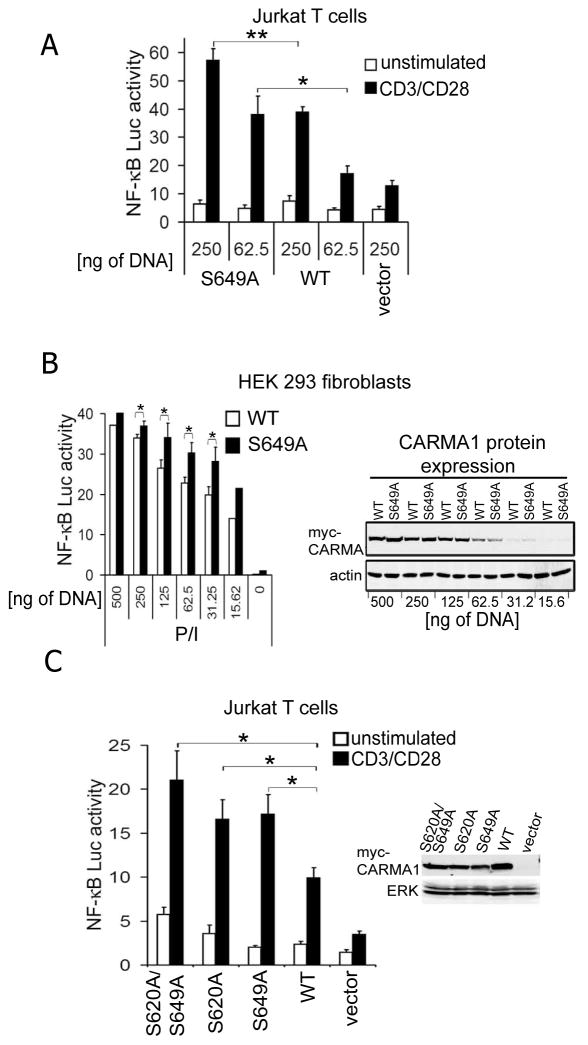
We consistently observed that, following stable transduction of CARMA1−/− DT40 cells using retroviral vectors, protein expression levels of myc-CARMA1-S649A were ~3–5 fold lower than those observed for wt myc-CARMA1, or myc-CARMA1 with other serine mutations (Fig. 1). Shinohara et al. reported a similar phenomenon with the analogous serine residue in the chicken CARMA1 sequence, S660 (22). Notably, despite its reduced expression level, myc-CARMA1-S649A reconstituted IKK and JNK activation to levels similar to that induced by the WT CARMA1 protein. This observation strongly suggested that the S649A mutation might enhance the signaling activity of CARMA1. Although we and others have previously reported that the S649A mutation has no effect on NF-κB reporter gene expression in transient transfection assays (20), we retested this possibility using a range of doses of the transfection plasmid. Vectors expressing myc-tagged CARMA1 WT or S649A were transiently transfected, along with NF-κB-luciferase and TK-Renilla reporter plasmids, into Jurkat T cells followed by CD3/CD28 stimulation. CARMA1-S649A-transfected cells consistently exhibited a ~30% to ~50% increase in CD3/CD28-induced NF-κB activation 24h post-transfection (Fig. 2A). The activity of the myc-CARMA1 and myc-CARMA1-S649A proteins was also evaluated in transiently transfected and P/I-stimulated HEK-293 cells (Fig. 2B). Comparison of NF-κB reporter gene activation between various doses of myc-CARMA1 or myc-CARMA1-S649A expression vectors showed that myc-CARMA1-S649A expressing cells consistently exhibited ~30% more NF-κB activity than cells transfected with myc-CARMA1. Anti-myc immunoblotting (Fig. 2B, right panel), showed equivalent levels of WT and mutant protein expression in transfected cells, indicating that differential protein expression cannot account for these observed differences in NF-κB activation.
FIGURE 2.
NF-κB activation is enhanced in Jurkat T cells and HEK 293 fibroblasts transiently expressing the CARMA1-S649A and -S620A mutants in AR and P/I stimulated cells respectively. A, Jurkat cells were co-transfected for 24h with the Igκ2-Luciferase/pRL-TK reporter vectors plus 250 and 62 ng of expression vectors for myc-CARMA1 (WT) and myc-CARMA1-S649A or 250 ng of empty vector DNA. Cells were harvested and left unstimulated (white bars) or stimulated (black bars) with plate coated anti-CD3ε/CD28, and luciferase activity was analyzed as a measurement of NF-κB activation. B, HEK 293 fibroblasts were transfected with different concentrations of myc-CARMA1 (white bars) or myc-CARMA1-S649A (black bars) expression vectors, and Igκ2-Luciferase/pRL-TK reporter vectors. Cells were treated with or without P/I and luciferase activity was analyzed to measure NF-κB activation. Protein levels of myc-CARMA1 and CARMA1-S649A mutant were analyzed by immunoblotting using anti-myc. Anti-actin was used as a loading control. C, Jurkat T cells were co-transfected for 24h with the Igκ2-Luciferase/pRL-TK reporter vectors plus 250 ng of expression vectors for myc-CARMA1, myc-CARMA1-S649A, myc-CARMA1-S620A, myc-CARMA1-S620A/S649A or empty vector. Cells were harvested and left unstimulated (white bars) or stimulated (black bars) with plate coated anti-CD3ε/CD28, and luciferase activity was analyzed as a measurement of NF-κB activation. Protein levels of myc-CARMA1 proteins were analyzed by immunoblotting using anti-myc in transfected HEK 293 cells with 250 ng of DNA. Anti-ERK was used as the loading control. (A and C). Graphs show mean ± SEM of three to four experiments. Student’s t-test comparing activated cells expressing myc-CARMA1 vs. CARMA1-S649A, -S620A, or S620A/S649A; p values: * ≤ 0.05; ** ≤ 0.01. The relative amounts of luciferase to transfection control Renilla are expressed in each experiment.
A recent study by Bidère et al. showed that human CARMA1 activity was decreased by phosphorylation of S608, possibly by casein kinase1α (24). To test if mutation of this phosphorylation target in murine CARMA1 (S620, the murine analogue of human S608) could additively or synergistically enhance down-regulation of CARMA1 activity with S649A, we compared the ability of CARMA1 with single serine mutants vs. double mutant myc-CARMA1-S(620, 649)A to enhance NF-κB reporter gene expression 24 h post-transfection in Jurkat T cells (Fig. 2C). Myc-CARMA1-S620A promoted enhanced NF-κB activation to levels similar to that of myc-CARMA1-S649A. However, combining these individual serine mutations did not lead to an additive increase in NF-κB reporter gene expression. The double-mutant myc-CARMA1-S(620,649)A led to NF-κB activity similar to that observed with the single serine mutants. These data suggest that CARMA1 activation status can be down-modulated by distinct phosphorylation events downstream of alternative signaling pathways (e.g. CK1α vs. PKC); and imply that S620 and S649 phosphorylations are likely to target the same mechanism for CARMA1 down-regulation.
DT40 B cells expressing myc-CARMA1-S649A exhibit reduced growth and survival
Several alternative hypotheses could explain why the S649A mutation led to the observed reduction in CARMA1 expression levels in stably transduced cells: the S649A mutation may directly destabilize the CARMA1 protein: enhanced downstream signaling events may directly promote CARMA1 degradation; or cells that express higher levels of the S649A mutant may be counter-selected due to an alteration in cell growth parameters. As protein stability assays showed no difference in protein half-life between CARMA1 WT and S649A (data not shown), we elected to track the selection of ectopic CARMA1 expression in CARMA1−/− DT40 B cells using an IRES-driven cis-linked GFP marker.
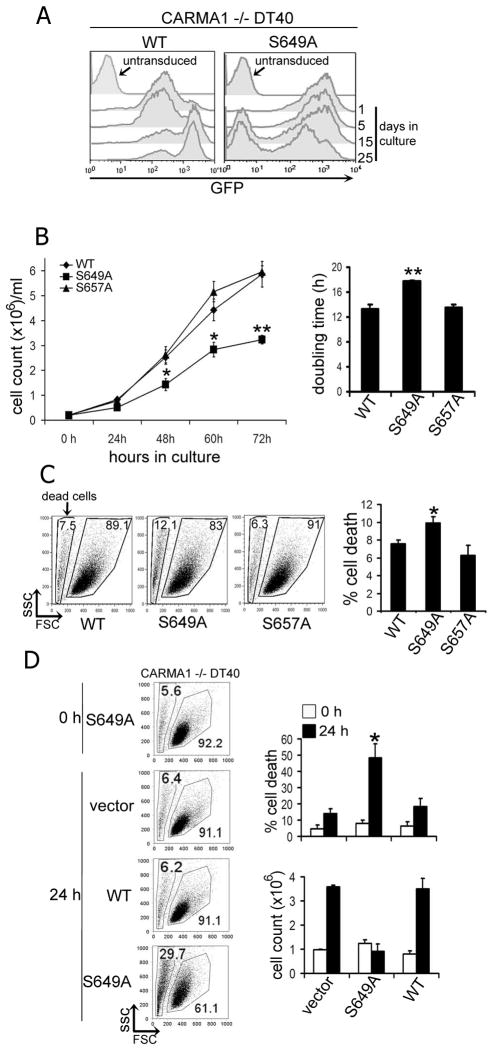
Cells were stably transduced using retroviral vectors expressing myc-CARMA1 or myc-CARMA1-S649A and FACS sorted to obtain homogeneous GFP-positive populations. In vivo selection was subsequently visualized by flow-cytometric analysis of GFP at multiple time points post-sorting (Fig. 3A). WT myc-CARMA1 expressing cells exhibited a progressive increase in relative GFP expression over a 25 day culture period (Fig 3A). In contrast, DT40 cells expressing myc-CARMA1-S649A exhibited counter-selection leading to lower GFP expression levels; including, ultimately, out-growth of GFP null populations at later cell passages (Fig 3A).
FIGURE 3.
CARMA1−/− DT40 cells expressing myc-CARMA1-S649A are counter-selected and show reduced survival and cell growth. A, CARMA1 −/− DT40 cells stably expressing myc-CARMA1 (WT) or CARMA1-S649A were FACS-sorted to obtain populations expressing homogenous, high levels of the IRES-driven cis-marker GFP. Cells were subsequently cultured at 37°C and 5% CO2 for 25 days in standard media (RPMI plus 10% FCS, 1% chicken Serum and supplements). Cells were collected at the indicated time points and GFP levels analyzed by flow cytometry. Figure shows overlapped histograms at successive time points. Untransduced, refers to cells untreated with retrovirus. B, GFP-sorted myc-CARMA1, -S649A or S657A-expressing CARMA1−/− DT40 cells were cultured at 2×105 cells/ml for 0–72h. Cells were harvested at the indicated time points and counted using a hemocytometer. Graph show cells number (106) vs. time in hours. Right graph shows doubling-time analyses for myc-CARMA1 vs. myc-CARMA1-S649A or –S657A expressing cells calculated using the logarithmic least squares fitting technique (http://www.doubling-time.com). Graphs show the mean ± SEM for three independent experiments. C, Percentage of dead cells was analyzed using cell forward and side-scatter analysis, dead cells were gated in dot-plots as indicated (arrow). Right graph show the percentages of cell death in myc-CARMA1 vs. myc-CARMA1-S649A or –S657A expressing populations. Data shown are mean ± SEM of seven independent experiments. D, Fresh retroviral supernatants for vector alone, myc-CARMA1 or myc-CARMA1-S649A, were used to transduce CARMA1−/− DT40 cells; and transduced populations were analysed at 0 and 24 hours for cell survival by flow cytometry as described in (C), or trypan blue exclusion. Right graphs show the percentage of cell death (top) or cell growth ×106 (bottom), at 0 h (white bars) vs. 24 h (black bars) post-transduction. Graphs show the mean ± SEM of three independent experiments. Student’s t-test comparing cells expressing myc-CARMA1-S649A vs. myc-CARMA1, myc-CARMA-S657A, or vector. p values: * ≤ 0.05; ** ≤ 0.01.
Counter-selection results from cell growth or survival deficits secondary to the expressed transgene. To determine whether CARMA1 S649A expression led to decreased cell growth and/or survival, CARMA1−/− DT40 cells stably expressing myc-CARMA1, myc-CARMA1-S649A or myc-CARMA1-S657A, were cultured at identical initial cell numbers (2×105 cells/ml), and their growth was tracked by counting cell numbers at various time points over 72 h (Fig. 3B). Cell populations expressing myc-CARMA1-S649A consistently exhibited reduced cell growth with ~2-fold lower cell counts at each time point analyzed. Similarly, cells expressing myc-CARMA1-S649A exhibited a delay of ~4h in doubling-time as compared with myc-CARMA1 or myc-CARMA1-S657A expressing cells (Fig. 3B, right). Additionally, we consistently observed that myc-CARMA1-S649A expressing cells showed a modest but significant increase in basal cell death as assessed by forward- and side-scatter analysis (Fig. 3C).
Supporting these observations, experiments were also performed in CARMA1−/− DT40 cells acutely transduced with freshly prepared retrovirus expressing either empty-vector, myc-CARMA1 or myc-CARMA1-S649A, and survival and cell numbers were analyzed at 0 and 24 hrs post-transduction (Fig. 3D). While cells transduced with the empty vector or myc-CARMA1 showed similar cell numbers and viability after 24 hours, cells transduced with myc-CARMA1-S649A failed to increase in cell number and exhibited a concomitant ~30% change in dead cells as assessed by either flow cytometry or trypan blue exclusion (Fig. 3D left and right panels, respectively).
Thus, reduced cell survival, leading to counter-selection against myc-CARMA1-S649A, likely accounts for the lower expression levels of this protein mutant in avian DT40 B cell lines.
Increased JNK signaling in B-cell lines expressing the CARMA1-S649A mutant
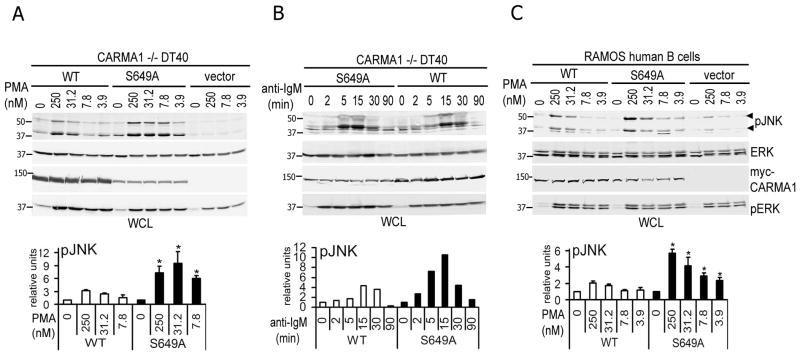
JNK signals have been implicated in the induction of cell death in DT40 B cells, and also in the reduction of cell growth in many other cell types (25, 26). As it is well-documented that CARMA1 activation induces both NF-κB and JNK activation in B and T cells (7, 8, 22), we hypothesized that the myc-CARMA1-S649A mutation enhanced JNK activation. To test this idea, we directly evaluated JNK phosphorylation in CARMA1−/− DT40 B cells (Fig. 4). In our initial studies, we did not detect differences in JNK activation in myc-CARMA1 vs. myc-CARMA1-S649A expressing cells using 1μM of PMA (Fig. 1), likely a result of signal saturation. Thus, for these studies we stimulated CARMA1 −/− DT40 cells stably expressing either myc-CARMA1, myc-CARMA1-S649A or empty vector using a range of PMA doses and a fixed Ionomycin dose (Fig. 4). As shown in Fig. 4A, while JNK phosphorylation was absent in vector-transduced CARMA1 −/− DT40 cells, cells expressing myc-CARMA1 or the -S649A mutant exhibited reconstituted JNK phosphorylation. Myc-CARMA1-S649A-expressing cells, however, consistently exhibited higher phospho-JNK levels than WT expressing cells (Fig. 4A); and increased activity was observed even at the lowest PMA doses tested. In contrast, both cell populations showed similar levels of ERK phosphorylation, implying that this altered activity was specific to JNK signaling.
FIGURE 4.
Phosphorylation of S649 negatively regulates CARMA1-dependent JNK activation. A, CARMA1−/− DT40 cells (2×106 cells/condition) expressing myc-CARMA1 (WT), myc-CARMA1-S649A or empty vector were left unstimulated or stimulated with decreasing low doses of PMA (250, 31.2, 7.8 and 3.9 nM) and 1 μg/ml Ionomicin for 15 min. p-JNK was analyzed by immunoblotting. B, CARMA1−/− DT40 cells expressing myc-CARMA1 or myc-CARMA1-S649A were left unstimulated or were stimulated with 5 μg/ml of IgM for the indicated time points. CARMA1 activity was assessed by p-JNK immunoblotting from 2×106 cells/condition in WCL. C, Ramos human B cells stably expressing myc-CARMA1, myc-CARMA1-S649A or empty vector (1×106 cells/condition) were unstimulated or stimulated with the indicated doses of PMA and 1 μg/ml Ionomicin for 15 min at 37°C. Whole cell lysates were prepared immnoblotted as described in A. A–C, p-ERK, ERK and myc (CARMA1) blots were used as activation and loading controls, respectively. Band densities of JNK phosphorylation were quantified and are expressed relative to those of the protein loading control myc-CARMA1; 0 time points were set as 1 and used to normalize p-JNK levels from stimulated cells. Graphs shown in A and C show the mean ± SEM from 3 independent experiments. The Student’s t-test was used to compare JNK activation between cells expressing CARMA1 vs. CARMA1-S649A. * p values ≤0.05.
To determine whether these events also occurred following direct AR stimulation, CARMA1−/− DT40 B cells expressing CARMA1-WT vs. -S649A were activated with anti-chicken IgM (5 μg/ml) for specific time points and p-JNK levels were assessed by immunoblotting (Fig. 4B). Under these conditions, cells expressing the CARMA1-S649A mutant exhibited more rapid and slightly enhanced JNK activation (peaking at 5 min) compared to myc-CARMA1 (peaking at 15 min); although these differences were more subtle than P/I driven responses. To determine whether these findings were relevant to mammalian B cells, the human B cell line (Ramos) was identically transduced and activated with low P/I doses (Fig. 4C). Consistent with the results using DT40 cells, JNK phosphorylation was significantly increased in Ramos cells expressing myc-CARMA1-S649A compared to that induced by myc-CARMA1 expressing cells; and relative ERK phosphorylation remained indistinguishable between the cell lines. When JNK phosphorylation levels were quantified in relation to CARMA1 expression levels, JNK phosphorylation was consistently ~2 to 3-fold higher in myc-CARMA1-S649A than in myc-CARMA1 expressing cells (Fig. 4A–C, bottom panels).
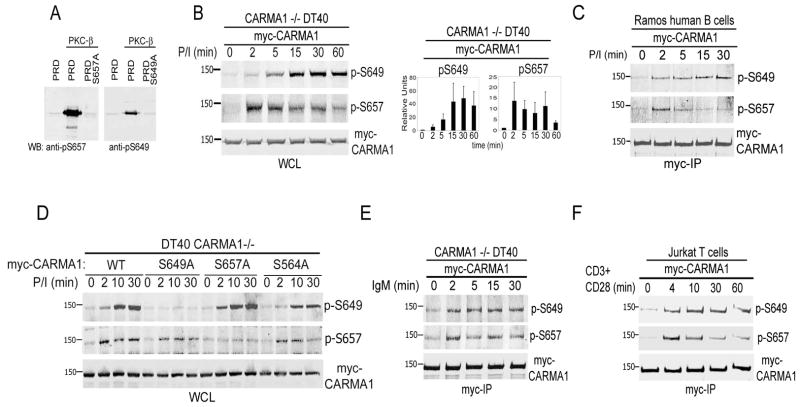
Analysis of CARMA1 phosphorylation in vivo using phospho-serine specific antibodies
To understand whether S649 has a physiologically relevant regulatory role in the AR signaling cascade, we directly assessed whether it was indeed phosphorylated in response to AR or P/I stimulation in vivo. We generated polyclonal antibodies against p-S649, as well as p-S657 as a positive control (analogous to chicken S668; (22)) to measure in vivo phosphorylation of these residues. To verify their specificity, the resulting antibodies were first used to immunoblot bacterially expressed CARMA1 PRD protein fragments with and without in vitro phosphorylation by recombinant PKCβ. As shown in Fig. 5A, both antibodies reacted only with phosphorylated WT-PRD, and not with un-phosphorylated WT, or with phosphorylated PRDs with serine-to-alanine mutations at S649 or S657, respectively.
FIGURE 5.
CARMA1 residues S649 and S657 exhibit differential phosphorylation kinetics as detected by phospho-specific polyclonal antibodies. A, GST-PRD (PKC-regulated domain), GST-PRD-S649A, and GST-PRD-S657A purified from recombinant bacteria were incubated with or without purified PKCβ in the presence of diacyl glycerol, calcium, and non-radiolabeled ATP. In vitro phosphorylation of S649 and S657 was detected by immunoblotting using phosphor-specific antibodies. B, CARMA1−/− DT40 cells stably expressing myc-CARMA1 were stimulated with P/I (1 μM and 1 μg/ml, respectively) for the indicated time points. Cells were lysed with RIPA buffer, and lysates resolved by SDS-PAGE. Right, graph of p-S649 and p-S657 levels relative to myc-CARMA1 levels in P/I stimulated DT40 cells. Data represents the mean ± SD of four independent experiments. C, Ramos B cells (15×106 cells/condition), were stimulated with P/I as in (B) for the indicated time points. CARMA1 was immunoprecipitated using anti-myc antibodies (myc-IP). D, CARMA1−/− DT40 cells (2×106 cells/condition) expressing myc-CARMA1 or myc-CARMA1 with single mutations (S649A, S657A, and S564A) were stimulated with P/I for the indicated time points, lysed with RIPA buffer, and lysates resolved by SDS-PAGE. E and F, CARMA1−/− DT40 cells (E) and Jurkat T cells (F) stably expressing myc-CARMA1 were activated for the indicated time points with anti-IgM (5 μg/ml) or anti-CD3/CD28 (10/1 μg/ml), respectively (25–30 ×106 cells/condition). CARMA1 was immunoprecipitated from the resulting cell lysates using anti-myc antibodies (myc-IP), followed by SDS-PAGE. B–F, Proteins from WCL (whole cell lysates) or myc-IPs were transferred to PVDF membranes and p-S649, p-S657 and myc-CARMA1 were detected by immunoblotting.
To analyze the phosphorylation of S649 and S657 downstream of AR signals in vivo, lymphoid cell lines including B-cell (CARMA1−/− DT40, and human Ramos B cells) and T-cell (Jurkat) lines were retrovirally transduced to stably express myc-CARMA1 or myc-CARMA1 containing specific mutations of these serine residues. Cells were then stimulated with P/I (Fig. 5B–D), or AR specific antibodies (Fig. 5E and F). CARMA1 phosphorylation was detected by Western blotting using the phospho-serine antibodies in either whole cell lysates (WCL) in CARMA1 −/− DT40 cells (Fig. 5B and F) or in myc- immunoprecipiates (myc-IP, Fig. 5C, E and F). P/I induced phosphorylation of the two serine residues with distinct kinetics in both B-cell lines. While p-S657 was rapidly induced, showing a maximum peak 2–5 min post-stimulation and a steady reduction at later time points, p-S649 was delayed but more sustained, reaching a maximum between 15 and 30 min post-stimulation. Similarly, we analyzed these phosphorylation events induced by AR signaling in B and T cells lines (Fig. 5E–F). Anti-IgM in DT40 CARMA1−/− B-cells or anti-CD3/CD28 in Jurkat T-cells induced p-S657 and p-S649 with almost identical kinetics to that induced by P/I.
Due to the substantial disparity between the kinetics of S649 and S657 phosphorylation, we investigated whether there was a hierarchical dependence of phosphorylation between these and other serine residues. Phosphorylation of CARMA1 with single S-to-A mutations of S649 and/or S657 was tested in P/I-stimulated cells. Constructs with S564A mutations were also included in the analysis, as S564 is also required for NF-κB activation in lymphocytes (20). As shown in Fig. 5D, phosphorylation of either S649 or S657 were each unaffected by the absence of the other serine residues, indicating that phosphorylation of these individual residues can occur independently from each other.
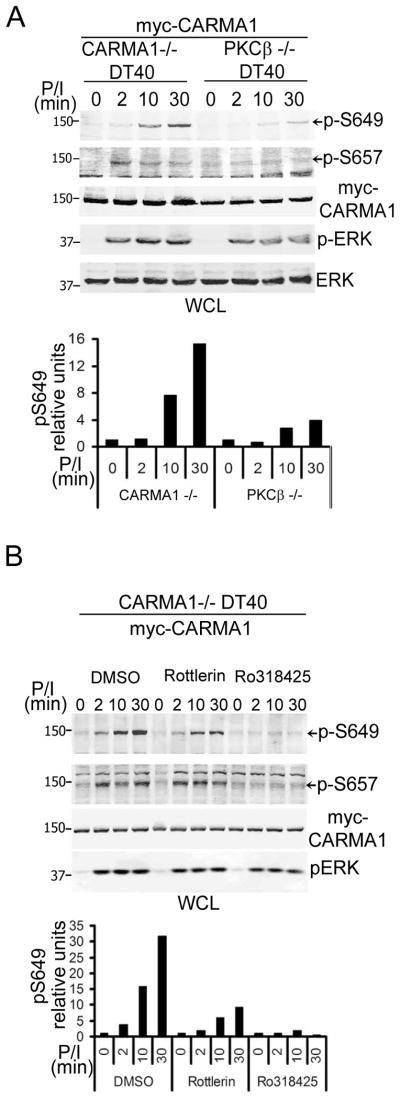
Requirement for PKC activity in CARMA1 phosphorylation at S649 and S657
Previous studies have shown that CARMA1 phosphorylation, detected in cells lines using total anti-phospho serine antibodies or in vitro using radioactive phospho-labeling of bacterially expressed proteins, required the activity of specific PKC isoforms; PKCβ (found in B cells) and PKCθ (in T cells) (19, 20, 23). Nevertheless, whether phosphorylation of individual serine residues requires the exclusive activity of a particular PKC isoform or kinase in vivo has not been fully determined. It has been reported that chicken S668 (mouse S657) requires PKCβ activity in DT40 B-cell line (22); however, whether S649 phosphorylation requires the same kinase is unknown. To assess whether PKCβ is essential for phosphorylation of both S649 and S657 in B cells, PKCβ−/− DT40 cells stably expressing myc-CARMA1 were activated and CARMA1 phosphorylation was analyzed and compared to that observed in myc-CARMA1-reconstituted CARMA1−/− DT40 cells (Fig. 6A). As anticipated, both p-S657 and p-S649 were markedly reduced in PKCβ−/− DT40 cells. However, while p-S657 was abolished in the PKCβ−/− cells, a low-level of p-S649 was clearly present at later time points. This result suggested that other kinases or alternative PKC isoforms may be involved in CARMA1-phosphorylation of S649. To test this hypothesis, CARMA1−/− DT40 cells expressing myc-CARMA1 were pre-incubated with either a pan-PKC or a novel-PKC inhibitor and subsequently stimulated with P/I (Fig. 6B). While the pan-PKC inhibitor abrogated both p-S657 and p-S649, the novel PKCs inhibitor (Rottlerin) exhibited no effect on p-S657 but partially decreased p-S649, indicating that, while PKCβ is the major effector for CARMA1 phosphorylation and activation in B cells, other kinases (likely including novel PKC isoforms) also participate in S649 phosphorylation.
FIGURE 6.
PKCβ expression and activity is required for S657 phosphorylation, but a novel PKC activity also participates in S649 phosphorylation in DT40 B cells. A, CARMA1−/− and PKC-β−/− DT40 cells stably expressing myc-CARMA1 (2×106 cells/condition) were stimulated with P/I (1 μM and 1 μg/ml, respectively) for the indicated time points. B, CARMA1−/− DT40 cell expressing myc-CARMA1 (2×106 cells/condition) were pre-incubated with 5 μM of Rottlerin (novel PKC inhibitor), Ro318425 (general PKC inhibitor), or DMSO (vehicle) then stimulated with P/I for the indicated time points. P-S649, p-S657, myc-CARMA1, p-ERK and ERK were assessed by immunoblot. A and B, Graph show the amounts of pS649 relative to myc. WCL, whole cell lysates.
Discussion
The current study clearly demonstrates that S649 is an in vivo kinase target downstream of AR engagement, and that loss of this serine has a positive effect on CARMA1 activity, as measured by both IKK activation and JNK phosphorylation. Thus, in the wt protein, phosphorylation at S649 likely results in CARMA1 down-regulation. These events were conserved in both chicken and human B-cell lines, as well as in a human T-cell line. Aided by the generation of polyclonal phospho-specific antibodies, we found that S649 was phosphorylated in lymphocytes with a distinct kinetic pattern upon cell stimulation. While the CARMA1 activating phosphorylation at S657 occurs rapidly and is transitory (Fig. 5 and (22)), the inhibitory phosphorylation at S649 is both delayed and more sustained. Interestingly, our data also show that phosphorylation of CARMA1 does not follow a hierarchical process. Phosphorylation of S649 occurred independently of phosphorylation at other residues including S564 and S657 (Fig. 5). As PKCβ and θ are implicated in both rapid, activating phosphorylations and later-occurring de-activating phosphorylation (Fig 5), the mechanism governing the selective phosphorylation kinetics remains to be determined.
It is well-established that PKC isoforms may have both positive and negative regulatory effects on AR signaling in lymphocytes (27, 28). Additionally, it is well-established that signaling proteins can be targeted by multiple kinases that may either positively or negatively affect the target protein activity. For example, Src protein tyrosine kinases and the Tec family kinase Btk are activated by specific tyrosine kinases, and de-activated by other serine/threonine kinases (29, 30). In this study, we demonstrate that CARMA1 activation is both positively and negatively regulated by at least 3 independent, PKC-mediated phosphorylation events in the PKC regulated domain (PRD) induced by AR cross-linking (and by signals that mimic AR engagement) in B and T cell lines. In B cells, PKCβ is required for S657 phosphorylation, and is a major kinase targeting S649 (Fig. 6). However, we observed that the p-S649 signal is partially intact in the absence of PKCβ. Because Rottlerin, a specific inhibitor of novel PKCs, partially blocked p-S649 (but not p-S657), we suspect that a novel PKC isoform also contributes to the phosphorylation of CARMA1 at residue S649. Interestingly, previous studies have shown that genetic deletion of the novel PKC isoform PKCδ, which is expressed in B cells, results in B cell hyper-activation and increased proliferation, which under tolerogenic conditions leads to increased NF-κB and JNK activation upon antigenic stimulation (31, 32). Further investigation will be required to determine whether CARMA1 is a target for PKCδ, or has a role in this phenotype.
It is also now well established that, while PKCβ/θ phosphorylation is required for CARMA1 activity, other pathways can target CARMA1 phosphorylation, influencing the strength of its signal. Other kinases that have been shown to target CARMA1 (both within and outside of the PRD domain) include IKKβ, CK1α and calmodulin-dependent protein kinase II (CaMK-II) (22, 24, 33)(Fig. 7D). Interestingly, Bidère et al. (24) have shown that although CK1α promotes CARMA1 activation, it also functions as a negative regulator of CARMA1-dependent NF-κB activation. CK1α can phosphorylate S608 in human CARMA1 in vitro, and this modification may trigger CARMA1 degradation in vivo. In contrast, we have not been able to identify a defect in turnover using the murine CARMA1-S649A mutant (data not shown), suggesting that other negative regulatory events are more likely triggered by this phosphorylation. Surprisingly, S620 and S649 phosphorylation also do not additively increase CARMA1 activation (Fig. 2). While in vivo phosphorylation of S608/620 (human/mouse, respectively) by CK1α remains to be directly demonstrated, together these data suggest that at least two alternative phosphorylation events (at S620 or S649) can mediate down-regulation of CARMA1 signaling, and imply these signals promote negative regulation via a similar pathway.
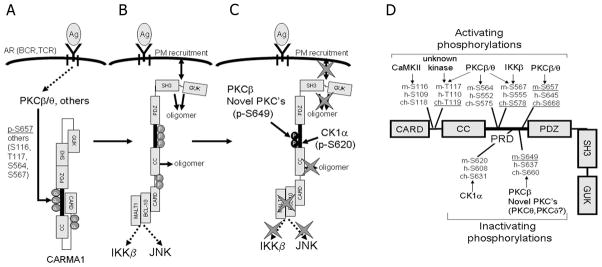
FIGURE 7.
Working model for CARMA1 regulation; and detailed schema of a candidate activating/inactivating phosphorylation sites and kinases. A, CARMA1 exists primarily in a inactive closed conformation in the cytosol. Antigen (Ag) recognition promotes PKCβ and PKCθ activation in B- and in T-cells, respectively. These PKC isoforms and probably other kinases (including CK1α, CamKII or IKKβ) rapidly phosphorylate CARMA1 (2–5 min) at specific residues, thereby releasing conformational interference. B, This conformational change promotes CARMA1 plasma membrane (PM) recruitment and oligomerization and its interaction with BCL-10 and MALT1 and subsequent activation of the IKK and JNK signaling cascades. C, Reaching maximum phosphorylation levels between 15–30 min post-activation, other residues including S649 and, likely, S620 are phosphorylated by classical or novel PKC isoforms and CK1α, respectively. These later phosphorylation events negatively regulate CARMA1 activation. D, Schema showing the location of the activating and inactivating phosphorylation sites and candidate enzymes that regulate their modification. m-, h-, ch- refer to mouse, human and chicken sites, respectively. Underlined residues indicate the phosphorylation sites that have been directly demonstrated in vivo.
Although significant, p-S649 down-regulation of CARMA1 activity was incomplete, and was most evident when low doses of activators were used. We hypothesize that p-S649 may set a threshold for AR-driven IKK and JNK activation. We expect that additional signaling events are likely to be required to fully turn off the CARMA1 signal. Indeed, other negative regulatory processes that have been shown to de-activate the CARMA1 signalosome as a whole include BCL10 degradation (34–37); de-ubiquitination of substrates including MALT1 and TRAFs by A20 (or CYLD) (38–40); and direct CARMA1 ubiquitination and degradation (MMG, KS, and DJR, manuscript submitted). Furthermore, phosphorylation of some activating residues in CARMA1 is rapid and transitory (Fig. 5); possibly suggesting involvement of phosphatase(s) in CARMA1 inactivation.
Genetic knock-out models in mice have demonstrated that loss of CARMA1 has no effect on early stages of B and T cell development. However, CARMA1 is required for the development of B1 cells, for maintenance of mature B2 B cells, and for AR signaling of both B and T cells (6–9). Although it is not known if over-active or allelic variants of CARMA1 have any role in pathologies of early B or T cells, there is mounting evidence to suggest that CARMA1 signaling enhances B cell lymphomagenesis in humans (41, 42). A subset of poor prognosis, diffuse large B cell lymphomas (ABC-DLBCL) requires basal CARMA1 expression and activity in order to promote NF-κB activation and survival (42). Furthermore, mutations in the coiled-coil domain of CARMA1, which produces a constitutive active CARMA1 molecule, have been described in nearly 10% of these ABC-DLBCLs. Thus, our finding that expression of the over-active CARMA1-S649A mutant is counter-selected in CARMA1−/− DT40 B cells appears to contradict these earlier findings. We hypothesize that, since CARMA1 activates both NF-κB (associated with cell survival programs) and JNK (associated with apoptotic programs), over-active CARMA1 may differentially affect a cell depending on how important these pathways are to its growth and survival. It should be noted that JNK activation is strongly linked with apoptosis of DT40 cells. Additionally, while DT40 cells constitutively activate NF-κB, they down-regulate their BCR over time, indicating a counter-selection for the AR pathway. Taken together, the results presented here further support the idea that the survival and growth of B lymphoma cells, chicken DT40 cells in this case, are sensitive to the activation status of CARMA1.
In summary, the data presented in this study, combined with previous work, suggest the following working model for CARMA1 activation and subsequent deactivation via specific phosphorylation signals (Fig. 7). In resting cells, most CARMA1 is maintained in an inactive closed-conformation (Fig. 7A). AR stimulation activates PKCβ or PKCθ in B or T cell, respectively (as well as other kinase pathways), leading to rapid phosphorylation at specific residues including, most notably, S657 (Fig. 7A) and S564 in murine CARMA1. These modifications promote a conformational change in CARMA1, triggering its oligomerization, plasma membrane recruitment, and interaction with critical proteins (including BCL10, MALT1 and others) involved in both IKK and JNK activation (Fig. 7B). While these activation events occur early in the activation of CARMA1, a delayed wave of PKCβ-, novel-PKC-, and possibly, CK1α-dependent phosphorylation targets residues S649 and possibly S620. These latter and sustained events down-regulate CARMA1 activation through currently unclear mechanisms that may include limiting BCL10 and MALT1 interaction, reduction of CARMA1 oligomerization or plasma membrane recruitment, or control of CARMA1 turnover (Fig. 7C). As it is apparent that multiple phosphorylation signals regulate the extent of CARMA1 activation and de-activation; it will be important to fully elucidate the kinase targets in CARMA1 and the mechanisms controlling CARMA1 down-regulation, in order to understand how the lymphocyte fine-tunes its response to antigenic challenges.
Acknowledgments
We wish to thank Angel Hui and members of the Rawlings lab for assistance and thoughtful discussions.
Abbreviations
- CARMA1
CARD-MAGUK containing protein 1
- AR
Antigen receptors
- P/I
PMA/Ionomycin
Footnotes
This work supported by NIH R01-HD037091, NIH Fogarty International Center grant R03-TW007322, and CDRF Grant UKB2-2831-KV-06.
Conflict of Interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 2.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 3.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 5.Rincon M, Davis RJ. Regulation of the immune response by stress-activated protein kinases. Immunol Rev. 2009;228:212. doi: 10.1111/j.1600-065X.2008.00744.x. [DOI] [PubMed] [Google Scholar]
- 6.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O’Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol. 2003;13:1252. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 7.Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D’Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 8.Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, Domashenz H, Hong NA, Glynne RJ, Nelms KA, Goodnow CC. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 9.Newton K, V, Dixit M. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003;13:1247. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 10.Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:836. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:830. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 12.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. Embo J. 2002;21:5184. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 14.Gaide O, Martinon F, Micheau O, Bonnet D, Thome M, Tschopp J. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-kappaB activation. FEBS Lett. 2001;496:121. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 15.McAllister-Lucas LM, Inohara N, Lucas PC, Ruland J, Benito A, Li Q, Chen S, Chen FF, Yamaoka S, Verma IM, Mak TW, Nunez G. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-kappaB induction. J Biol Chem. 2001;276:30589. doi: 10.1074/jbc.M103824200. [DOI] [PubMed] [Google Scholar]
- 16.Tanner MJ, Hanel W, Gaffen SL, Lin X. CARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-kappaB activation. J Biol Chem. 2007;282:17141. doi: 10.1074/jbc.M700169200. [DOI] [PubMed] [Google Scholar]
- 17.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 18.McCully RR, Pomerantz JL. The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol Cell Biol. 2008;28:5668. doi: 10.1128/MCB.00418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Moreno-Garcia ME, Sommer KM, Bandaranayake AD, Rawlings DJ. Proximal signals controlling B-cell antigen receptor (BCR) mediated NF-kappaB activation. Adv Exp Med Biol. 2006;584:89. doi: 10.1007/0-387-34132-3_7. [DOI] [PubMed] [Google Scholar]
- 22.Shinohara H, Maeda S, Watarai H, Kurosaki T. IkappaB kinase beta-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med. 2007;204:3285. doi: 10.1084/jem.20070379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinohara H, Yasuda T, Aiba Y, Sanjo H, Hamadate M, Watarai H, Sakurai H, Kurosaki T. PKC beta regulates BCR-mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J Exp Med. 2005;202:1423. doi: 10.1084/jem.20051591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidere N, V, Ngo N, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, Vazquez A, Sakai K, Zhang J, Meng Z, Veenstra TD, Staudt LM, Lenardo MJ. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009;458:92. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 26.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 27.Su TT, Guo B, Rawlings DJ. Emerging roles for PKC isoforms in immune cell function. Mol Interv. 2002;2:141. doi: 10.1124/mi.2.3.141. [DOI] [PubMed] [Google Scholar]
- 28.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17:555. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 30.Kang SW, Wahl MI, Chu J, Kitaura J, Kawakami Y, Kato RM, Tabuchi R, Tarakhovsky A, Kawakami T, Turck CW, Witte ON, Rawlings DJ. PKCbeta modulates antigen receptor signaling via regulation of Btk membrane localization. Embo J. 2001;20:5692. doi: 10.1093/emboj/20.20.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mecklenbrauker I, Saijo K, Zheng NY, Leitges M, Tarakhovsky A. Protein kinase Cdelta controls self-antigen-induced B-cell tolerance. Nature. 2002;416:860. doi: 10.1038/416860a. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, Nakayama KI. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 33.Ishiguro K, Green T, Rapley J, Wachtel H, Giallourakis C, Landry A, Cao Z, Lu N, Takafumi A, Goto H, Daly MJ, Xavier RJ. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol Cell Biol. 2006;26:5497. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol. 2004;24:3860. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobry C, Lopez T, Israel A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci U S A. 2007;104:908. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H, Di L, Fu G, Chen Y, Gao X, Xu L, Lin X, Wen R. Phosphorylation of Bcl10 negatively regulates T-cell receptor-mediated NF-kappaB activation. Mol Cell Biol. 2007;27:5235. doi: 10.1128/MCB.01645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu S, Du MQ, Park SM, Alcivar A, Qu L, Gupta S, Tang J, Baens M, Ye H, Lee TH, Marynen P, Riley JL, Yang X. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J Clin Invest. 2006;116:174. doi: 10.1172/JCI25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun SC. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- 39.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 40.Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P, Krappmann D. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 41.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Fisher RI, Chan WC, Staudt LM. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 42.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]