Abstract
Objective
The authors tested for genetic linkage of DSM-IV-diagnosed major depressive disorder in families that were ascertained for cigarette smoking.
Method
Within a study that targeted families characterized by a history of smoking, analyses derived a subset of 91 Australian families with two or more offspring with a history of DSM-IV major depressive disorder (affected sibling pairs, N=187) and 25 Finnish families (affected sibling pairs, N=33). Within this affected sibling pair design, the authors conducted nonparametric linkage analysis.
Results
In the Australian heavy smoking families, the authors found a genome-wide significant multipoint LOD score of 4.14 for major depressive disorder on chromosome 3 at 24.9 cM (3p26-3p25).
Conclusions
Genome-wide significant linkage was detected for major depressive disorder on chromosome 3p in a sample ascertained for smoking. A linkage peak at this location was also observed in an independent study of major depressive disorder.
Introduction
Genetic linkage studies of major depressive disorder have found suggestive evidence across multiple genomic regions with little convergence of findings 1. Genetic association, including genomewide association studies (GWAS), findings have been similarly disparate 2–7. These inconsistencies raise the question of whether major depressive disorder encompasses a number of poorly understood subtypes (e.g., depression in smokers). Among smokers seeking cessation treatment, lifetime rates of major depression have been estimated at over 60%8. Cigarette smokers with a history of depression tend to report more severe nicotine withdrawal symptoms 9–11, are more likely to relapse to smoking after a quit attempt8, and may be at increased risk for a recurrent episode of depression after smoking cessation12. Models describing the relationship between symptoms of depression and smoking include examples of shared genetic risk13. We carried out genetic linkage analyses of DSM-IV diagnosed major depressive disorder in two samples that are part of the Nicotine Addiction Genetics project14;15, an international consortium focused on tobacco dependence. We used an affected sibling pair design, in which at least two adult offspring per family reported a history of DSM-IV major depressive disorder, and tested for linkage. Results appear to confirm a genome-wide significant linkage signal at 3p26-3p25, also reported in an independent linkage study of major depressive disorder16.
Method
Samples
The Nicotine Addiction Genetics linkage project enrolled participants at the Queensland Institute of Medical Research in Australia and the University of Helsinki in Finland. Both sites utilized twin registries and targeted families of index cases who previously reported a history of cigarette smoking in interview or questionnaire surveys; the Australian site using a higher threshold measure when recruiting index cases with a history of heavy smoking14. These original linkage samples included microsatellite marker data and telephone diagnostic interviews in 289 families from the Australian site (offspring, N=917; parents, N=392) and 161 families from the Finnish site (offspring, N=522; parents, N=19). More than 90% of the participants from the Australian site were of Anglo-Celtic or Northern European ancestry, and all of the participants from the Finnish site were of Finnish ancestry. The assessment included a diagnostic telephone interview, adapted from the Semi-Structured Assessment for the Genetics of Alcoholism17;18, which obtained a comprehensive assessment of lifetime DSM-IV19 major depressive disorder as well as a tobacco use and dependence assessment derived from the Composite International Diagnostic Interview20. Additional details on the original samples and assessments have been described elsewhere 14;15;21. For the present study, we conducted genetic linkage analyses using the 91 Australian families who had one or more affected sibling pairs concordant for history of major depressive disorder (N=187). In the Finnish sample, only 25 families had one or more affected sibling pairs (N=33), and thus the primary analyses focused on the Australian sample. A summary of the two major depressive disorder linkage samples is presented in Table 1 of the data supplement.
Analyses
For both the Australian and Finnish samples, 381 autosomal microsatellite markers were genotyped and spaced at approximately 10 cM across the genome, positioned using the deCODE genetic map22. Details of genotyping, including quality control procedures are described elsewhere14,15. Single- and multi-point affected sibling pair nonparametric linkage (the latter using a 2 cM grid) was conducted in MERLIN (Multipoint Engine for Rapid Likelihood Inference)23, which generated LOD scores 24, 25. Upon detection of LOD scores > 3, genome-wide corrected p values were calculated from 1,000 replicates simulated in MERLIN26;27.
Follow-up fine mapping included the addition of 290 single nucleotide polymorphism (SNPs) on chromosome 3 that were available for a portion of the Australian major depressive disorder linkage sample (84 out of 91 families), from other Australian projects with overlapping samples that have obtained GWAS data (see reference 28). Through the use of Snagger software 29, these SNPs were selected to be in low linkage disequilibrium (maximum pairwise r2= 0.2), to have a minor allele frequency > 40%, and to have a minimum distance between two tags of 450 kb in order to optimize information content.
Results
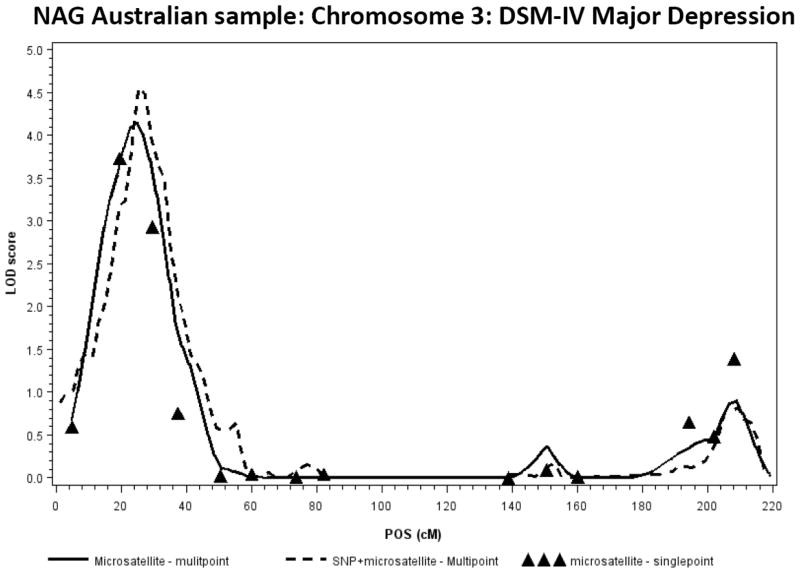
For the Australian linkage sample, a multipoint LOD of 4.14 for major depressive disorder was found on chromosome 3 (at 24.9 cM). The highest single-point linkage signal emerged at microsatellite marker D3S1304, with a LOD score of 3.7. The LOD of 4.14 at 3p in this sample met genomewide significance (p = 0.004). A complete report of these single- and multipoint findings, along with marker positions by chromosome, is provided in Table 2 of the data supplement. The addition of 290 SNPs on chromosome 3 narrowed the linkage locus and increased the LOD score to 4.55 (at 25.3 cM) (see Figure 1). For the Finnish major depressive disorder sample, a multipoint LOD score of 2.10 was found on chromosome 20 (at 90.9 cM).
Figure 1.
Chromosome 3: DSM-IV Major Depression NAG Australian sample; the LOD of 4.14 meets genome-wide significance at p = 0.004 with 1000 simulations in MERLIN;
Given that no linkage signals over 1.5 overlapped across both samples, we did not conduct a combined analysis. However, if we were to correct conservatively for testing in both samples by doubling the p value generated from the 1,000 simulations, we would still maintain a genomewide significant p value of 0.008.
Discussion
Overall, major depressive disorder was associated with a significant genetic linkage peak (a LOD score of 4.14) on chromosome 3 in the Nicotine Addiction Genetics Australian sample, replicating the linkage reported at the same location by Breen et al 16. These converging findings suggest that the genomic region spanning 3p26-3p25 is an important area for further investigation in genetic research on major depressive disorder. Given the small number of Finnish affected sibling pairs, the lack of confirmation in this particular sample is not unexpected. The genetic variants accounting for this linkage signal have not yet been convincingly identified. Although our highest single-point microsatellite marker (D3S1304 [LOD score =3.7]) lies within the metabotropic glutamate receptor 7 gene (GRM7), subsidiary association analyses within a one-LOD support interval, using GWAS data available in the Australian sample (28), found only nominal association for two SNPs within GRM7 (p < .05). Even our strongest association effect, with was a p value of 0.00014 for rs6765537 (a nonsynonymous SNP within C3or20 at 33.87 cM), did not replicate in the Finnish families, nor did it replicate in the Genetic Association Information Network major depressive disorder sample from the Netherlands30, 2. Thus, while others have found suggestive association between SNPs in GRM7 and Major Depressive Disorder (7;31), genome-wide significant effects have not been reported. Further, because linkage implicates very broad regions, GRM7 is among many genes that might be hypothesized to explain our signal.
In terms of other linkage findings, one for quantity smoked in samples ascertained for depression has been reported near our chromosome 3 finding for major depressive disorder 32, raising the possibility of common genetic influences across major depressive disorder and smoking-related behavior or of gene-by-environment (i.e., smoking) interaction effects on major depressive disorder.
There are important limitations associated with our results. Our sample of 91 families is small by standards of modern genomic efforts. Thus, we cannot exclude the possibility that our finding is a false positive that coincidentally appears to replicate Breen et al.16, whose sample is much larger (> 800 families). Additionally, our finding at 3p26-3p25 (highest peak at 24.9 cm) does not align with the meta-analyses results reported by McMahon et al.33, who suggest that variants located at 3p21.1 (at approximately 70–72 cM) are associated with mood disorders. Our future efforts to localize the genetic variants influencing major depressive disorder will entail confirmatory analyses in other samples and additional genotyping in this region on 3p.
Supplementary Material
Acknowledgments
Dr. Pergadia receives funding from a K08 award (grant DA019951) from the National Institute on Drug Abuse. Dr. Glowinski is the principal investigator for the National Institutes of Health grant MH073151 and a co-investigator of grants AA015210 and R49CE001510; she also serves on the advisory board for the Klingenstein Third Generation Foundation. Dr. Wray has received speaker’s honorarium from Eli Lilly; she also receives funding from an Australian Research Council Future Fellowship. Dr. Agrawal receives funding from the National Institutes of Health and has served as a principal investigator for the Alcoholic Beverage Medical Research Foundation/The Foundation for Alcohol Research. Dr. Saccone receives funding from the National Institutes of Health (grant DA024722). Dr. Loukola receives funding from an Academy of Finland postdoctoral fellowship. Dr. Broms has received funding from the Doctoral Programs of Public Health, University of Helsinki; and she has received a consultation fee from Pfizer (for nicotine dependence measurements). Dr. Grant receives funding from the National Institutes of Health. Dr. Nelson has received research support from the National Institutes of Health and the U.S. Department of Defense. Ms. Chou receives salary funding through a National Institutes of Health grant. Dr. Vink receives funding from the Netherlands Organisation for Scientific Research (VENI grant 451–06-004). Dr. MacGregor receives funding from an Australian National Health and Medical Research Council fellowship. Mr. Liu receives funding from the Australian National Health and Medical Research Council (project grant 496675). Dr. Medland receives funding from a National Health and Medical Research Council (Australia) Sidney Sax Fellowship. Dr. Montgomery receives funding from an Australian National Health and Medical Research Council fellowship. Dr. Goate has received research funding and speaker’s honoraria from AstraZeneca, Genentech, and Pfizer; she has also received advisory panel payment from AstraZeneca. Dr. Heath has received funding from the National Institutes of Health (grants AA07728, AA07580, AA11998, AA13320, AA13321, and AA017688). Dr. Kaprio has received a consultation fee from Pfizer (for pharmacogenetics for smoking cessation) as well as an unrestricted grant from Pfizer for the Global Research Awards for Nicotine Dependence; he has also received funding from the Academy of Finland Center of Excellence for Complex Disease Genetics. Dr. Madden is principal investigator for the National Institutes of Health grants DA12854 and DA027995; she is also coinvestigator for grants AA011998, AA017915, DA018267, and DA23668. All other authors report no financial relationships with commercial interests. Supported by a Center for Inherited Disease Research grant to fund genome-wide association studies (to the late Dr. Richard Todd, former principal investigator of grant AA13320) and the European Union (contract number QLG2-CT-2002-01254 [Dr. Kaprio]). Funding for the Genetic Association Information Network (GAIN) major depressive disorder study (parent studies were the Netherlands Study of Depression and Anxiety and the Netherlands Twin Register) was provided by the Netherlands Scientific Organization (grants 904–61-090, 904–61-193, 480–04-004, 400–05-717, and 911–09-032); the Netherlands Organisation for Scientific Research Genomics program (grant SPI-56–464-14192); the Centre for Neurogenomics and Cognitive Research; the Genetics of Mental Illness (grant ERC-230374); the European Union (grant EU/W LRT-2001–01254); ZonMW (geestkracht program grant 10–000-1002); the National Institute of Mental Health (grant R01 MH059160); and matching funds from participating institutes of the Netherlands Study of Depression and Anxiety and the Netherlands Twin Register. The genotyping of GAIN samples was provided through grant MH081802. The data set can be found online at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000020.v2.p1.
The Nicotine Addiction Genetics project is an international collaborative study that includes the following three sites: Queensland Institute of Medical Research, Queensland, Australia (principal investigator, Dr. Martin); the University of Helsinki, Helsinki (principal investigator, Dr. Kaprio); and Washington University, St. Louis (principal investigator, Dr. Madden). Data collection is conducted at the Queensland Institute of Medical Research and the University of Helsinki, with Washington University serving as the coordinating site and lead institution. Genotyping and data analysis are conducted at all three sites. The authors acknowledge the important roles of Drs. Richard Todd and Leena Peltonen, two recently deceased senior investigators, for their contributions to the present project and, more broadly, to the field of psychiatric genetics. The authors also thank the Australian and Finnish families for their participation and cooperation as well as the staff from all three Nicotine Addiction Genetics project sites.
Reference List
- 1.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, de Geus EJ, Willemsen G, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis CM, Ng MY, Butler AW, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 4.Bosker FJ, Hartman CA, Nolte IM, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- 5.Wray NR, Pergadia ML, Blackwood DH, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Potash JB, Knowles JA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2010 doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyn SI, Shi J, Kraft JB, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glassman AH, Stetner F, Walsh BT, et al. Heavy smokers, smoking cessation, and clonidine. Results of a double-blind, randomized trial. JAMA. 1988;259:2863–2866. [PubMed] [Google Scholar]
- 9.Covey LS, Glassman AH, Stetner F. Depression and depressive symptoms in smoking cessation. Compr Psychiatry. 1990;31:350–354. doi: 10.1016/0010-440x(90)90042-q. [DOI] [PubMed] [Google Scholar]
- 10.Madden PA, Bucholz KK, Dinwiddie SH, et al. Nicotine withdrawal in women. Addiction. 1997;92:889–902. [PubMed] [Google Scholar]
- 11.Pergadia ML, Agrawal A, Heath AC, Martin NG, Bucholz KK, Madden PA. Nicotine withdrawal symptoms in adolescent and adult twins. Twin Res Hum Genet. 2010;13:359–369. doi: 10.1375/twin.13.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glassman AH, Covey LS, Stetner F, Rivelli S. Smoking cessation and the course of major depression: a follow-up study. Lancet. 2001;357:1929–1932. doi: 10.1016/S0140-6736(00)05064-9. [DOI] [PubMed] [Google Scholar]
- 13.Kendler KS, Neale MC, MacLean CJ, Heath AC, Eaves LJ, Kessler RC. Smoking and major depression. A causal analysis. Arch Gen Psychiatry. 1993;50:36–43. doi: 10.1001/archpsyc.1993.01820130038007. [DOI] [PubMed] [Google Scholar]
- 14.Saccone SF, Pergadia ML, Loukola A, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. American Journal of Human Genetics. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loukola A, Broms U, Maunu H, et al. Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics J. 2008;8:209–219. doi: 10.1038/sj.tpj.6500464. [DOI] [PubMed] [Google Scholar]
- 16.Breen, et al. A genomewide significant linkage for severe depression on chromosome 3: the Depression Network Study. American Journal of Psychiatry. 2011 doi: 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- 17.Bucholz KK, Cadoret R, Cloninger CR, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 18.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 19.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington, DC: American Psychiatric Association; 1994. Revised ed. [Google Scholar]
- 20.Cottler LB, Robins LN, Grant BF, et al. The CIDI-core substance abuse and dependence questions: cross-cultural and nosological issues. The WHO/ADAMHA Field Trial. Br J Psychiatry. 1991;159:653–658. doi: 10.1192/bjp.159.5.653. [DOI] [PubMed] [Google Scholar]
- 21.Pergadia ML, Agrawal A, Loukola A, et al. Genetic linkage findings for DSM-IV nicotine withdrawal in two populations. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:950–959. doi: 10.1002/ajmg.b.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 24.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994;50:118–127. [PubMed] [Google Scholar]
- 26.North BV, Curtis D, Sham PC. A note on the calculation of empirical P values from Monte Carlo procedures. Am J Hum Genet. 2002;71:439–441. doi: 10.1086/341527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.North BV, Curtis D, Sham PC. A note on calculation of empirical P values from Monte Carlo procedure. Am J Hum Genet. 2003;72:498–499. doi: 10.1086/346173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medland SE, Nyholt DR, Painter JN, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edlund CK, Lee WH, Li D, Van Den Berg DJ, Conti DV. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC Bioinformatics. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boomsma DI, Willemsen G, Sullivan PF, et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 31.Muglia P, Tozzi F, Galwey NW, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 32.Vink JM, Beem AL, Posthuma D, et al. Linkage analysis of smoking initiation and quantity in Dutch sibling pairs. Pharmacogenomics J. 2004;4:274–282. doi: 10.1038/sj.tpj.6500255. [DOI] [PubMed] [Google Scholar]
- 33.McMahon FJ, Akula N, Schulze TG, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.