Abstract
Members within the paramyxovirus subfamily Paramyxovirinae constitute a large number of highly virulent human and animal pathogens. The glycoproteins present on these viruses are responsible for mediating host cell attachment and fusion and are key targets for the design of antiviral entry inhibitors. In the present review, we discuss recent structural studies which have led to a better understanding of the various mechanisms by which different paramyxoviruses use their attachment glycoproteins to hijack specific protein and glycan cell-surface receptors to facilitate viral entry. It is observed that the paramyxovirus attachment glycoprotein consists of a conserved overall structure which includes an N-terminal six-bladed β-propeller domain which is responsible for cell receptor binding. Crystal structures of this domain from different biomedically important paramyxoviruses, including measles, Nipah, Hendra, Newcastle disease and parainfluenza viruses, alone and in complex with their functional cell-surface receptors, demonstrate three contrasting mechanisms of receptor engagement that paramyxoviruses have evolved to confer discreet protein- and glycan-receptor specificity. This structural information highlights the adaptability of the paramyxovirus attachment glycoprotein surface and the potential for the emergence of new and potentially harmful viruses in human hosts.
Keywords: paramyxovirus, protein crystallography, structural virology, viral glycoprotein, virus entry
Abbreviations: DANA, 2,3-dehydro-2-deoxy-N-acetylneuraminic acid; F, fusion glycoprotein; G, attachment glycoprotein; H, haemagglutinin; HeV, Hendra virus; HN, haemagglutinin-neuraminidase; HNV, henipavirus; MV, measles virus; NDV, Newcastle disease virus; PIV, parainfluenza virus; NiV, Nipah virus; r.m.s.d., root mean square deviation; SCR, short consensus repeat; SLAM, signalling lymphocyte activation molecule
Paramyxovirus entry
The subfamily Paramyxovirinae belongs to the Paramyxoviridae family and includes five genera: Rubulavirus, Avulavirus, Respirovirus, Henipavirus and Morbillivirus. This group of viruses constitutes over 31 vertebrate-borne viruses, many of which are currently of worldwide biomedical importance. The negative-polarity single-stranded genome of viruses within this group encodes seven to nine proteins, four of which are structural: the attachment glycoprotein, F (fusion) glycoprotein (a class-I fusion architecture [1]), matrix protein and nucleocapsid protein. The attachment and F glycoproteins extend outwards from a surrounding lipid bilayer envelope and assemble as a higher-order hetero-oligomer. This glycoprotein complex orchestrates the paramyxoviral entry process of cellular attachment and subsequent pH-independent fusion of the host cell and viral membranes, ultimately leading to the release of viral RNA into the host cell.
The ability of a paramyxovirus to undergo host cell entry is highly dependent on the capacity of the attachment glycoprotein to specifically bind receptors which are expressed on the cell surface. As a result of the varying specificity of paramyxoviruses for different receptors, the attachment glycoprotein falls into one of three classes: HN (haemagglutinin-neuraminidase), H (haemagglutinin) or G (attachment glycoprotein). HN glycoproteins [e.g. from NDV (Newcastle disease virus)] have the ability to haemagglutinate red blood cells and cleave sialic acid linkages (to protect against viral self-agglutination), whereas H glycoproteins [e.g. from MV (measles virus)] haemagglutinate, but lack neuraminidase activity, and G glycoproteins [e.g. from NiV (Nipah virus)] lack both haemagglutination and neuraminidase activity. The cell-surface receptors utilized by paramyxoviruses for attachment can be either protein or carbohydrate. HN glycoproteins such as PIVs (parainfluenza viruses) and NDV [2] use sialic acid (N-acetylneuraminic acid), a terminal saccharide present on cell glycans. H and G glycoproteins, on the other hand, use glycoprotein cell-surface receptors such as CD46 [3,4] and SLAM (signalling lymphocyte activation molecule) (or CD150) [5,6] for MV, and ephrinB2 and ephrinB3 for the highly pathogenic NiV and HeV (Hendra) viruses [7–9].
In the present review, we focus on the recent structural studies which have revealed a conserved basic architecture of the paramyxovirus attachment glycoprotein as well as the differential mechanisms by which each class of attachment glycoprotein is known to interact with its host cell receptor.
HN glycoprotein structure
As for all classes of paramyxovirus attachment glycoproteins, HNs have a type-II integral membrane protein architecture consisting of an N-terminal cytoplasmic and transmembrane region (~70 amino acids), a stalk region which has been implicated as being responsible for glycoprotein oligomerization and interaction with the F glycoprotein (~100 amino acids), and a six-bladed β-propeller domain which is involved in receptor binding (~400 amino acids) (Figures 1–3).
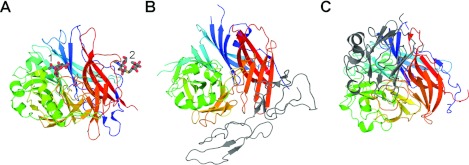
Figure 1. Representative structures of the three Paramyxovirinae attachment glycoprotein classes in complex with their human cell-surface receptors.
All attachment glycoproteins are superimposed in the same orientation and shown as cartoons coloured as a rainbow with the N-terminus in blue and the C-terminus in red. (A) NDV-HN in complex with DANA at the primary receptor-binding site (1) and thiosialoside at the putative secondary receptor-binding site (2) (PDB code 1USX). DANA and thiosialoside are shown as sticks with carbon atoms coloured grey, oxygen atoms red, nitrogen atoms blue and sulfur atoms yellow. (B) MV-H in complex with the SCR1 and SCR2 domains of CD46 (PDB code 3INB). CD46 is shown as a grey cartoon. (C) NiV-G in complex with the receptor-binding domain of ephrinB2 (PDB code 2VSM). EphrinB2 is shown as a grey cartoon.
Crystal structures of the globular HN receptor-binding β-propeller have been solved from three viral species: NDV (an Avulavirus) [10,11], PIV5 (a Rubulavirus) [12] and PIV3 (a Respirovirus) [13]. The β-propeller consists of six blades (each consisting of four antiparallel β-strands) assembled in a torroidal arrangement around a central axis and is well conserved among all the three known HN structures [average of 1.6 Å (1 Å=0.1 nm) r.m.s.d. (root mean square deviation) between equivalent Cα positions] [12]. In the case of PIV5-HN, the glycoprotein was crystallized in the presence of the tetramerization-inducing stalk domain (amino acids 37–117). However, no electron density for this region was observed, suggesting that its structure may be conformationally variable in the protein crystal [12]. Crystallized NDV-HN and PIV3-HN constructs, on the other hand, lacked the stalk domain and were demonstrated to be monomeric in solution [10,11,13]. Despite differences between the constructs and solution-state measurements, NDV-HN (both a dimer and tetramer were observed), PIV3-HN (dimer observed) and PIV5-HN (tetramer) crystals were composed of either dimers or tetramers, where the overall mode of association between β-propeller subunits was broadly comparable, suggestive of a common mode of HN glycoprotein association on the envelope surface [12].
HN glycoprotein receptor binding
The receptor-binding site of the viral HN is located deep in the centre of the β-propeller (Figures 1A and 2A). The crystal structures of NDV-HN, PIV3-HN and PIV5-HN have all been solved in complex with carbohydrate ligands including sialic acid (β-anomer), the inhibitors DANA (2,3-dehydro-2-deoxy-N-acetylneuraminic acid) and Zanamivir (4-guanidino-Neu5Ac2en), and the trisaccharide sialyl-lactose [10–13]. These structures have demonstrated that the receptor-binding sites in paramyxoviral HN glycoproteins rely on a surface of seven well-conserved residues [10–13]. In contrast with the rigid sialic acid-binding cleft observed in influenza-HN [14–16], the binding site in PIV3-HN, PIV5-HN and NDV-HN is architecturally plastic and adaptable to different sialic acid analogues. This plasticity is manifested in the movement of not only receptor-binding residues, but also entire loops located in the first and sixth blades of the β-propeller. It was suggested by Crennell et al. [10] that this protein flexibility is required for the multifunctional properties of the HN-binding site as it is responsible for binding and destroying its carbohydrate receptor as well as activating the associated F glycoprotein [10]. Thus the distinct conformational states observed in these structures may reflect a switch between a receptor engagement and an activated catalytic state [10,11].
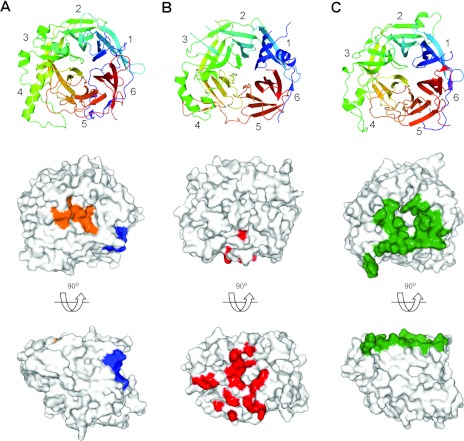
Figure 2. Comparison of the receptor-binding sites for HN, H and G Paramyxovirinae attachment glycoproteins.
In the top panel, (A) NDV-HN (PDB code 1USX), (B) MV-H (PDB code 3INB) and (C) NiV-G (PDB code 2VSM) are shown as β-propellers and are coloured as in Figure 1. The β-propeller blades are numbered from one to six according to standard nomenclature [10]. In the bottom panels, surface representations for each of the molecules are shown. Cell-surface receptor-binding footprints were calculated using PDBsum [48] and plotted on to the surface of each viral glycoprotein. In (A), the DANA-binding site on NDV-HN is shown in orange and the thiosialoside-binding site is shown in blue. In (B), the CD46-binding site on MV-H is shown in red. In (C), the ephrinB2-binding site is shown in green.
The hypothesis of a multifunctional HN-binding site was later confirmed by site-directed mutagenesis of amino acids in the carbohydrate-binding cavity of NDV-HN which resulted in reduced enzymatic activity, carbohydrate-binding affinity and overall fusogeneity of the associated NDV-F glycoprotein [17,18]. A second sialic acid site has also been observed near the dimerization interface of NDV-HN β-propellers (Figures 1A and 2A). The presence of such a secondary binding site has been suggested to be important for maintaining virus cell avidity during structural changes which may occur to the attachment–fusion complex following initial receptor engagement [11]. This binding site, however, has not been observed in the analogous PIV3-HN and PIV5-HN structures, and, as a result, its functionality with regard to all Paramyxovirinae HN glycoproteins is still to be confirmed [12,13].
Attachment by H glycoproteins
In contrast with sialic acid-binding HN glycoproteins, G and H glycoproteins attach to protein cell-surface receptors using a carbohydrate-independent mechanism. Morbilliviruses such as MV have two established functional receptors: CD46 and SLAM [3–6]. CD46 is an ubiquitously expressed type-I membrane glycoprotein consisting of four extracellular SCR (short consensus repeat) domains (SCR1–SCR4) and is involved in the negative regulation of complement activation [19]. The two most N-terminal of these (SCR1 and SCR2) have been predicted to be involved in MV-H binding [20]. SLAM is a type-I membrane glycoprotein and is expressed on haemopoietic cells such as B-cells, T-cells, dendritic cells and macrophages [21,22]. The extracellular portion of SLAM consists of two Ig-like domains, the most N-terminal of which is suspected to be involved in MV-H binding [23,24]. The usage of SLAM and CD46 receptors, however, differs between MV strains. Whereas Edmonston and related vaccine strains have been shown to use both SLAM and CD46 as entry receptors, SLAM, but not CD46, has proved to be the receptor for the majority of wild-type isolates [3,4,25,26].
It has been suggested that MV utilizes additional functional ligands, including an as yet unidentified receptor on polarized epithelial cells [27,28]. Another recently identified receptor includes the extracellular matrix metalloproteinase inducer (CD147/EMMPRIN) which, rather than binding to MV-H, binds to virion-associated cyclophilin [29]. The capacity of MV to use these alternative receptors helps to rationalize the virus's broad tissue tropism as, in addition to infecting haemopoietic cells, MV also infects endothelial, epithelial and neuronal cells [30].
The crystal structure of the unbound C-terminal head domain of MV-H has been independently determined by multiple groups [31,32] and reveals that this receptor-binding domain consists of a six-bladed β-propeller (Figures 1B and 2B) which is most closely related to the β-propeller fold of the known NDV-HN, PIV3-HN and PIV5-HN glycoprotein structures. Similar to many of the HN glycoprotein structures described above, MV-H β-propellers were observed to pack in a dimeric arrangement [32]. Structural comparison of the MV-H β-propeller with other HN glycoprotein β-propellers indicates, however, that there is a large structural divergence between the Morbillivirus, Avulavirus, Respirovirus and Rubulavirus attachment glycoproteins (average of 3.2 Å r.m.s.d. between MV-H and other paramyxoviral HN glycoproteins over 295 equivalent Cα atoms) (Figure 3) [32]. These architectural differences are most visible when looking down the β-propeller fold. Whereas the known HN structures are quite globular and rounded in shape, the MV-H propeller has been described to form a more cubic arrangement [31,32].
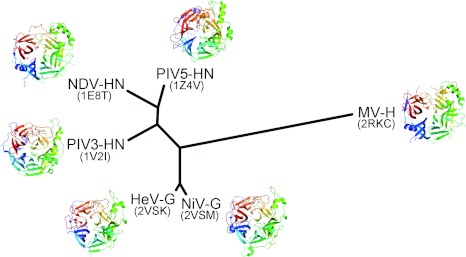
Figure 3. Structural phylogeny of representative receptor-binding β-propeller domains from the Paramyxovirinae.
Superpositions were performed using SHP [49] to optimize the probability of equivalence between Cα residue pairs. Following alignment, a pairwise evolutionary distance matrix was constructed and converted into an unrooted tree using PHYLIP [50]. PDB codes are shown in parentheses.
H glycoprotein receptor interactions
In the centre of the MV-H β-propeller there is a pocket analogous to the sialic acid-binding site in HN glycoproteins. However, despite the presence of this similar HN landscape, none of the seven essential sialic acid-binding residues is conserved. Additionally, the secondary sialic acid-binding site that is located in one of the NDV-HN structures [11] is not present, and the equivalent surface appears to be inaccessible due to the presence of a nearby N-linked carbohydrate. These data were indicative that morbilliviruses are not likely to utilize sialic acid as a secondary low-affinity receptor.
Residues implicated in binding to SLAM, CD46 and the unknown receptor on polarized epithelial cells have been mapped on to crystal structures of MV-H and are suggestive of engagement occurring at the fourth and fifth propeller blades away from the analogous sialic acid-binding site in HN glycoproteins [28,31,32]. The structure of MV-H in complex with the SCR1 and SCR2 domains of CD46 has recently been determined and confirms some of these predictions (Figure 1B) [33,34]. In this crystal, MV-H forms a dimer similar to that observed in unbound MV-H with one CD46 molecule engaging each MV-H. The SCR1 and SCR2 domains of CD46 were observed to bind to MV-H at the side of the fourth and fifth blades of the β-propeller burying a surface of over 2100 Å2 in the protein–protein interface (Figure 2B) [34]. In contrast with the relatively deep sialic acid-binding site found on HN glycoproteins, the CD46-binding site, although extensive, is quite shallow. Additionally, by sequence comparison of SLAM with CD46, Santiago et al. [34], have identified a Pro-Pro motif in the MV-H-CD46 interface which is common to both SLAM and CD46 MV-H receptor-binding domains [34]. These observations, in addition to previous site-directed mutagenesis studies which compared MV-H-SLAM and MV-H-CD46 specificity [35,36], indicate that this site may be used to bind both receptors [34]. Such results underscore the ability of MV-H to adapt to new cell-surface receptors, and thus new hosts.
Attachment by G glycoproteins
NiV and HeV [collectively referred to as HNV (henipavirus)] are the sole members of the genus Henipavirus, a group of viruses which are zoonotic and are associated with high mortality rates [37]. HNV use the flying fox as a natural host, and infections have been reported in a number of other vertebrate species including humans, bats, pigs, cats, dogs and goats [37,38]. Clinical symptoms of human HNV infection include fever, hypotension, dizziness, acute encephalitis and respiratory illness. The time from disease onset to death is rapid (approx. 7–10 days) [37]. The broad respiratory and neurological pathology of these viruses can be directly attributed to their use of ephrinB2 and ephrinB3 as functional entry receptors during viral attachment [7–9]. EphrinB2 and ephrinB3 are well conserved among HNV-susceptible species (<95% conserved) and are widely expressed in many cell types, including neurons, bone, stem cells and epithelial cells, where they are responsible for fundamental signalling processes which underlie axon guidance, vascular development and osteogenesis [39,40].
Henipavirus is the only genus within the Paramyxovirinae subfamily which attach to their cell-surface receptors through a G glycoprotein which lacks both haemagglutination and neuraminidase activity. Multiple structures of the receptor-binding domains of NiV-G and HeV-G have been solved in their unbound state and reveal that, similar to other Paramyxovirinae attachment glycoproteins, the G glycoprotein consists of a C-terminal six-bladed β-propeller fold [41–43]. The β-propeller structures superimpose most closely on to NDV-HN and PIV-HN (average 2.3 Å r.m.s.d. over 376 Cα atoms) structures, but are much less similar to that of the cubic arrangement observed in MV-H (3.3 Å r.m.s.d. over 320 equivalent Cα atoms) (Figure 3) [44]. The contrasting architectures of HNV attachment glycoproteins and MV-H reflect the independent evolution of distinct mechanisms by which these two classes of glycoproteins have acquired the ability to bind cell-surface protein receptors [44].
G glycoprotein receptor binding
As observed in MV-H, HNV-G has a noticeable cavity in the centre of the β-propeller which is reminiscent of the sialic acid-binding site observed in HN glycoproteins. Despite its presence and the overall structural similarity of HNV-G to HN glycoproteins, only one of the seven sialic acid-binding residues which are conserved in the HNs are present in HNV-G. These observations, analogous to those made in the MV-H structure [31,32], are suggestive that this cleft has been left as a vestigial feature marking the evolutionary departure of Henipavirus from other sialic acid-binding paramyxoviruses, and that the use of high-affinity interactions with the low-abundance cell-surface ephrins removes the need for a receptor releasing enzyme to facilitate effective virus spread from cell to cell.
Mutagenesis studies have identified putative HNV-G residues which confer specificity to ephrinB2 and ephrinB3 binding [45–47]. When mapped on to the unbound NiV-G structure, these sites localize to the top of the β-propeller near the HN sialic acid-binding site and distant from the MV-H CD46-binding site. Co-crystal structures of NiV-G and HeV-G in complex with ephrinB2 confirmed many of these functional predictions [44]. EphrinB2, which consists of a Greek key β-barrel fold, engages HeV-G and NiV-G in nearly identical positions at the centre of the β-propeller, directly above the analogous HN sialic acid-binding site, in a 1:1 interaction (Figures 1C and 2C). The relatively shallow protein–protein interactions between the viral and human proteins (in comparison with HN–sialic acid interactions) in these two co-crystal structures are extensive (more than 2600 Å2 of buried surface) and dominated by hydrophobic contacts between residues in an ephrinB2 (GH) loop and residues located at the top surface of the β-propeller. Similar structural information was gained by study of NiV-G in complex with ephrinB3 [42]. The binding mode of ephrinB3 is very similar to that of ephrinB2, with the exception of minor residue differences in the ephrinB3 GH loop.
Comparison of bound and unbound structures of NiV-G and HeV-G also provides evidence for an induced-fit mechanism of ephrin binding, where loops in the first and sixth blades of the HNV-G β-propeller undergo conformational changes upon receptor engagement [41–43]. Such conformational changes facilitate the formation of an ephrin-binding pocket which accommodates a phenylalanine residue, Phe120ephrinB2. Phe120ephrinB2 was confirmed as being crucial to the interaction by site-directed mutagenesis and binding experiments using a F120AephrinB2 mutant which was found to be incapable of binding NiV-G [44]. It is also important to note that the conformational changes observed between the unbound and bound HNV-G may also be important for activation of the fusion mechanism. As in all of the Paramyxovirinae, the exact mechanism by which HNV-F are activated is unknown, and it is likely that ephrin-induced conformational changes of HNV-G may be important for activation of HNV-F, thereby facilitating fusion of the viral and host membranes.
Conclusions
In the present review, we have illustrated the various known mechanisms by which biomedically important viruses within the Paramyxovirinae subfamily use their envelope-associated attachment glycoproteins to hijack host cell receptors to facilitate attachment. Through comparison of structural data resulting from studies of viruses from different genera, we observe that all Paramyxovirinae utilize a receptor-binding six-bladed β-propeller domain to adapt to their corresponding host cell receptors. Whereas the sialic acid-binding HN glycoproteins of the avulaviruses, rubulabiruses and respiroviruses rely on a strictly conserved and multifunctional pocket deep in the centre of the β-propeller that can be used for both glycan binding and cleavage, protein-binding Morbillivirus H and Henipavirus G glycoproteins utilize shallow, yet extensive, single-purpose binding sites.
Structural comparison of H and G glycoprotein β-propellers reveals, however, that, despite having common protein-binding capacities, these two classes of attachment glycoprotein are markedly different in both their architecture and mode of receptor engagement. The six-bladed β-propeller of MV-H, for example, is more cubic in shape than its more globular HNV-G counterpart. Equally, MV-H engages its CD46 and SLAM entry receptors at overlapping binding sites which lie along the outside of the β-propeller, whereas HNV-G binds to ephrinB2 and ephrinB3 at the top and centre of the β-propeller.
Assuming a common evolutionary Paramyxovirus precursor, the structural and functional differences of the HN, H and G glycoproteins studied in the present review highlight the wide mechanistic diversity of the Paramyxovirus attachment. This underscores the ability of the six-bladed β-propeller scaffold to adapt to different host cell receptors and in turn suggests that this architecture provides a molecular platform by which potentially dangerous but currently unknown paramyxoviruses could change cellular tropism and thus emerge in new hosts.
Acknowledgements
We thank all those who have contributed to the structural characterization of HNV attachment glycoproteins: A. Radu Aricescu, David J. Harvey, Jonathan M. Grimes and Robert J. Gilbert.
Funding
D.I.S. is an MRC (Medical Research Council) Professor of Structural Biology, E.Y.J. is a Cancer Research UK Principal Research Fellow, T.A.B. is a Sir Henry Wellcome Postdoctoral Fellow, and M.C. is a Fellow of Oriel College, Oxford. This work was funded by the Wellcome Trust [grant number 075491/Z/04], Medical Research Council, Cancer Research UK, the Oxford Glycobiology Institute and Spine2 Complexes [grant number FP6-RTD-031220].
References
- 1.Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amonsen M., Smith D.F., Cummings R.D., Air G.M. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2–3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 2007;81:8341–8345. doi: 10.1128/JVI.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorig R.E., Marcil A., Chopra A., Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 4.Naniche D., Varior-Krishnan G., Cervoni F., Wild T.F., Rossi B., Rabourdin-Combe C., Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatsuo H., Ono N., Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J. Virol. 2001;75:5842–5850. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsuo H., Ono N., Tanaka K., Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 7.Negrete O.A., Levroney E.L., Aguilar H.C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 8.Bonaparte M.I., Dimitrov A.S., Bossart K.N., Crameri G., Mungall B.A., Bishop K.A., Choudhry V., Dimitrov D.S., Wang L.F., Eaton B.T., Broder C.C. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negrete O.A., Wolf M.C., Aguilar H.C., Enterlein S., Wang W., Muhlberger E., Su S.V., Bertolotti-Ciarlet A., Flick R., Lee B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLos Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crennell S., Takimoto T., Portner A., Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutininneuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 11.Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan P., Thompson T.B., Wurzburg B.A., Paterson R.G., Lamb R.A., Jardetzky T.S. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence M.C., Borg N.A., Streltsov V.A., Pilling P.A., Epa V.C., Varghese J.N., McKimm-Breschkin J.L., Colman P.M. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Varghese J.N., Laver W.G., Colman P.M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 15.Colman P.M., Varghese J.N., Laver W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 16.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discovery. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 17.Iorio R.M., Field G.M., Sauvron J.M., Mirza A.M., Deng R., Mahon P.J., Langedijk J.P. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 2001;75:1918–1927. doi: 10.1128/JVI.75.4.1918-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connaris H., Takimoto T., Russell R., Crennell S., Moustafa I., Portner A., Taylor G. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 2002;76:1816–1824. doi: 10.1128/JVI.76.4.1816-1824.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemper C., Atkinson J.P. Measles virus and CD46. Curr. Top. Microbiol. Immunol. 2009;329:31–57. doi: 10.1007/978-3-540-70523-9_3. [DOI] [PubMed] [Google Scholar]
- 20.Manchester M., Gairin J.E., Patterson J.B., Alvarez J., Liszewski M.K., Eto D.S., Atkinson J.P., Oldstone M.B. Measles virus recognizes its receptor, CD46, via two distinct binding domains within SCR1–2. Virology. 1997;233:174–184. doi: 10.1006/viro.1997.8581. [DOI] [PubMed] [Google Scholar]
- 21.Leonard V.H., Sinn P.L., Hodge G., Miest T., Devaux P., Oezguen N., Braun W., McCray P.B., Jr, McChesney M.B., Cattaneo R. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 2008;118:2448–2458. doi: 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocks B.G., Chang C.C., Carballido J.M., Yssel H., de Vries J.E., Aversa G. A novel receptor involved in T-cell activation. Nature. 1995;376:260–263. doi: 10.1038/376260a0. [DOI] [PubMed] [Google Scholar]
- 23.Ono N., Tatsuo H., Tanaka K., Minagawa H., Yanagi Y. V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J. Virol. 2001;75:1594–1600. doi: 10.1128/JVI.75.4.1594-1600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno S., Seki F., Ono N., Yanagi Y. Histidine at position 61 and its adjacent amino acid residues are critical for the ability of SLAM (CD150) to act as a cellular receptor for measles virus. J. Gen. Virol. 2003;84:2381–2388. doi: 10.1099/vir.0.19248-0. [DOI] [PubMed] [Google Scholar]
- 25.Ono N., Tatsuo H., Hidaka Y., Aoki T., Minagawa H., Yanagi Y. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 2001;75:4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckland R., Wild T.F. Is CD46 the cellular receptor for measles virus? Virus Res. 1997;48:1–9. doi: 10.1016/s0168-1702(96)01421-9. [DOI] [PubMed] [Google Scholar]
- 27.Takeda M., Tahara M., Hashiguchi T., Sato T.A., Jinnouchi F., Ueki S., Ohno S., Yanagi Y. A human lung carcinoma cell line supports efficient measles virus growth and syncytium formation via a SLAM- and CD46-independent mechanism. J. Virol. 2007;81:12091–12096. doi: 10.1128/JVI.01264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahara M., Takeda M., Shirogane Y., Hashiguchi T., Ohno S., Yanagi Y. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 2008;82:4630–4637. doi: 10.1128/JVI.02691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe A., Yoneda M., Ikeda F., Terao-Muto Y., Sato H., Kai C. CD147/EMMPRIN acts as a functional entry receptor for measles virus on epithelial cells. J. Virol. 2010;84:4183–4193. doi: 10.1128/JVI.02168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffin D.E. Measles virus. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Staus S.E., editors. Fields Virology. 5th edn. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1551–1585. [Google Scholar]
- 31.Colf L.A., Juo Z.S., Garcia K.C. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 32.Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santiago C., Gutierrez-Rodriguez A., Tucker P.A., Stehle T., Casasnovas J.M. Crystallization and preliminary crystallographic analysis of the measles virus hemagglutinin in complex with the CD46 receptor. Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 2010;66:91–94. doi: 10.1107/S1744309109050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santiago C., Celma M.L., Stehle T., Casasnovas J.M. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 35.Santiago C., Bjorling E., Stehle T., Casasnovas J.M. Distinct kinetics for binding of the CD46 and SLAM receptors to overlapping sites in the measles virus hemagglutinin protein. J. Biol. Chem. 2002;277:32294–32301. doi: 10.1074/jbc.M202973200. [DOI] [PubMed] [Google Scholar]
- 36.Masse N., Ainouze M., Neel B., Wild T.F., Buckland R., Langedijk J.P. Measles virus (MV) hemagglutinin: evidence that attachment sites for MV receptors SLAM and CD46 overlap on the globular head. J. Virol. 2004;78:9051–9063. doi: 10.1128/JVI.78.17.9051-9063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eaton B.T., Broder C.C., Middleton D., Wang L.F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bossart K.N., Tachedjian M., McEachern J.A., Crameri G., Zhu Z., Dimitrov D.S., Broder C.C., Wang L.F. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372:357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Pasquale E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 40.Pasquale E.B. Eph–ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Bowden T.A., Crispin M., Harvey D.J., Aricescu A.R., Grimes J.M., Jones E.Y., Stuart D.I. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J. Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu K., Rajashankar K.R., Chan Y.P., Himanen J.P., Broder C.C., Nikolov D.B. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowden T.A., Crispin M., Harvey D.J., Jones E.Y., Stuart D.I. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J. Virol. 2010;12:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowden T.A., Aricescu A.R., Gilbert R.J., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 45.Bishop K.A., Stantchev T.S., Hickey A.C., Khetawat D., Bossart K.N., Krasnoperov V., Gill P., Feng Y.R., Wang L., Eaton B.T., et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J. Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guillaume V., Aslan H., Ainouze M., Guerbois M., Wild T.F., Buckland R., Langedijk J.P. Evidence of a potential receptor-binding site on the Nipah virus G protein (NiV-G): identification of globular head residues with a role in fusion promotion and their localization on an NiV-G structural model. J. Virol. 2006;80:7546–7554. doi: 10.1128/JVI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negrete O.A., Chu D., Aguilar H.C., Lee B. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J. Virol. 2007;81:10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski R.A. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart D.I., Levine M., Muirhead H., Stammers D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 50.Felsenstein J. PHYLIP: Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]