We wish to let CBE—Life Sciences Education readers know about a portal to a set of curricular lab modules designed to integrate genomics and bioinformatics into commonly taught courses at all levels of the undergraduate curriculum. Through a multi-year, collaborative process, we developed, implemented, and peer-reviewed inquiry-based, integrated instructional units (I3Us) adaptable to a range of teaching settings, with a focus on both model and nonmodel systems. Each of the products is built on vetted design principles: 1) they have clear pedagogical objectives; 2) they are integrated with lessons taught in the lecture; 3) they are designed to integrate the learning of science content with learning about the process of science; and 4) they require student reflection and discussion (Figure 1; National Research Council [NRC], 2005). Eleven I3Us were designed and implemented as multi-week modules within the context of an existing biology course (e.g., microbiology, comparative anatomy, introduction to neurobiology), and three I3Us were incorporated into interdisciplinary biology/computer science classes. Our collection of genomics instructional units, together with extensive supporting material for each module, is accessible on a dedicated website (http://serc.carleton.edu/genomics/activities.html) that also provides links to bioinformatics tools and online assessment and pedagogical resources for teaching genomics.
Figure 1.
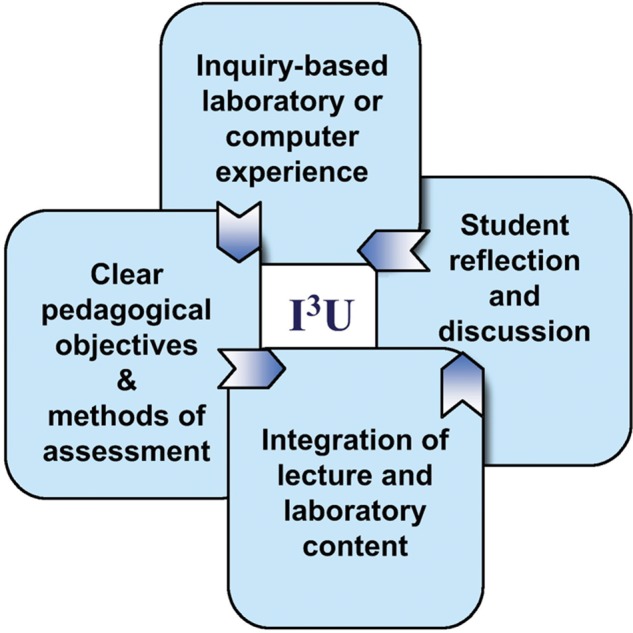
Pedagogical elements of the I3U, which was based on the findings of America's Lab Report (NRC, 2005) and was used as the primary curricular design framework for this project.
Rapid advances in genome sequencing and analysis offer unparalleled opportunity and challenge for biology educators. More data are being generated than can be analyzed and contextualized in traditional teaching or research models. Indeed, this explosion of data has spawned rapid growth in the discipline of bioinformatics, which is focused on development of the computational tools and approaches for extracting biologically meaningful insights from genomic data. At the same time, access to vast quantities of genomic data stored in publicly available databases can offer educators ways to engage undergraduates in authentic research and to democratize research that was previously possible only at research-intensive universities with massive instrumentation infrastructures. The integration of genomic and bioinformatic approaches into undergraduate curricula represents one response to the national calls for biology teaching that is more quantitative and promotes deeper understanding of biological systems through interdisciplinary analyses (National Academy of Sciences, 2003; Association of American Medical Colleges and Howard Hughes Medical Institute [HHMI], 2009; NRC, 2009; American Association for the Advancement of Science, 2011). Yet relatively few faculty members who teach undergraduate biology have expertise in the fields of genomics or bioinformatics, and they may therefore feel inadequately prepared to develop their own new curricular modules capitalizing on this dispersed abundance of available resources.
Our Teagle Foundation–funded genomics education initiative, Bringing Big Science to Small Colleges: A Genomics Collaboration, was designed to address the challenges of helping faculty members integrate genome-scale science into the undergraduate classroom. The goal of the project was to create and disseminate self-contained curricular units that stimulate students and faculty alike to think in new ways and at different scales of biological inquiry. To this end, a series of three workshops over 3 yr brought together a total of 34 faculty participants from 19 institutions and a diverse array of disciplines—including biology, computer science, and science education—to develop a set of lab modules containing a substantial genomics component. We believe that these modules are suitable for integration into existing courses in the biology curriculum and are adaptable to a variety of teaching settings.
The project website serves as a portal to activity sheets describing each I3U, complete with learning goals, teaching tips, and links to teaching materials, as well as downloadable resources and assessment tools (Figure 2), that can be customized by any interested educator. Each I3U was peer-reviewed by fellow participants, as well as by a professional project consultant who has extensive experience with Web-based description of teaching materials using this format (Manduca et al., 2006). The goals of this review process were to ensure that each I3U met the design criteria articulated above, and to evaluate whether the activity sheet provided both an easily accessible overview of the content and enough detailed information for other instructors to adapt and implement the material and its associated assessment strategies. This peer review was complemented by each participant's own explicitly framed evaluation of his/her I3U through a formal reflection form (accessible at http://serc.carleton.edu/genomics/workshop09/index.html). Although these I3Us were designed for courses currently taught by the project participants within the specific institutions’ curricula, we propose that they can be inserted into other courses encompassing similar content (Tables 1 and 2) and/or learning goals (e.g., Figure 2). We have received many communications from colleagues at other institutions who have adapted our I3Us for their courses.
Figure 2.
Excerpt from an activity sheet from the Genomics Instructional Units Minicollection describing one of the curricular modules developed within the Bringing Big Science to Small Colleges program (for the complete activity sheet, see http://serc.carleton.edu/genomics/units/19163.html).
Table 1.
List of I3Us generated in the Bringing Big Science to Small Colleges collaborative project, grouped by the general level in the curriculum in which they were originally taught
| ID | I3U title | Conceptual content |
|---|---|---|
| Introductory level | ||
| A | Reconstructing the Evolution of Cauliflower and Broccoli | Plant development Evolution Bioinformatics |
| B | Human Single Nucleotide Polymorphism Determination | Genetics Human evolution Bioinformatics |
| C | Local Population Structure and Behavior of the Wood Frog Rana sylvatica | Population genetics Behavioral ecology |
| Intermediate level | ||
| D | Comparison of Protein Sequences: BLAST Searching and Phylogenetic Tree Construction | Molecular biology Molecular evolution Bioinformatics |
| E | Phylogenetic Analysis of Bony Fishes: Morphological and mtDNA Sequence Comparisons | Phylogenetics Vertebrate biology Bioinformatics |
| F | Molecular Evolution of Gene Families | Genetics Molecular evolution Bioinformatics |
| G | Exploring the Chamaecrista fasciculata Gene Space | Plant genetics Molecular evolution Bioinformatics |
| H | Metagenomic Analysis of Winogradsky Columns | Microbial metabolism Community ecology Ecosystems studies Bioinformatics/programming |
| I | Behavior, Neuroanatomy, Genomics: What Can We Learn from Mouse Mutants? | Neurobiology Behavioral genetics Bioinformatics |
| J | Expression of Gerontogenes in Neurons: A Comparative Genomic Approach to Studying the Role of the Nervous System in Lifespan/Aging (Raley-Susman and Gray, 2010) | Molecular evolution Behavioral neuroscience Bioinformatics |
| K | Comparison of a Highly Polymorphic Olfactory Receptor Gene Subfamily in Genetically Diverse Dog Breedsa | Molecular evolution Phylogenetics Sensory biology Bioinformatics/programming |
| Advanced level | ||
| L | Integrative Activities to Study the Evolution of Nervous System Function | Neurobiology Evolution Bioinformatics |
| M | Modeling Molecular Evolutiona | Molecular evolution Bioinformatics/programming Computer science |
| N | Constructing and using a PAM-Style Scoring Matrixa | Molecular evolution Bioinformatics/programming Computer science |
aI3U was implemented in an interdisciplinary biology/computer science course.
Table 2.
Pedagogical attributes (scale of biological organization, genomic level of analysis, and bioinformatic skills taught) of I3Us developed in this project and disseminated on the project's website
| Questions asked at the level of | Analysis | Bioinformatics skills/tools | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I3Ua | Focal taxa | Evolution | Behavior | Physiology | Morphology | Cell | Molecular | Biochemistry | Functional | Comparative | Metagenomic | Species-specific DNA database | Multi-species genome browser | Protein modeling | BLAST/alignment | Phylogenetics | Programming |
| A | Brassica | √ | √ | √ | √ | ||||||||||||
| B | Human | √ | √ | √ | |||||||||||||
| C | Wood frog | √ | √ | √ | √ | ||||||||||||
| D | Fish/vertebrates | √ | √ | √ | √ | √ | √ | ||||||||||
| E | Fish/vertebrates | √ | √ | √ | √ | √ | √ | ||||||||||
| F | Xenopus | √ | √ | √ | √ | √ | √ | ||||||||||
| G | Pea/various | √ | √ | √ | √ | √ | |||||||||||
| H | Eubacteria | √ | √ | √ | √ | √ | √ | ||||||||||
| I | Mouse | √ | √ | √ | √ | √ | √ | √ | |||||||||
| J | Worm | √ | √ | √ | √ | √ | |||||||||||
| K | Dog | √ | √ | √ | √ | √ | √ | √ | |||||||||
| L | Various | √ | √ | √ | √ | √ | √ | √ | |||||||||
| M | Various | √ | √ | √ | √ | √ | √ | ||||||||||
| N | N/A | √ | √ | √ | √ | ||||||||||||
aLetters denote I3U units as follows: A: Reconstructing the Evolution of Cauliflower and Broccoli; B: Human Single Nucleotide Polymorphism Determination; C: Local Population Structure and Behavior of the Wood Frog Rana sylvatica; D: Comparison of Protein Sequences: BLAST Searching and Phylogenetic Tree Construction; E: Phylogenetic Analysis of Bony Fishes: Morphological and mtDNA Sequence Comparisons; F: Molecular Evolution of Gene Families; G: Exploring the Chamaecrista fasciculata Gene Space; H: Metagenomic Analysis of Winogradsky Columns; I: Behavior, Neuroanatomy, Genomics: What Can We Learn from Mouse Mutants?; J: Expression of Gerontogenes in Neurons: A Comparative Genomic Approach to Studying the Role of the Nervous System in Lifespan/Aging; K: Comparison of a Highly Polymorphic Olfactory Receptor Gene Subfamily in Genetically Diverse Dog Breeds; L: Integrative Activities to Study the Evolution of Nervous System Function; M: Modeling Molecular Evolution; N: Constructing and Using a PAM-Style Scoring Matrix.
One fundamental characteristic of each I3U in our collection is the focus on guided inquiry. The benefits to an undergraduate of hands-on participation in research are well documented (Nagda et al., 1998; Gafney, 2001; Hunter et al., 2007; Kardash et al., 2008; Lopatto, 2009). Integrating authentic research experiences into the undergraduate curriculum allows this powerful learning model to be scaled from use with only a few students to use with entire laboratory sections (Lopatto 2009; Lopatto et al. 2008). Like other national participatory genomic teaching initiatives (Campbell et al., 2006, 2007; Ditty et al., 2010; Shaffer et al., 2010; HHMI, 2011), our model for curriculum development in genomics emphasizes synergies between student-centered research and education. However, in contrast with some of these other projects, our grassroots approach leveraged a wealth of existing expertise by providing opportunities for individual faculty members to develop, implement, modify, evaluate, and share undergraduate teaching modules that stem from their own research and/or teaching interests. In this regard, our project most closely resembles the Genome Consortium for Active Teaching, which provides faculty members and their undergraduates access to microarrays from a variety of organisms, allowing participants to define their own research questions in a model system with which they are already familiar (Campbell et al., 2006, 2007).
Our collaborative effort among biologists, computer scientists, and science educators has yielded a collection of pedagogical resources that can be adapted for use in a wide variety of educational settings. We invite other biologists to begin building on our work by using and providing feedback on our I3Us. Faculty who have tested materials that exemplify our design principles are encouraged to add them to our collection. For further information, please contact the corresponding author.
ACKNOWLEDGMENTS
This project was supported by a Teagle Foundation Fresh Thinking grant, along with supplemental support from Williams College, Vassar College, and Ismail Kola of Schering-Plough.
Footnotes
§Present address: Washington State University, School of Biological Sciences, Pullman, WA 99164.
†These authors contributed equally to the writing of this letter.
REFERENCES
- American Association for the Advancement of Science (AAAS) Vision and Change in Undergraduate Biology Education. Washington, DC: 2011. [Google Scholar]
- Association of American Medical Colleges (AAMC) and Howard Hughes Medical Institute (HHMI) Scientific Foundations for Future Physicians. Washington, DC: 2009. [Google Scholar]
- Campbell AM, Eckdahl TT, Fowlks E, Heyer LJ, Mays Hoopes LL, Ledbetter ML, Rosenwald AG. Genome Consortium for Active Teaching (GCAT) Science. 2006;311:1103–1104. doi: 10.1126/science.1121955. [DOI] [PubMed] [Google Scholar]
- Campbell AM, Ledbetter MLS, Hoopes LLM, Eckdahl TT, Heyer LJ, Rosenwald AG, Fowlks E, Tonidandel S, Bucholtz B, Gottfried G. Genome Consortium for Active Teaching: Meeting the goals of BIO2010. CBE Life Sci Educ. 2007;6:109–118. doi: 10.1187/cbe.06-10-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditty JL, et al. Incorporating genomics and bioinformatics across the life sciences curriculum. PLoS Biol. 2010;8:e1000448. doi: 10.1371/journal.pbio.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafney L. The impact of research on undergraduates’ understanding of science. Counc Undergrad Res Q. 2001;21:172–176. [Google Scholar]
- HHMI. Science Education Alliance. 2011. www.hhmi.org/grants/sea (accessed 3 May 2012) [Google Scholar]
- Hunter A-B, Laursen SL, Seymour E. Becoming a scientist: the role of undergraduate research in students’ cognitive, personal, and professional development. Sci Educ. 2007;91:36–74. [Google Scholar]
- Kardash C-A, Wallace M, Blockus L. Science undergraduates’ perceptions of learning from undergraduate research experiences. In: Miller RL, Rycek RF, Balcetis E, Barney ST, Beins BC, Burns SR, Smith R, Ware ME, editors. Developing, Promoting, and Sustaining the Undergraduate Research Experience in Psychology. Washington, DC: Society for the Teaching of Psychology; 2008. http://teachpsych.org/resources/e-books/ur2008/ur2008.php (accessed 3 May 2012) [Google Scholar]
- Lopatto D. Science in Solution: The Impact of Undergraduate Research on Student Learning. Tuscon, AZ: Research Corporation for Science Advancement; 2009. [Google Scholar]
- Lopatto D, et al. Genomics Education Partnership. Science. 2008;322:684–685. doi: 10.1126/science.1165351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manduca CA, Fox S, Iverson ER. Digital library as network and community center. D-Lib Magazine. 2006;12 doi:10.1045/december2006-manduca. [Google Scholar]
- Nagda BA, Gregerman SR, Jonides J, von Hippel W, Lerner JS. Undergraduate student-faculty research partnerships affect student retention. Rev Higher Educ. 1998;22:55–72. [Google Scholar]
- National Academy of Sciences. BIO 2010: Transforming Undergraduate Education for Future Research Biologists. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- National Research Council (NRC) America's Lab Report. Washington, DC: National Academies Press; 2005. [Google Scholar]
- NRC. A New Biology for the 21st Century: Ensuring the United States Leads the Coming Biology Revolution. Washington, DC: National Academies Press; 2009. [PubMed] [Google Scholar]
- Raley-Susman KM, Gray JM. Exploration of gerontogenes in the nervous system: a multi-level neurogenomics laboratory module for an intermediate neuroscience and behavior course. J Undergrad Neurosci Educ. 2010;8:A108–A115. [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, et al. The Genomics Education Partnership: successful integration of research into laboratory classes at a diverse group of undergraduate institutions. CBE Life Sci Educ. 2010;9:55–69. doi: 10.1187/09-11-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]



