Abstract
Background
Hybridisation is presumed to be an important mechanism in plant speciation and a creative evolutionary force often accompanied by polyploidisation and in some cases by apomixis. The Potentilla collina group constitutes a particularly suitable model system to study these phenomena as it is morphologically extensively variable, exclusively polyploid and expresses apomixis. In the present study, the alpine taxon Potentilla alpicola has been chosen in order to study its presumed hybrid origin, identify underlying evolutionary processes and infer the discreteness or taxonomic value of hybrid forms.
Results
Combined analysis of AFLP, cpDNA sequences and ploidy level variation revealed a hybrid origin of the P. alpicola populations from South Tyrol (Italy) resulting from crosses between P. pusilla and two cytotypes of P. argentea. Hybrids were locally sympatric with at least one of the parental forms. Three lineages of different evolutionary origin comprising two ploidy levels were identified within P. alpicola. The lineages differed in parentage and the complexity of the evolutionary process. A geographically wide-spread lineage thus contrasted with locally distributed lineages of different origins. Populations of P. collina studied in addition, have been regarded rather as recent derivatives of the hexaploid P. argentea. The observation of clones within both P. alpicola and P. collina suggested a possible apomictic mode of reproduction.
Conclusions
Different hybridisation scenarios taking place on geographically small scales resulted in viable progeny presumably stabilised by apomixis. The case study of P. alpicola supports that these processes played a significant role in the creation of polymorphism in the genus Potentilla. However, multiple origin of hybrids and backcrossing are considered to produce a variety of evolutionary spontaneous forms existing aside of reproductively stabilised, established lineages.
Keywords: AFLP, Apomixis, cpDNA, Hybridisation, Introgression, Polyploidy, Reproduction mode, Potentilla, Rosaceae
Background
Interspecific hybridisation has long been considered a potentially innovative evolutionary force playing an important role in speciation and phenotypic diversification e.g. [1-3]. Hybridisation between two (or more) distantly related species may be accompanied by doubling of the genome thus overcoming the common sterility in hybrids by providing each chromosome with a pairing partner (also referred to as allopolyploidy; [4]). Furthermore, hybridisation is also believed to be fundamental to the occurrence of apomixis (asexual reproduction through seeds), which is found almost exclusively in polyploids and highly heterozygous species [5].
Hybridisation is an important mechanism in the formation of species in the highly polymorphic genus Potentilla. Possible hybrid origins of several taxa associated with morphological variability, intermediacy and consequent taxonomic complexity were a concern already in the 19th century (e.g. [6,7]). Later on, the presence of apomixis [8-10] and extensive intraspecific ploidy variation [11-13] supported this view and added to the understanding of the evolutionary pathways followed by the genus.
The Potentilla collina group from the series Argenteae Th. Wolf. [14] seems to be a particularly suitable model system for studying the contribution of hybridisation, polyploidisation and apomixis to the evolution of the genus. At least fifteen species [15,16] belonging to this group are considered either locally to regionally distributed microspecies and represent a taxonomically complicated Eurasian hybrid complex. The observed morphological variability and exclusive polyploidy (x = 7; 2n = 5–12x), with occasional observation of chromosome aberrations [17], are explained by the hybrid origin of the group involving taxa from the series Aureae Th. Wolf and Argenteae Th. Wolf [14,18,19]. Within the group, the development of both female and male gametophytes was reported to be disturbed or the offspring originated through apomictic pathways. Apomixis by means of apospory and pseudogamy was obligate or close to obligate [20-22]. Full or partial male sterility (9–44 %) has also been found in several studied individuals [17,21]. Furthermore, in a hexaploid P. collina biotype only uni- and bivalents, but no tri- or tetravalents were observed [23], which suggests the presence of at least two different genomes. Experimental hybridisations confirmed interfertility between presumed parents (e.g. [18,24]) and fertilisation of reduced (BII-hybrids) and unreduced egg cells (BIII-hybrids) [24,25] have also been reported.
One example from the P. collina group is the Potentilla alpicola de La Soie, a microspecies restricted to the western and central Alps [15,26]. It occupies montane to subalpine habitats and is often found in sympatry with P. argentea L. and P. pusilla Host from the P. verna group [27] on population or local scales. Chromosome numbers reported so far revealed polyploidy in this taxon (2n = 5x, 6x, 12x; [26]). Concerning the morphology, P. alpicola is usually intermediate in most morphological characters, with some individuals tending to P. argentea.
The following study is based on the assumption of an autochthonous origin of P. alpicola and various potential parental taxa occurring in sympatry were included. We combined ploidy data with amplified fragment length polymorphisms (AFLP) and chloroplast (cp) DNA sequencing and asked four main questions: (i) Is P. alpicola of hybrid origin? (ii) If yes, which taxa have been involved in its formation? (iii) Did P. alpicola arise at several localities independently (polytopically), or did it arise in one locality and spread afterwards throughout the Central Alps? (iv) Are hybrid forms reproductively stabilised, i.e. discrete? Finally, we comment (v) on resulting taxonomic implications.
Material and methods
Plant material
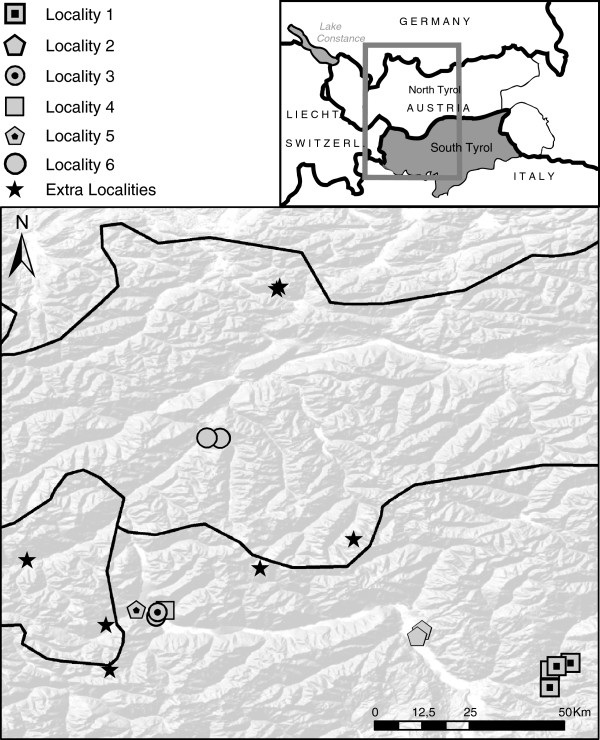
Plant material was collected from six broader localities within the central Alps (South Tyrol, Switzerland and North Tyrol) (Figure 1; Table 1; Additional file 1). Potentilla alpicola was sampled together with the sympatrically co-occurring possible parental taxa P. argenteaP. pusillaP. incana G.Gaertn., B.Mey. & Scherb]. Putative hybrid populations, morphologically deviating from P. alpicola, but belonging to the P. collina group were also sampled (referred to as “P. collina”). Species present in the Central Alps P. aurea L., P. brauneana Hoppe, P. crantzii (Crantz) Beck ex Fritsch, P. frigida Vill., P. thuringiaca Bernh. ex Link, P. pusilla × thuringiaca, P. aff. verna and potentially involved in the genesis of P. alpicola were sampled from additional six localities. Individuals were collected from a distance of at least 5 m from each other. In total, 293 accessions representing 30 populations and 11 taxa were investigated, 5–27, but mostly 10 samples per population. Herbarium vouchers from plants collected during field trips as well as from transplanted plants are deposited in HEID herbarium. In order to present the geographical data ArcGIS v9.1 (ESRI, USA) software with the Hillshade WMS-layer [28] was used.
Figure 1 .
Geographic origin of the studied populations. A full list of localities and population codes are given in Additional file 1.
Table 1.
Sampling localities of studied taxa
| Locality/PopID | Taxon | Locality |
|---|---|---|
|
LOC_1 |
|
Völs/Seis am Schlern (NE from Bozen) |
| Pop088 |
P. argentea |
ITA; SW of Seis am Schlern |
| Pop085 |
P. pusilla |
ITA; Völs am Schlern, 0.5 km N |
| Pop089 |
P. incana |
ITA; Völs am Schlern, Mongadui |
| Pop086 |
P. alpicola |
ITA; Völs am Schlern, St. Konstantin |
| Pop087 |
P. alpicola/pusilla |
ITA; Völs am Schlern, St. Konstantin |
|
LOC_2 |
|
Burgstall/Lana-Burgstall (SE from Meran) |
| Pop093 |
P. argentea |
ITA; Burgstall, western slope |
| Pop094 |
P. pusilla |
ITA; Burgstall, western slope |
| Pop095 |
P. collina |
ITA; Burgstall/Lana, railway station |
|
LOC_3 |
|
Glurns (Vinschgau) |
| Pop098 |
P. argentea |
ITA; Glurns, sedimentation tank |
| Pop099 |
P. pusilla |
ITA; Glurns, settlement Sölles |
| Pop097 |
P. collina |
ITA; Glurns, sedimentation tank |
|
LOC_4 |
|
Schluderns (Vinschgau) |
| Pop100 |
P. argentea |
ITA; Schluderns, Kalvarienberg |
| Pop101 |
P. pusilla |
ITA; Schluderns, Kalvarienberg |
| Pop102 |
P. alpicola |
ITA; Schluderns, Kalvarienberg |
|
LOC_5 |
|
Laatsch/Münstertal valley |
| Pop198 |
P. argentea |
ITA; exit of the Münstertal valley/Laatsch |
| Pop199 |
P. pusilla |
ITA; exit of the Münstertal valley/Laatsch |
| Pop200 |
P. alpicola |
ITA; exit of the Münstertal valley/Laatsch |
|
LOC_6 |
|
Kauns (Northern Tyrol) |
| Pop206 |
P. argentea |
AUT; Ötztaleralpen, Kauns, W of the church |
| Pop205 |
P. pusilla |
AUT; Ötztaleralpen, Kauns, ESE of the church |
| Pop204 |
P. collina |
AUT; Ötztaleralpen, Kauns, ESE of the church |
|
EXTRA LOCALITIES |
|
|
| Pop202 |
P. argentea |
CHE; Münstertal valley, NW Müstair |
| Pop074 |
P. aurea |
AUT; Northern Limestone Alps |
| Pop075 |
P. brauneana |
AUT; Northern Limestone Alps |
| Pop080 |
P. crantzii |
AUT; Obergurgl, Rotmoostal valley |
| Pop096 |
P. frigida |
AUT; Ötztaleralpen, summit Fineiljoch |
| Pop103 |
P. frigida |
ITA; Southern Tyrol, Stilfserjoch pass |
| Pop201 |
P. pusilla |
CHE; Münstertal valley, NW Müstair |
| Pop190 |
P. pusilla × thuringiaca |
CHE; Engadin, Ftan, above the railway station |
| Pop189 |
P. thuringiaca |
CHE; Engadin, Ftan, above the railway station |
| Pop203 | P. aff. verna | CHE; Münstertal valley, NW Müstair |
Populations are ordered according to broader localities (LOC_1 – LOC_6), country codes follow ISO 3166–1 Alpha-3.
Chromosome counts and DNA ploidy level estimation
The DNA ploidy levels were determined by flow cytometry from fresh leaf petioles using the Partec Ploidy Analyser PA (Partec, Germany) at the IPK, Gatersleben and at the Department of Pharmacognosy, University of Vienna. The samples were prepared according to the two-step (Otto) protocol [29] with the internal standard Lycopersicon esculentum cv. Stupické polní tyčkové rané [30]; Potentilla incana Ptl4311] and the nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). Sample/standard ratios were calculated from the means of the sample and standard fluorescence histograms, and only those with coefficients of variation (CVs) < 5 % for the G0/G1 peak of the analysed sample were considered. In order to obtain a reliable reference for the DNA ploidy estimation, chromosome numbers of individuals of the studied taxa were counted following Murín [31] or Dobeš [13] (see Additional file 1). In case of P. argenteaP. incana and P. thuringiaca individuals have been already karyotyped elsewhere ([32,33]). The DNA ploidy level has been attributed for each species separately based on the regression of sample/standard fluorescence ratios against the ratios of the counted individuals.
DNA extraction, cpDNA amplification and sequencing
The total DNA was isolated from freshly-collected, silica gel-dried leaf tissue from single individuals using the procedure of Dobeš and Paule [34]. The plastid trnH(gug)-psbA intergenic spacer (IGS) was amplified using the primers: trnH(gug) 5’-CGC GCA TGG TGG ATT CAC AAT CC-3’ and psbA 5’-GTT ATG CAT GAA CGT AAT GCT C-3’ [35] and the PCR reactions were performed as described in Paule et al. [32]. The cycle sequencing was accomplished on both strands. All sequences were edited and a consensus was made of forward and reverse sequences using the software SeqMan v4.0 (DNASTAR, USA).
AFLP analysis
The AFLP analyses were performed using the protocol established by Vos et al. [36] with few modifications as applied by Paule et al. [32]. Three differentially fluorescence labelled PCR products of the same sample were multiplexed and diluted and the fragments were separated on a MegaBase 500 DNA capillary-sequencer together with an ET-ROX 550 size standard (Amersham Biosciences, USA). In each run, a total of 48 samples were analysed, including one standard sample applied to each run, one negative control, one repeat within the runs and several other repeats (altogether 5 %). Raw data were visualised and the fragments manually scored using GeneMarker v1.8 (SoftGenetics, USA). Processed data were exported as a presence/absence matrix.
Data analyses
The DNA-sequences were multiply aligned by means of the ClustalX v1.83 [37] and the alignments were manually refined using the GeneDoc v2.7 [38]. Two regions were excluded from the alignment due to repeated sequence motifs (poly-A stretches) and three indels were manually coded for presence and absence. Phylogenetic relationships among the cpDNA haplotypes were evaluated by means of the network analysis using the TCS v1.2 [39] with a default connection limit of 95 %.
The following statistical parameters were computed using the R-script AFLPdat ([40]; R v2.9.2 environment [41]) for the whole dataset, taxa or lineages revealed by later analyses: total number of the fragments, proportion of polymorphic fragments, number of private fragments and proportion of shared fragments among lineages. The number of different AFLP genotypes and Nei´s genotype diversity [42] in the P. alpicola populations were estimated using the programs Genotype v1.1 and Genodive v1.2 [43]. The functions allow entering a threshold/error rate, estimated from the observed differences among the replicates or alternatively from the observed pairwise differences between the genotypes.
In order to visualise the phylogenetic relationships among the genotypes (in a sense of AFLP phenotype as used in the following), a Neighbor-Net analysis (as implemented in SplitsTree4 v4.5; [44]) based on Jaccard distance matrix calculated beforehand with DistAFLP (accessible at http://pbil.univ-lyon1.fr/ADE-4/microb/) has been carried out. Since the relationship between hybrid taxa and their parents is not hierarchical, the similarity among AFLP genotypes was presented in a two-dimensional ordination using EUKLID [45]. EUKLID differs from alternative ordination methods in maximizing the distances among predefined groups in the mapping of data. The analysis is based on pairwise Euclidean distances. A mapping error has been calculated estimating the difference in the distance of objects in two-dimensional presentation relative to the distances of objects in the original multidimensional data matrix [46].
Results and discussion
DNA ploidy levels
In total, 212 individuals from 27 populations of nine studied taxa have been investigated by means of flow cytometry (see Additional file 1). Hundred and forty-one samples were measured at the IPK, Gatersleben and 71 samples at the University of Vienna [33,47]. The CVs for the G0/G1 peak of the analysed sample ranged from 1.50 to 5.13 (x[bar] = 2.70). Reference chromosome numbers were obtained for individuals of all except three taxa. The ploidy level has not been determined for P. frigida and P. crantzii populations as reference chromosome counts failed. However, based on the previously published data [48], both may be tetraploid or P. crantzii possibly of higher ploidy. One DNA ploidy level has been determined for P. collina. As no reference chromosomes were counted, the ratios were regressed with P. argentea counts (because of the high genetic affinity of these taxa; see later). Flow cytometric analysis also revealed that two P. alpicola individuals (Ptl4146, Ptl4148) could possibly be aneuhexaploid. Results for all studied taxa are summarised in Table 2.
Table 2.
Summary of the chromosome counts and flow cytometric analyses
| Taxon | DNA ploidy | N | Chromosome number: individual(s) counted | DNA ploidy determination |
|---|---|---|---|---|
|
P. alpicola |
5x |
4 |
2n = 35: Ptl4081, Ptl4149 |
CC regression |
| |
6x |
36 |
2n = 42: Ptl4026, Ptl4141, Ptl4881, |
CC regression |
| |
|
|
Ptl4887, Ptl4911, Ptl4913 |
|
|
P. argentea |
2x |
13 |
see Paule et al. 2011 |
CC regression |
| |
6x |
29 |
see Paule et al. 2011 |
CC regression |
|
P. aurea |
2x |
5 |
2n = 14: Ptl3961 |
CC regression |
|
P. brauneana |
2x |
3 |
2n = 14: Ptl3973 |
CC regression |
|
P. collina |
6x |
20 |
– |
CC regression |
|
P. crantzii |
possibly 4x |
|
– (Dobeš & Vitek 2000) |
Lit. review |
|
P. frigida |
possibly 4x |
|
– (Dobeš & Vitek 2000) |
Lit. review |
|
P. incana |
4x |
9 |
see Scherbatin 2009 |
CC regression |
|
P. pusilla |
4x |
26 |
2n = 28: Ptl4048 |
CC regression |
| |
5x |
11 |
2n = 35: Ptl4184 |
CC regression |
| |
6x |
1 |
2n = 48: Ptl4132 |
CC regression |
| |
7x- |
1 |
2n = 49: Ptl4133, Ptl4187, Ptl4188 |
CC count |
| |
7x |
6 |
|
CC regression |
| |
|
|
2n = 42: Ptl4491, Ptl4497, Ptl4500 |
|
|
P. pusilla × P. thuringiaca |
6x |
8 |
|
CC regression |
| |
|
|
2n = 63: Ptl4571 |
|
| |
9x |
10 |
|
CC regression |
|
P. thuringiaca |
|
|
2n = 49: Ptl4325, Ptl4328 |
|
| P. aff. verna | 7x | 10 | CC regression |
CC regression: regression against counted individuals; CC count: chromosome count; Lit. review: literature review.
AFLP analyses
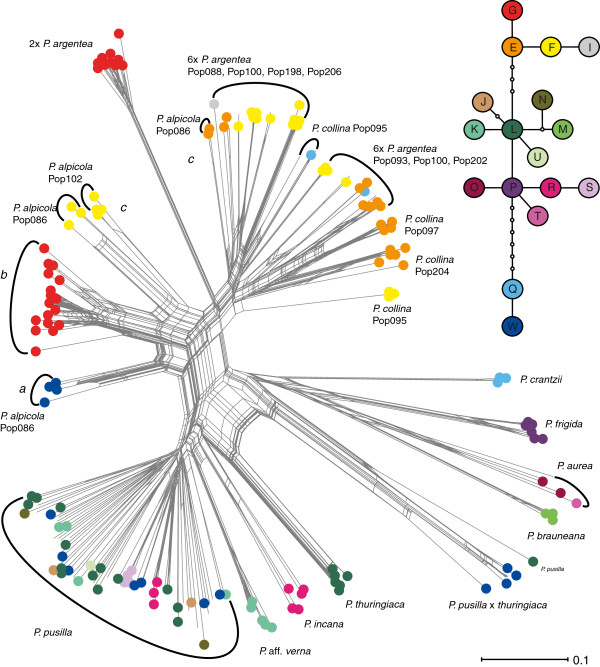
In total, 283 accessions from 30 populations were investigated; 4–27, but mostly 10 individuals per population (see Additional file 1). Three AFLP primer combinations resulted in 241 clearly scorable fragments sized from 63–537 bp and 96.68% of them were polymorphic. The number of fragments within species ranged from 58 to 205. The repeatability of the data was 98.33–100% (x[bar] =98.76%). The major splits in the Neighbor-Net separated three groups of AFLP genotypes (Figure 2): 1. P. verna group (including P. thuringiaca), 2. P. argentea, P. collina, and P. alpicola, 3. the Aureae Alpestres and the Aureae Frigidae including P. pusilla × P. thuringiaca.
Figure 2 .
Phylogenetic relationships inferred on the basis of AFLP data using the Neighbor-Net as implemented in SplitsTree4. Colour-coding refers to the trnH-psbA cpDNA sequences resolved in the parsimony network depicted next to the Neighbor-Net diagram. Small empty circles represent haplotypes that are not present, but necessary to link all observed haplotypes to the network. All haplotypes are separated from the nearest haplotype by one nucleotide difference. The scale bar indicates genetic distance.
CpDNA sequence data and haplotype distribution
The cpDNA sequences were obtained for a total of 175 individuals, for at least three samples per population (see Additional file 1). The length of the trnH-psbA IGS ranged from 439 bp to 487 bp. Sixteen nucleotide substitutions, eight indels and two poly-A stretches were detected. The length of the alignment was 550 bp. After manual coding of the indels for the presence and absence and removal of the poly-A stretches, the total length of the alignment was reduced to 443 bp and 23 parsimony informative sites were considered. The sequences are deposited at NCBI GenBank (see Additional file 1).
Altogether, seventeen trnH-psbA cpDNA haplotypes were identified and the TCS network analysis revealed three groups of haplotypes (Figure 2) separated from each other by 4–12 mutations. Haplotypes E, F, G, and I representing the first group were carried by P. argentea[32] and the most individuals of P. alpicola and P. collina (see Additional file 1). The second group was composed of haplotypes J, K, L, M, N, O, P, R, S, T, and U and included P. thuringiaca, the taxa from the P. verna group (P. pusillaP. incanaP. aff. verna) as well as members of Wolf’s [14]Aureae Alpestres (P. aurea) and Aureae Frigidae (P. brauneanaP. frigida). Haplotypes J, K, L, N, R, S, and U were observed in individuals of AFLP genotype-group 1 only. Haplotypes M; O and T; and P were specific to P. brauneana; P. aurea; and P. frigida, respectively. In the third group haplotype Q was observed in hexaploid P. argentea (see also [32]), one individual of P. collina and in P. crantzii. Haplotype W was found in P. pusillaP. pusilla × thuringiaca, and P. alpicola.
Identity of Potentilla alpicola and P. collina individuals
The Neighbor-Net analysis revealed different positions of P. alpicola in the phylogenetic network suggesting different evolutionary fates for particular populations. A majority of the individuals representing three localities (Localities 1, 4 and 5), formed a separate single cluster (Figure 2). Population Pop102 clustered with hexaploid P. argentea, similarly, as did the three studied populations of P. collina. In combination with the haplotype and cytotype data, three lineages of P. alpicola have been defined: pentaploids carrying haplotype W (lineage a), hexaploids carrying haplotype G (lineage b), and hexaploids grouped with hexaploid P. argentea carrying haplotypes E/F (lineage c) (Figure 2).
The three P. alpicola lineages possessed 115–138 AFLP fragments (Table 3). The highest proportion of fragments among all studied species was shared with P. pusilla (92.75–94.66 %) and hexaploid P. argentea (87.79–92.03 %) by all three lineages (Table 3). Similar pattern was observed for Potentilla collina which carried 164 fragments, 90.85 % and 87.80 % of which shared with hexaploid P. argentea and P. pusilla, respectively.
Table 3.
AFLP fragments shared among taxa
| Nb shared fragments/ % shared fragments | P.alpicola5x (a), (115) | P.alpicola6x (b), (131) | P. alpicola(c), (138) | P. collina(164) |
|---|---|---|---|---|
|
P. argentea 2x (78) |
58/50.43 |
68/51.91 |
66/47.83 |
71/43.29 |
|
P. aurea (74) |
47/40.87 |
54/41.22 |
57/41.30 |
62/37.80 |
|
P. brauneana (58) |
44/38.26 |
47/35.88 |
45/32.61 |
49/29.88 |
|
P. frigida (66) |
49/42.61 |
55/41.98 |
53/38.41 |
53/32.32 |
|
P. crantzii (76) |
56/48.70 |
59/45.04 |
58/42.03 |
60/36.59 |
|
P. incana (130) |
88/76.52 |
96/73.28 |
92/66.67 |
102/62.20 |
|
P. argentea 6x (177) |
104/90.43 |
115/87.79 |
127/92.03 |
149/90.85 |
|
P. pusilla (205) |
107/93.04 |
124/94.66 |
128/92.75 |
144/87.80 |
|
P. thuringiaca (156) |
92/80.00 |
99/75.57 |
99/71.74 |
113/68.90 |
|
P. pusilla × thuringiaca (136) |
87/75.65 |
87/66.41 |
88/63.77 |
101/61.59 |
| P. aff. verna (118) | 85/73.91 | 88/67.18 | 85/61.59 | 92/56.10 |
The number (Nb) of fragments observed for each taxon is given in brackets right to its name. Nb of shared fragments and % of shared fragments (Nb of shared fragments/Nb of fragments in P. alpicola lineages, and P. collina, respectively) for the taxon combinations, are provided in the field of conjunction.
The following taxa had unique fragments: diploid P. argentea – 1 specific fragment, hexaploid P. argentea – 6, P. pusilla – 6, P. thuringiaca – 4, and P. pusilla × thuringiaca – 1. Out of these specific fragments, each P. alpicola lineage contained two fragments of P. pusilla. Additionally, P. alpicola lineage c contained one fragment of hexaploid P. argentea. Potentilla collina carried five specific fragments of hexaploid P. argentea. One specific fragment was observed for P. collina and P. alpicola. Prior to the comparisons of shared bands, individuals found in the clusters of differing taxonomy have been excluded from the analysis (P. argentea: Ptl4408 − 4410; P. pusilla: Ptl4464). These individuals were either taxonomically misidentified when collected in the field or the morphology was not reflected by the molecular data.
Genotypic/Clonal assignment analysis
We have assumed that the same AFLP genotype represents a “clone”. If taken strictly, clones with no difference in banding patterns have been recognised in several populations of P. alpicola (Pop87, Pop102 and Pop200) and P. collina (Pop95, Pop97). However, based on the data repeatability and the pairwise differences between genotypes, a threshold of 4 and 5, respectively, has been suggested. Hence, 5 differences have been chosen as a threshold in the clonal assignment analysis (Table 4) and the analyses have been carried out for each P. alpicola and P. collina population.
Table 4.
Indices of clonal diversity forP. alpicolaandP. collinapopulations
| Nb | Nbgeno | Dg | |
|---|---|---|---|
|
P. alpicola |
|
|
|
| Pop86 |
8 |
8 |
1.000 |
| Pop87 |
9 |
3 |
0.417 |
| Pop102 |
27 |
3 |
0.373 |
| Pop200 |
9 |
5 |
0.806 |
|
P. collina |
|
|
|
| Pop95 |
10 |
3 |
0.600 |
| Pop97 |
9 |
2 |
0.500 |
| Pop204 | 7 | 4 | 0.714 |
Measures are based on the AFLP data and computed using Genotype and Genodive. Nb, number of samples; Nbgeno, number of genotypes considering a threshold of 5 fragments; Dg, genotypic diversity.
The majority of the P. alpicola populations consisted of 1 or 2 abundant clones (Table 4), with an exception of a diverse population Pop86 (Dg = 1.00). Most of the identified clones were population specific, but one clone was shared by populations Pop102 and Pop200. Three studied P. collina populations were composed of 2 and 3 clones. Accordingly, we have assumed that the observed population structure can be attributed to the apomictic mode of reproduction in both P. alpicola and P. collina.
Evolutionary origin of Potentilla alpicola and P. collina
The presence of four different chloroplast haplotypes (E, F, G, W) from two distinct haplotype groups (12 mutation steps apart; Figure 2) indicates that P. alpicola did not arise through a gradual differentiation but rather via other evolutionary processes. This pattern agrees with the assumed hybrid origin of P. alpicola. Due to the fact that the chloroplast genome is maternally inherited in the majority of angiosperms [49], the cpDNA bears on directionality of hybridisation. The three following taxa were thus identified as mothers: P. pusilla, diploid, and hexaploid P. argentea. In the case of P. collina, the cpDNA data suggest that only hexaploid P. argentea may have served as a mother. This finding is supported by the fact that P. alpicola as well as P. collina was found in close spatial proximity to these taxa.
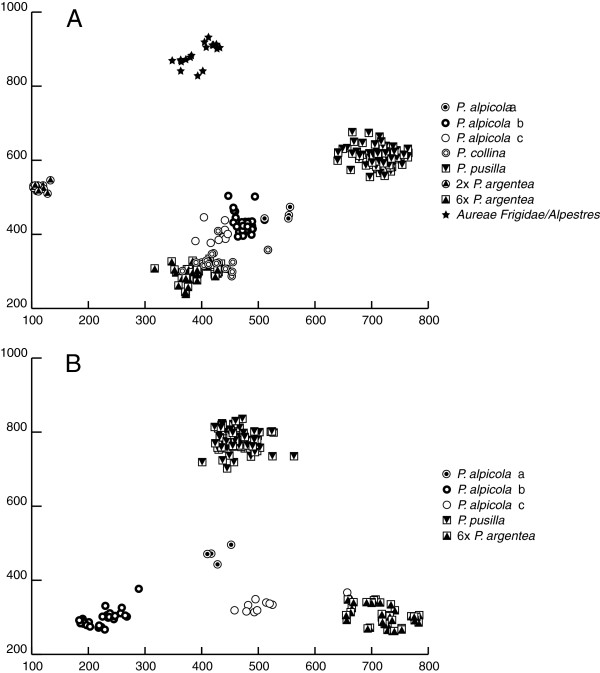
For further verification of this assumption, the genetic similarity of the hybrid taxa relative to these four groups was simultaneously mapped using the EUKLID ordination together with a control group Aureae Alpestres/Aureae Frigidae. Both, P. collina and P. alpicola, were genetically intermediate between hexaploid P. argentea and P. pusilla, but with most of the individuals closer to P. argentea or in case of P. collina partly overlapping with the P. argentea cluster (Figure 3a). This result supports parentage of hexaploid P. argentea and P. pusilla as a most likely scenario. In a taxonomically more focused EUKLID analysis using in addition to these two groups the P. alpicola-specific lineage as reference group, the remaining P. alpicola individuals were genetically intermediate between hexaploid P. argentea and the P. alpicola-cluster and P. pusilla and the P. alpicola-cluster, respectively (Figure 3b).
Figure 3 .
Two-dimensional mapping of AFLP-based Euclidean distances using EUKLID. A:Potentilla pusilla, diploid, and hexaploid P. argentea, and the combined group Aureae Frigidae (P. brauneana, P. frigida)/Aureae Alpestres (P. aurea, P. crantzii) are reference groups located at the outer limits of the plot. The putative hybrids P. alpicola (lineages a–c) and P. collina are placed at an approximately intermediate position between P. pusilla and hexaploid P. argentea. B: Graphical representation of the genetic relationships of P. alpicola lineage a and lineage c to the reference taxa P. pusilla, hexaploid P. argentea, and P. alpicola lineage b (see text for details). The mapping error of the two-dimensional presentations is 0.187 (a) and 0.083 (b), respectively.
Proportions of shared AFLP fragments substantiated this finding indicated by a major nuclear contribution of hexaploid P. argentea and P. pusilla (91.72 and 89.81 % of the total shared fragments; Table 2) to the P. alpicola genome. The highest proportion of fragments from a diploid taxon recovered by diploid P. argentea also supported a contribution from this cytotype. Concerning the species-specific fragments identified in the putative parental taxa (Table 5), P. alpicola revealed 2 fragments from P. pusilla, one fragment from hexaploid P. argentea and no fragments from other taxa. Hence, P. alpicola lineages combine alleles of putative parents with an exception of one fragment, similarly as in the synthetic F1 allohexaploid between the tetraploid Triticum turgidum ssp. dicocoides and the diploid Aegilops tauschii[50], where the majority of the bands were additive, 17 % of both parental fragments were absent and, 2.4 % appeared de novo. The combined data thus suggest parentage of P. pusilla and both diploid and hexaploid P. argentea but with varying contributions to the P. alpicola genome.
Table 5.
Summary of the molecular relationships among hybrid lineages and identified parents
| Lineage | Haplotype | Total % of AFLP bands/Taxon | Species-specific AFLP bands | Geographic origin |
|---|---|---|---|---|
|
P. alpicola 5x (a) |
W |
93.04/P. pusilla |
2/6 P. pusilla |
Pop86/Loc1 |
| 90.43/6x P. argentea |
|
|
||
| 50.43/2x P. argentea |
|
|
||
|
P. alpicola 6x (b) |
G |
94.66/P. pusilla |
2/6 P. pusilla |
Pop86/Loc1 |
| 87.79/6x P. argentea |
|
Pop87/Loc1 |
||
| |
|
Pop102/Loc4 |
||
| 51.91/2x P. argentea |
|
Pop200/Loc5 |
||
|
P. alpicola 6x (c) |
F |
92.03/P. pusilla |
2/6 P. pusilla |
Pop86/Loc1 |
| 92.75/6x P. argentea |
1/6 6x P. argentea |
Pop87/Loc1 |
||
| |
|
Pop102/Loc4 |
||
| 47.83/2x P. argentea |
|
|
||
|
P. collina |
F, E |
87.80/P. pusilla |
5/6 6x P. argentea |
Pop95/Loc2 |
| |
|
90.85/6x P. argentea |
|
Pop97/Loc3 |
| |
|
|
|
Pop204/Loc6 |
| 43.29/2x P. argentea |
Potentilla collina shared 90.85 % (149/164) of the fragments with the hexaploid P. argentea. Fourteen fragments were shared with P. pusilla. However, this is also the case in several hexaploid P. argentea individuals (e.g. Ptl4331–32, Ptl4335). Hence, we do not consider it an indication for a recent hybrid origin, but rather a reflection of possible introgression, which is in agreement with predominance of P. argentea-specific haplotypes in P. collina.
Multiple versus single hybrid formation and complexity of the evolutionary process
Within our data, there is only little evidence that P. alpicola has a common ancestor. A majority of the studied populations possess different haplotypes and AFLP genotypes and clones were mostly population specific. The only subgroup of possibly common origin is the lineage b composed of individuals from three different localities (Locality 1, 4 and 5; Figure 1). The lineage possesses both a single cytotype and chloroplast haplotype and individuals from the populations Pop102 and Pop200, 8 km afar, share one AFLP genotype.
In order to verify this possibility, we asked the question, if the origin of this lineage can be explained by a single evolutionary event or if a more complex scenario should be considered. In case of a single evolutionary event, the disjunct distribution lineage b can be explained by dispersal. In the second case, directional selection for the observed genotypes at multiple localities has to be assumed. For that purpose, we tried to infer the ploidy of the gametes put out by the identified most likely parental species of P. alpicola: diploid and hexaploid P. argentea and P. pusilla. Based on the distribution of genotypic pairwise differences within the populations, a flow cytometric seed screen (Dobeš et al. unpublished research), the occurrence of anorthoploidy (whose maintenance is concomitantly coupled to asexual reproduction in Potentilla), and the literature record e.g. [22,51,52], regular formation of reduced egg cells via meiosis (followed by sexual fertilisation) was inferred for diploid P. argentea as well as tetraploid P. pusilla. In contrast, facultative apomeiotic origins of unreduced egg cells were found for hexaploid P. argentea and high polyploid (5x, 6x, 7x) P. pusilla cytotypes. As pollen - in contrast to female gametogenesis – in both sexual and apomictic Potentillas is almost exclusively produced via meiosis [20,53-55], the following likely ploidies are expected for male / female gametes: 1x / 1x diploid P. argentea; 3x / 6x hexaploid P. argentea; and 2x, 3x / 2x, 3x, 5x, 6x, and 7x P. pusilla.
Based on this set of parental male and female gamete ploidies, we determined possible gamete combinations resulting by fusion in the pentaploid and hexaploid P. alpicola genomes. Interestingly, the origin of the pentaploids (lineage a) only could be explained by a single crossing event as both the observed cpDNA haplotype and genetic similarities with the donating parents have met expectations. In contrast, the formation of none of the hexaploid P. alpicola populations (lineages bc) could be explained by a single event as individuals either carried a haplotype incompatible with the proposed cross and/or their genetic composition did not reflect proportions of the contributed parental genomes. This line of arguments supports the idea of a complex evolutionary history, in particular for lineage b. This lineage alternatively may have a single origin and subsequently dispersed to its present places of occurrence or have originated multiple times under the assumption of directional selection. Both interpretations are theoretically compatible with the recognition of these specific P. alpicola forms as a species. Obviously, the lineage consists of individuals characterised by a coherent combination of molecular and karyological characters. Furthermore, the origin of the pentaploid lineage a and the hexaploid lineage c may be explained by backcrosses with P. pusilla and P. argentea as suggested by intermediate position in the EUKLID (Figure 3). Hence, we consider these individuals as products of introgression of P. alpicola into P. pusilla and hexaploid P. argentea, respectively, or vice versa. Such rare sexual events have been documented by Holm and Ghatnekar for hexaploid apomictic P. argentea[51].
Taxonomic comments
A final decision on the taxonomic status of the P. alpicola lineages depends on further studies of its constancy through the reproductive process and comparative autecological studies. Clonality observed within each population of P. alpicola (except Pop86) assumably can be attributed to the apomictic mode of reproduction as already observed in other taxa of the P. collina group [21,22], hexaploid P. argentea[51], and high polyploid P. pusilla (Dobeš et al. unpublished research). Autogamy associated with homozygosity may alternatively explain the pattern, but seems unlikely for the pentaploids at least as anorthoploids cytotype should not be maintained by sexuality in Potentilla. In any case, the observed levels of clonality suggest stable inheritance of the hybrid forms.
The lack of unique AFLP fragments and its limited geographic distribution, suggest a recent origin of P. alpicola. Although a coherent evolutionary lineage may be recognised among the studied P. alpicola-forms and accepted as a taxonomic unit following the cohesion species model [56], the widespread existence of individuals formed by backcrosses with the parents, strongly complicates the species limits. A solution to the problem may be achieved by additional efforts to complete the sampling and by the molecular approach followed here. Nevertheless, this aim is hampered by conservation issues resulting from a serious decline of populations. In Switzerland, P. alpicola is critically endangered and recently known from two or three localities [26], including the locus classicus in the Wallis [16].
Conclusions
Combined analysis of AFLP, cpDNA sequences and ploidy levels suggested a hybrid origin of P. alpicola and P. collina populations in the South Tyrol. Diploid and hexaploid P. argentea and P. pusilla have been identified as parental taxa in different hybridisation giving rise to three lineages of P. alpicola. In contrast, P. collina populations have been regarded rather as recent derivatives of hexaploid P. argentea. Although the identified lineages of P. alpicola had different evolutionary origins, individuals from three geographically dispersed populations could possibly carry one name.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JP carried out the molecular work, flow cytometric analyses, statistical analyses, and drafted the manuscript. AS carried out flow cytometric analyses and the chromosome counting. CD designed the study, provided the samples, carried out flow cytometric analyses and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of studied accessions and experiments. AFLP – amplified fragment length polymorphism, FCM – flow cytometry (cc – chromosomes counted, p – presumed ploidy level based on the genetic data 1, - – aneuploidy). Samples are ordered according to broader localities (LOC_1 – LOC_6), country codes follows ISO 3166-1 Alpha-3.
Contributor Information
Juraj Paule, Email: jpaule@senckenberg.de.
Antonia Scherbantin, Email: antoniascherbantin@hotmail.com.
Christoph Dobeš, Email: christoph.dobes@univie.ac.at.
Acknowledgements
We are grateful to DFG for funding (DO792/1-1 held by Ch.D). Further thanks go to Marcus A. Koch for supervising J.P.’s PhD studies at the former Heidelberg Institute for Plant Science (HIP), now COS Heidelberg; the Botanic Garden Heidelberg, in particular to Torsten Jakob, for cultivation and taking care of the living plant collection, Johannes Saukel (Department of Pharmacognosy of the University of Vienna) for performing the Euklid analyses and Markus Kiefer (COS Heidelberg) for lab and computer advice. We also express our gratitude to Thomas Wilhalm (Bolzano) for giving directions to several localities.
References
- Lotsy JP. Evolution by means of hybridization. M. Nijhoff, The Hague; 1916. [Google Scholar]
- Anderson E. Introgressive Hybridization. John Wiley & Sons, New York; 1949. [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annual Rev Pl Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Kihara H, Ono T. Chromsomenzahlen und systematische Gruppierung der Rumex-Arten. Z Zellf Mikroskop Anat. 1926;4:475–481. [Google Scholar]
- Savidan Y. Apomixis: genetics and breeding. Plant Breed Rev. 2000;18:13–86. [Google Scholar]
- Krašan F. Ueber einige Kulturversuche mit Potentilla verna und cinerea. Österr Bot Z. 1867;17:273–276. [Google Scholar]
- Čelakovský L. Ueber Potentilla Lindackeri Tausch und Potentilla radiata Lehm. Österr Bot Z. 1889;39:201–205. [Google Scholar]
- Gentscheff GJ. Über die pseudogame Fortpflanzung bei Potentilla. Genetica. 1938;20:398–408. [Google Scholar]
- Gustafsson Å. Apomixis in higher plants. Part III. Biotyp und species formation. Lunds Univ Årsskr NF Avd 2. 1947;43:183–370. [Google Scholar]
- Hunziker R. Beitrag zur Aposporie und ihrer Genetik bei Potentilla. Arch Julius Klaus Stift Vererbungsforsch Sozialanthropol Rassenhyg. 1954;29:135–222. [Google Scholar]
- Müntzing A, Müntzing G. Some new results concerning apomixis, sexuality and polymorphism in Potentilla. Bot Notiser. 1941;94:237–278. [Google Scholar]
- Ehrendorfer F. Mediterran-mitteleuropäische Florenbeziehungen im Lichte cytotaxonomischer Befunde. Feddes Rep. 1970;81:3–32. [Google Scholar]
- Dobeš C. Die Karyogeographie des Potentilla verna agg. (Rosaceae) in Österreich – mit ergänzenden Angaben aus Slowenien, Kroatien, der Slowakei und Tschechien. Ann Naturhist Mus Wien. 1999;101B:599–629. [Google Scholar]
- Wolf T. Monographie der Gattung Potentilla. Biblioth Bot. 1908. pp. 1–715.
- Kurtto A, Lampinen R, Junikka L, editor. Atlas Florae Europaeae 13. Distribution of vascular plants in Europe. Rosaceae (Spiraea to Fragaria, excl. Rubus) The Commitee for Mapping the Flora of Europe & Societas Biologica Fennica Vanamo, Helsinki; 2004. [Google Scholar]
- Gregor T. Typisierungen in der Potentilla collina-Gruppe (Potentilla subgrex Collinae Th. Wolf). 1. Teil: Sippen ohne Zackenhaare. Kochia. 2008;3:61–73. [Google Scholar]
- Müntzing A. Heteroploidy and polymorphism in some apomictic species of Potentilla. Hereditas. 1958;44:280–329. [Google Scholar]
- Asker S, Fröst S. The “Potentilla collina problem” – a chemotaxonomic approach. Hereditas. 1970;66:49–70. [Google Scholar]
- Gregor T, Rollik J, Weising K. RAPD-Untersuchungen und Chromosomenzählungen in der Potentilla-collina-Gruppe (Rosaceae) Ber Bayer Bot Ges. 2002;72:159–167. [Google Scholar]
- Müntzing A. Pseudogamie in der Gattung Potentilla. Hereditas. 1928. pp. 267–283.
- Gentscheff GJ, Gustafsson Å. Parthenogenesis and pseudogamy in Potentilla. Bot Notiser. 1940;1940:109–132. [Google Scholar]
- Håkansson A. Untersuchungen über die Embryologie einiger Potentilla-Formen. Lunds Univ Årssk NF Avd 2. 1946;42:1–70. [Google Scholar]
- Müntzing A, Müntzing G. Spontaneous changes in chromosome number in apomictic Potentilla collina. Hereditas. 1943;29:451–460. [Google Scholar]
- Rutishauser A. Pseudogamie und Polymorphie in der Gattung Potentilla. Arch Julius Klaus Stift Vererbungsforsch Sozialanthropol Rassenhyg. 1948;23:247–424. [Google Scholar]
- Asker S. The occurrence of aberrants in some apomictic Potentilla argentea biotypes. Hereditas. 1966;56:54–70. [Google Scholar]
- Käsermann C, Moser DM. Merkblätter Artenschutz – Blütenpflanzen und Farne. Bundesamt für Umwelt, Wald und Landschaft (BUWAL), Bern; 1999. [Google Scholar]
- Ehrendorfer F, editor. Liste der Gefäßpflanzen Mitteleuropas. Gustav Fischer, Stuttgart; 1973. [Google Scholar]
- Auer M, Fees M, Neubauer S, Over M, Zipf A. A Workflow for Processing a Hillshade WMS-Layer for entire Europe based on SRTM. Poster at AGIT, Salzburg; 2009. the case of www.osm-wms.de. [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murín A. Substitution of cellophane for glass covers to facilitate preparation of permanent squashes and smears. Stain Technol. 1960;35:351–353. [PubMed] [Google Scholar]
- Paule J, Sharbel TF, Dobeš C. Apomictic and sexual lineages of the Potentilla argentea L. group (Rosaceae) – cytotype and molecular genetic differentiation. Taxon. 2011;60:721–732. [Google Scholar]
- Scherbantin A. Methodenentwicklung zur durchflusszytometrischen Bestimmung der Chromosomenzahlen in der GattungPotentilla(Rosaceae). Diploma thesis. Department of Pharmacognosy, University of Vienna; 2009. [Google Scholar]
- Dobeš C, Paule J. A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol Phylogenet Evol. 2010;56:156–175. doi: 10.1016/j.ympev.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small R. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Ehrich D. AFLPdat: a collection of R functions for convenient handling of AFLP data. Mol Ecol Notes. 2006;6:603–604. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. , Vienna, Austria; 2009. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press, New York; 1987. [Google Scholar]
- Meirmans PG, van Tienderen PH. Genotype and Genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Saukel J, Anchev M, Guo Y-P, Vitkova A, Nedelcheva A, Goranova V, Konakchiev A, Lambrou M, Nejati S, Rauchensteiner F, Ehrendorfer F. Comments on the biosystematics of Achillea (Asteraceae-Anthemideae) in Bulgaria. Phytol Balcan. 2003;9:361–400. [Google Scholar]
- Hartung J, Elpelt B. Multivariate Statistik. Lehr- und Handbuch der angewandten Statistik. R. Oldenbourg, München & Wien; 1986. [Google Scholar]
- Scherbantin A, Paule J, Dobeš C. Karyogeography and hybrid origin of European Potentilla species (Rosaceae) Sci Pharm. 2009;77:s264. [Google Scholar]
- Dobeš C, Vitek E, editor. Documented Chromosome Number Checklist of Austrian Vascular Plants. Verlag des Naturhistorischen Museums Wien, Vienna; 2000. [Google Scholar]
- Mogensen HL. The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot. 1996;83:383–404. [Google Scholar]
- Dong YZ, Liu ZL, Shan XH, Qiu T, He MY, Liu B. Allopolyploidy in wheat induces rapid and heritable alterations in DNA methylation patterns of cellular genes and mobile elements. Russ J Genet. 2005;41:890–896. [PubMed] [Google Scholar]
- Holm S, Ghatnekar L. Apomixis and sexuality in hexaploid Potentilla argentea. Hereditas. 1996;125:53–60. [Google Scholar]
- Holm S, Ghatnekar L, Bengtsson BO. Selfing and outcrossing but no apomixis in two natural populations of diploid Potentilla argentea. J Evol Biol. 1997;10:343–352. [Google Scholar]
- Asker S. Apomixis and sexuality in the Potentilla argentea complex. I. Crosses with other species. Hereditas. 1970;66:127–144. [Google Scholar]
- Asker S. Polymorphism of Potentilla tabernaemontani and related taxa on Gotland. Hereditas. 1985;102:39–45. [Google Scholar]
- Rutishauser A. Konstante Art-und Rassenbastarde in der Gattung Potentilla. Mitt der Naturf Ges Schaffhausen. 1943;18:111–134. [Google Scholar]
- Hörandl E. Species concepts in agamic complexes: applications in the Ranunculus auricomus complex and general perspectives. Folia Geobot. 1998;33:335–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of studied accessions and experiments. AFLP – amplified fragment length polymorphism, FCM – flow cytometry (cc – chromosomes counted, p – presumed ploidy level based on the genetic data 1, - – aneuploidy). Samples are ordered according to broader localities (LOC_1 – LOC_6), country codes follows ISO 3166-1 Alpha-3.