Abstract
Background
This study was performed to develop a functional poly(D,L-lactide-co-glycolide)- poly(ethylene glycol) (PLGA-PEG)-bearing amino-active end group for peptide conjugation.
Methods and results
PLGA was preactivated following by copolymerization with PEG diamine. The resulting amphiphilic PLGA-PEG copolymer bearing 97.0% of amino end groups had a critical micelle concentration of 3.0 × 10−8 mol/L, and the half-effective inhibition concentration (IC50) of the prepared PLGA-PEG nanoparticles was >100 mg/mL, which was much higher than that of PLGA nanoparticles (1.02 ± 0.37 mg/mL). The amphiphilic properties of PLGA-PEG spontaneously formed a core-shell conformation in the aqueous environment, and this special feature provided the amino group on the PEG chain scattered on the surface of PLGA-PEG nanoparticles for efficient peptide conjugation. The peptide-conjugated PLGA-PEG nanoparticles showed three-fold higher uptake than peptide-free PLGA-PEG nanoparticles in a SKOV3 cell line with high expression of epidermal growth factor receptor. Both peptide-conjugated and peptide-free PLGA-PEG nanoparticles were used as nanocarriers for delivery of doxorubicin. Although the rate of release of doxorubicin from both nanoparticles was similar, drug release at pH 4.0 (500 U lipase) was faster than at pH 7.4. The IC50 of doxorubicin-loaded peptide-conjugated PLGA-PEG nanoparticles in SKOV3 cells (0.05 ± 0.03 μg/mL) was much lower (by 62.4-fold) than that of peptide-free PLGA-PEG nanoparticles (3.12 ± 1.44 μg/mL).
Conclusion
This in vivo biodistribution study in SKOV3 tumor-bearing mice was further promising in that accumulation of doxorubicin in tumor tissue was in the order of peptide-conjugated PLGA-PEG nanoparticles > peptide-free PLGA-PEG nanoparticles > doxorubicin solution.
Keywords: amphiphilic copolymer, peptide, nanoparticles, SKOV3 cell, doxorubicin
Introduction
Poly(D,L-lactic-co-glycolic acid) (PLGA) copolymers are widely used in drug delivery because of their good biocompatibility, ability to biodegrade into natural metabolites, and safety for human use.1,2 In recent decades, PLGA copolymers have been used in various medical devices and pharmaceutical dosage forms, including sutures, microparticles, nanoparticles, pellets, and implants for different types of drugs. They were approved by the US Food and Drug Administration for use in drug delivery systems, such as Lupron Depot®.3 The safety of PLGA in clinical use has been well established.4–7
Like colloidal drug delivery systems, when PLGA nanoparticles are intravenously administered, they are quickly eliminated from the systemic circulation by macrophages in the spleen and the mononuclear phagocyte system in Kupffer cells of the liver.8 Such rapid elimination limits the use of PLGA nanoparticles as drug delivery and targeting carriers. Thus, it is necessary to modify the PLGA nanoparticle surface so that it can escape uptake by the mononuclear phagocyte system and have a prolonged circulation time in the blood.9 This can be successfully achieved in PLGA nanoparticles when their surface is modified by hydrophilic poly(ethylene glycol) (PEG). PEG-coated nanoparticles avoid recognition by the mononuclear phagocyte system and elimination due to the shielding effect of the polymer chains in PEG.10
The biodistribution of nanoparticles is controlled mainly by their physical and biochemical properties, including particle size, the nature of the polymer and drug, and surface properties.11–14 The advantage of using nanoparticles in cancer therapy stems from the enhanced permeation and retention effect.15,16 Nanoparticles are passively targeting and accumulate in tumors via the enhanced permeation and retention effect after systemic administration.17–19 Drug concentration increases at the tumor site to improve their therapeutic efficacy and reduce nonspecific side effects.20,21 Moreover, polymeric nanoparticles have been used as an active targeting delivery system to conjugate a ligand for targeting to specific cancer cells.22–25 These ligands can be small molecules, proteins, antibodies, or peptides which bind specifically to cancer cells and provide an efficient and safe nanocarrier for drug delivery.26–29 Proteins and antibodies have a large molecular weight and bind to receptors with a steric structure and with high affinity. However, foreign antibodies are produced with biological methods, exhibit variations from batch to batch, and easily induce an unexpected immune response. The peptides are smaller than antibodies and can bind to receptors specifically, and rarely induce an immune response. Peptides are chemically synthesized on a large scale and the quality is more stable from batch to batch. Thus, using peptides as ligands can overcome the limitations of antibodies and enable them to retain their active targeting ability. The epidermal growth factor (EGF) receptor is highly expressed in human epithelial cancer cells, including breast, ovarian, and lung cancer.30–32 The EGF receptor is recognized as an important target for the development of treatment for cancer. Several EGF-targeting therapeutic agents, including cetuximab (Erbitux®) and erlotinib (Tarceva®), have already been approved by the US Food and Drug Administration.33 Kolonin et al identified 17 peptides that mimic EGF receptor ligands, including amphiregulin, heparin-binding EGF-like growth factor, and epiregulin.34 We further selected the NR7 peptide (NSVRGSR) based on alignment of the tripeptide motif with the EGF binding domain, and the NR7 peptide was the only peptide which not only appeared twice between these EGF receptor ligands but also contained tripeptide motifs relative to the mature EGF domain among these 17 peptides.35
The purpose of this study was to apply the core-shell conformation of the amphiphilic PLGA-PEG copolymer bearing amino-active end groups for peptide conjugation. The amphiphilic PLGA-PEG copolymer was synthesized by copolymerization of the preactivated PLGA-NHS with PEG diamine. The physicochemical properties of the resulting amphiphilic PLGA-PEG copolymer bearing amino end groups were characterized by 1H-NMR, and the yield, PEGylation efficiency, molecular weight, and critical micelle concentration were determined. PLGA-PEG nanoparticles were prepared using the solvent evaporation method, and their size, zeta potential, morphology, and cell viability were characterized. The NR7 peptide was selected as a peptide ligand to conjugate with our synthesized amphiphilic PLGA-PEG copolymer. Two approaches were investigated to prepare the peptide-conjugated PLGA-PEG nanoparticles. One was to conjugate the peptide with the amphiphilic PLGA-PEG copolymer followed by preparation of nanoparticles. The other was to conjugate the peptide with previously prepared PLGA-PEG nanoparticles exhibiting micelle core-shell conformation. The peptide conjugation ratios and performance of the peptide-conjugated nanoparticles were determined and characterized. The cellular uptake efficiency was evaluated in SKOV3 cells expressing high levels of the EGF receptor. Doxorubicin was used as the model anticancer drug. The doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles were characterized, and their SKOV3 cell growth inhibition efficiency and in vitro release at physiological pH 7.4 and simulated lysosomal pH 4.0 (500 U lipase) were investigated. Finally, an in vivo biodistribution study was performed in SKOV3 tumor-bearing mice.
Materials and methods
Materials
PLGA 50:50 (RG 503H, molecular weight 28 kDa, [η] = 0.32–0.44 dL/g) was purchased from Boehringer Ingelheim (Ingelheim, Germany). PEG diamine (NH2-PEG-NH 2; molecular weight 5 kDa) was from Laysan Bio Inc (Arab, AL). Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and pyrene were obtained from Tokyo Chemical Industry Co, Ltd (Tokyo, Japan). The NHS was from Acros Organics (Geel, Belgium). An L929 mouse fibroblast cell line and a SKOV3 human ovarian cancer cell line were purchased from the Bioresource Collection and Research Center (Hsinchu, Taiwan). Dulbecco’s modified Eagle’s medium, McCoy’s 5A medium, and fetal bovine serum were obtained from Biological Industries (Kibbutz Beit Haemek, Israel). 5-Carboxyfluorescein and 6-carboxyfluorescein succinimidyl ester (fluorescein-NHS) and fluorescein isothiocyanate (FITC) were from Pierce (Rockford, IL). Peptide-fluorescein (NSVRGSR, peptide-FTIC, 95% purity) was synthesized by Kelowna International Scientific Inc (Taipei, Taiwan).
Synthesis of amphiphilic PLGA-PEG copolymer
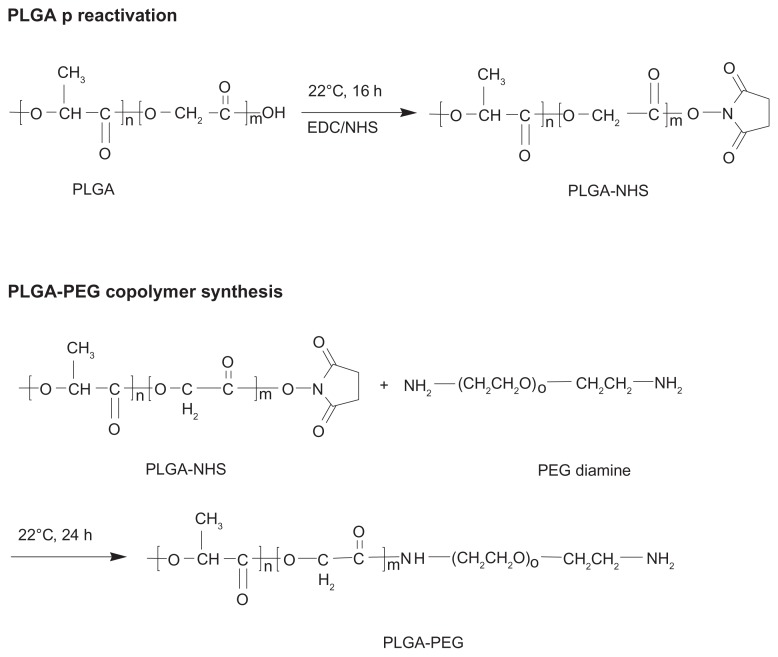
PLGA was preactivated before PEGylation with PEG diamine, and the reaction procedure is shown in Scheme 1.28 Briefly, PLGA polymer 5 g (178.5 μmol) was dissolved in 20 mL of dichloromethane. Subsequently, NHS 135 mg (1172.2 μmol) and EDC 230 mg (1198.3 μmol) were added to the PLGA solution and the mixture was reacted at 22°C for 16 hours under a nitrogen atmosphere. The preactivated PLGA-NHS was precipitated with ether and centrifuged at 4000 rpm for 5 minutes at room temperature. The residue was washed three times with an ice-cold mixture of an equal volume of ether and methanol. The PLGA-NHS obtained was dried in a desiccator under vacuum and stored at −20°C. For synthesis of PLGA-PEG, PLGA-NHS 750 mg (26.7 μmol) was dissolved in 4 mL of dichloromethane, and PEG diamine 133 mg (26.6 μmol) was added to the solution. The mixture was stirred at 22°C for 24 hours. The synthesized PLGA-PEG copolymer bearing the amino end group was precipitated with ice-cold methanol and centrifuged at 4000 rpm for 5 minutes at room temperature. The precipitate was washed three times with ice-cold methanol to remove unreacted PEG diamine. The PLGA-PEG copolymer bearing the amino end group was further purified by precipitation in cold methanol following by drying in a desiccator and storage at −20°C.
Scheme 1.
Synthesis of amphiphilic PLGA-PEG copolymer bearing amino-active end group.
Abbreviation: PLGA-PEG, poly(D,L-lactide-co-glycolide)-poly(ethylene glycol).
Characterization of PLGA-PEG amphiphilic copolymer
The yield of PLGA-PEG copolymer was calculated by equation (1).36 The PLGA-PEG copolymer was characterized by 1H-NMR (400 mHz, CDCl3) at δ 5.30–5.17 ppm for the CH of lactide, at δ 4.90–4.56 ppm for the CH2 of glycolide, at δ 3.61–3.55 ppm for the CH2 of ethylene glycol, and at δ 1.62–1.45 ppm for the CH3 of lactide. There were six and four protons in each D,L-lactide-co-glycolide and ethylene glycol monomer. The PEGylation efficiency was calculated by equation (2).37
| (1) |
| (2) |
The molecular weight of the polymer was determined by gel permeation chromatography with a refractive index detector (RI 2031 Plus, Jasco, Tokyo, Japan) and a Styragel® HR 4E column (7.8 × 300 mm, Waters, Milford, MA). The mobile phase was chloroform, the flow rate was set at 1 mL per minute, and the temperature was maintained at 35°C. Polymers were dissolved in chloroform, and filtered through a 0.2 μm syringe filter prior to injection. The molecular weight of PLGA-PEG was determined using polystyrene standards. The critical micelle concentration of PLGA-PEG copolymer-induced micelle formation was determined using a fluorescence spectrophotometer (SpectraMax Gemini XS, Mississauga, ON, Canada) and a pyrene as a fluorescence probe. The emission wavelength was set at 390 nm, and the excitation fluorescence was recorded at 336 nm and 333 nm. The critical micelle concentration value was determined from the plot of the ratio of I336/I333 versus logarithmic concentrations of PLGA-PEG copolymer. The concentration at the intersection of the horizontal and the steep parts of the curve corresponded to the critical micelle concentration value.38,39
Synthesis of PLGA-PEG-FITC copolymer
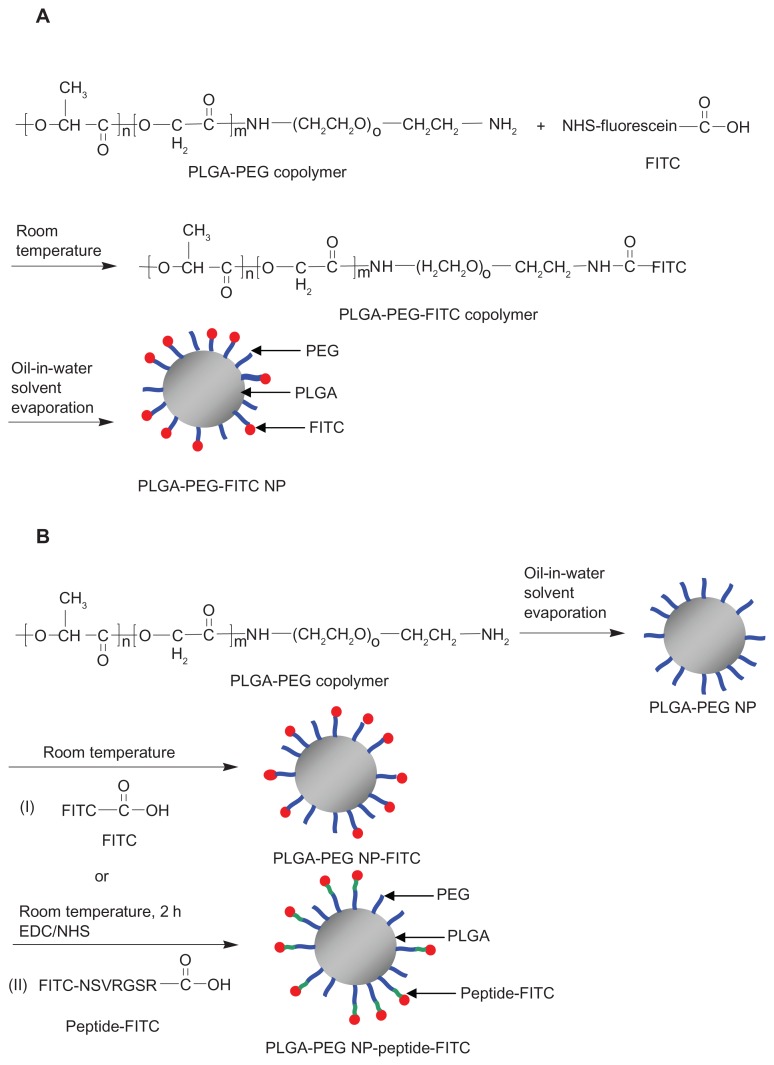
The procedure for synthesis of the PLGA-PEG-FITC copolymer is shown in Scheme 2A. PLGA-PEG 27.7 mg (2 μmol) was dissolved in 1 mL of dimethyl sulfoxide, and various amounts of FITC (1, 5, 10, 15, and 20 mg [2, 10, 20, 30, and 40 μmol]) were added into the solution separately. The mixture was stirred at room temperature in the dark for 2 hours. The molar feed ratio of PLGA-PEG: FITC 1:20 was further stirred for 4, 6, and 24 hours to investigate the effect of reaction time on conjugation of fluorescein. The PLGA-PEG-FITC synthesized was precipitated with ice-cold methanol and centrifuged at 10,000 rpm for 10 minutes at 4°C. The precipitate was collected and washed with ice-cold methanol three times followed by drying in the desiccator under vacuum. The obtained FITC-conjugated PLGA-PEG-FITC copolymers were stored at −20°C. The amount of fluorescein conjugated was determined using a fluorescence plate reader (SpectraMax Gemini XS) at an excitation wavelength of 485 nm and an emission wavelength of 538 nm. The fluorescein conjugation molar ratio was calculated by equation (3).40
Scheme 2.
Synthesis of (A) PLGA-PEG-FITC and (B) PLGA-PEG NP-FITC or PLGA-PEG NP-peptide-FITC nanoparticles.
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol); FITC, fluorescein isothiocyanate; NP, nanoparticle; NSVRGSR, NR7 peptide; EDC, ethyl- 3-(3-dimethylaminopropyl) carbodiimide hydrochloride; NHS, N-hydroxysuccinimide.
| (3) |
Preparation of nanoparticles
The nanoparticles were prepared using a solvent evaporation method. PLGA, PLGA-PEG, or PLGA-PEG-FITC 20 mg was dissolved in a 1 mL equal volume of acetone/ dichloromethane cosolvent and mixed well. The polymer solution was added into 10 mL of 0.25% polyvinyl alcohol solution and sonicated at 4°C for 2 minutes following by magnetic stirring for 3 hours. Polyvinyl alcohol was used to stabilize the emulsion in order to form particles of relatively small size and uniform size distribution.41 The remaining organic solvent was removed by rotary evaporator under reduced pressure at 35°C for 3 minutes. The nanoparticles were collected after centrifugation at 14,000 rpm at 4°C for 30 minutes and subsequently washed with deionized water three times. Finally, the nanoparticles were resuspended in 0.1 mL of deionized distilled water which contained 5 mg of sucrose as a cryoprotectant agent, and then freeze-dried and stored at −20°C. The particle size and zeta potential of nanoparticles were determined by zetasizer (Nano-ZS 90 Zetasizer, Malvern Instruments, Worcestershire, UK), and the surface morphology was observed by transmission electron microscopy (Tecnai G2, FEI Company, Hillsboro, OR) at 200 kV.
PLGA-PEG NP-FITC and PLGA-PEG NP-peptide-FITC nanoparticles
Scheme 2B(I) illustrates the conjugation of FITC with previous prepared PLGA-PEG nanoparticles to form PLGA-PEG NP-FITC. Next, 15.08 mg (1.09 μmol) of the PLGA-PEG nanoparticles were suspended in 0.624 mL of borated buffer (50 mM, pH 8.5). FITC 0.423 mg (1.09 μmol) was added into the suspension and stirred at room temperature in the dark for 2, 4, 6, and 24 hours. A similar procedure was used to prepare PLGA-PEG NP-peptide-FITC where the carboxyl group (C terminal) of the peptide-FITC was conjugated with the amino group of PLGA-PEG nanoparticles in the presence of carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), as shown in Scheme 2B(II). The details are as follows. The mixtures of PLGA-PEG nanoparticles 15.08 mg (1.09 μmol), peptide-FITC 1.39 mg (1.09 μmol), NHS 0.27 mg (2.2 μmol), and EDC 0.45 mg (2.2 μmol) were stirred at room temperature in the dark for 2 hours. The resulting PLGA-PEG NP-peptide-FITC was collected after centrifugation at 14,000 rpm for 30 minutes at 4°C and subsequently washed with deionized water three times. Finally, the nanoparticles were resuspended in 0.1 mL of deionized distilled water which contained 5 mg sucrose, and then freeze-dried and stored at −20°C. The particle size and zeta potential were measured as described previously. The amount of conjugated FITC was determined using a fluorescence plate reader at an excitation wavelength of 455 nm and an emission wavelength of 523 nm.
Cellular viability study
The cytotoxicity of PLGA and PLGA-PEG nanoparticles was studied using L929 mouse fibroblasts. L929 cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, and 0.1 mM nonessential amino acids, as well as 1.0 mM sodium pyruvate. The cells were seeded in a 96-well plate at a density of 8000 cells per well and maintained in a humidified incubator at 37°C in 5% CO2 for 24 hours. The cell viability was determined using a 3-(4,5-dimethylthiazol-2- yl)-2,5-diphenyl tetrazolium (MTT) assay.42 Serial dilutions of PLGA nanoparticles and PLGA-PEG nanoparticles were added in cultured medium into each well and incubated at 37°C for 72 hours. The medium was removed, and the cells were labeled with MTT solution and incubated at 37°C for 4 hours. The resulting formazan was solubilized in dimethyl sulfoxide, and the absorbance was measured using an enzyme-linked immunosorbent assay reader (Power Wave XS, BioTek, Winooski, VT) at 570 nm. The IC50 was calculated from the plot of cell viability versus nanoparticle concentration using SigmaPlot 10.0 software.43
Cellular uptake study
SKOV3 cells were seeded into 6-well plates at a density of 5 × 105 cells per well and cultured in 2 mL of complete McCoy’s 5A medium (McCoy’s 5A medium with 10% fetal bovine serum, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM nonessential amino acids, and 1.0 mM sodium pyruvate) at 37°C for 24 hours. The cells were then washed three times with phosphate-buffered saline, and incubated with several different fluorescein concentrations of peptide-free and peptide-conjugated PLGA-PEG nanoparticles at 37°C for 30 minutes. The fluorescein concentrations used for the peptide-conjugated PLGA-PEG nanoparticles were 0.27, 0.82, 2.47, 7.4, and 22.2 μM, and for peptide-free PLGA-PEG nanoparticles were 0.31, 0.94, 2.81, 8.43, and 25.3 μM. The nanoparticles were removed and the cells were washed with phosphate-buffered saline three times. The cells were harvested by trypsinization and centrifuged at 1400 rpm for 5 minutes. Finally, the cells were resuspended in phosphate-buffered saline, and cellular uptake of the fluorescein- conjugated nanoparticles was measured at an excitation wavelength of 488 nm and an emission wavelength of 530 nm (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ) and analyzed using BD CellQuest Pro software (version 6, Becton Dickinson). A total of 10,000 events were analyzed for each sample. The upper limit of background fluorescence was set to no more than 1% of the events with the autofluorescence controls (attributable to native cells).
Preparation and characterization of doxorubicin-loaded nanoparticles
Doxorubicin HCl (5.3 mg, 9.1 μmol) was dissolved in 5 mL of water, and triethylamine (3.1 mg, 30.6 μmol) was added to the solution. The mixture was stirred for 24 hours at room temperature under nitrogen. Doxorubicin was extracted three times with dichloromethane. The organic layer was collected and evaporated under reduced pressure. The doxorubicin obtained was dried in a desiccator under vacuum. Doxorubicin 0.5 mg and copolymer 20 mg was dissolved in 1 mL of acetone/dichloromethane (1/1, v/v). This solution was slowly added to 10 mL of 0.25% polyvinyl alcohol solution under sonication in an ice bath for 5 minutes following stirring at room temperature for 3 hours. The remaining organic solvent was removed by rotary evaporator under reduced pressure at 35°C for 10 minutes. The nanoparticles were collected after centrifugation at 14,000 rpm at 4°C for 30 minutes. Finally, the nanoparticles were resuspended in 0.1 mL of deionized distilled water containing 5 mg sucrose as a cryoprotectant agent, then freeze-dried and stored at −20°C. The yield, particle size, and zeta potential of the doxorubicin-loaded nanoparticles were measured. The amount of doxorubicin loaded into the nanoparticles in terms of drug loading (equation [4]) and encapsulation efficiency (equation [5]) were determined by dissolving 0.5 mg nanoparticles in 1 mL of acetone and measurement by fluorescence spectrophotometer (F-4500, Hitachi, Tokyo, Japan) at an excitation wavelength of 503 nm and an emission wavelength of 550 nm.44
| (4) |
| (5) |
In vitro release study
Doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles (5 mg) were resuspended in 1 mL of phosphate-buffered saline (pH 7.4, 0.01% sodium azide, 0.5% Tween 80) or 1 mL of acetic-buffered solution (pH 4.0, 500 U lipase, 0.01% sodium azide, 0.5% Tween 80) and then added into the dialysis bag (molecular weight cutoff 6000–8000 Da). The dialysis bag was placed in a 200 mL bottle containing 100 mL phosphate-buffered or acetic-buffered solution and stirred at 100 rpm and 37°C. Next, 1 mL of medium was withdrawn at each specific time interval and the same volume of fresh release medium was replaced. The fluorescence intensity of the doxorubicin released was measured at an excitation wavelength of 496 nm and an emission wavelength of 556 nm.
Cytotoxicity study
SKOV3 cells were seeded into a 96-well plate at a density of 9000 cells per well for 24 hours. Serial dilutions of doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles in 200 μL of McCoy5A medium were added into each well and incubated at 37°C for 72 hours in a humidified incubator containing 5% CO2. The medium was removed and the cells were washed three times with phosphate-buffered saline. The cells were labeled with MTT solution (0.5 mg/mL) at 37°C for 4 hours in a humidified incubator containing 5% CO2. The resulting formazan was solubilized in 200 μL of dimethyl sulfoxide and the absorbance was measured using an enzyme-linked immunosorbent assay reader (Power Wave XS) at 570 nm. The IC50 was calculated from the plot of cell viability versus doxorubicin concentration using SigmaPlot 10.0 software.43
Biodistribution of doxorubicin in SKOV3 tumor-bearing mice
Athymic mice (nude, female, 7–8 weeks old, 20–23 g) were maintained in an aseptic environment. The tumors were implanted by injecting 5 × 106 SKOV3 cells (in 50 μL McCoy5A medium and 50 μL Matrigel) into the subcutaneous region of each mouse. At predetermined time points, the major and minor axes of the tumors were measured using a caliper. The tumor volume was calculated using the formula a2b/2, where a and b are the lengths of the minor and major axes of the tumor, respectively.45,46 All animals were accommodated in a pathogen-free laboratory environment throughout the experiments. After implantation, the tumors were allowed to grow for 21 days, and the mean tumor volume was about 100 mm3. The animals received a single intravenous injection of free doxorubicin, doxorubicin-loaded peptide-free, or peptide-conjugated PLGA-PEG nanoparticles at an equivalent doxorubicin dose of 5 mg/kg. After 24 hours, blood samples were collected, the animals were sacrificed, and the brains, hearts, livers, kidneys, and tumor tissue were removed without perfusion. The organs were freeze-dried and weighed. Lyophilized tissue was homogenized in sodium phosphate buffer (pH 7.4) in an ice bath, and then extracted using a mixture of chloroform and isopropanol (3:1, v/v). The organic layer was collected after centrifugation, then completely dried under vacuum and redissolved in acetone. The solution was determined with a fluorescence detector at an excitation wavelength of 503 nm and an emission wavelength of 546 nm.
Results and discussion
Characterization of PLGA-PEG copolymer
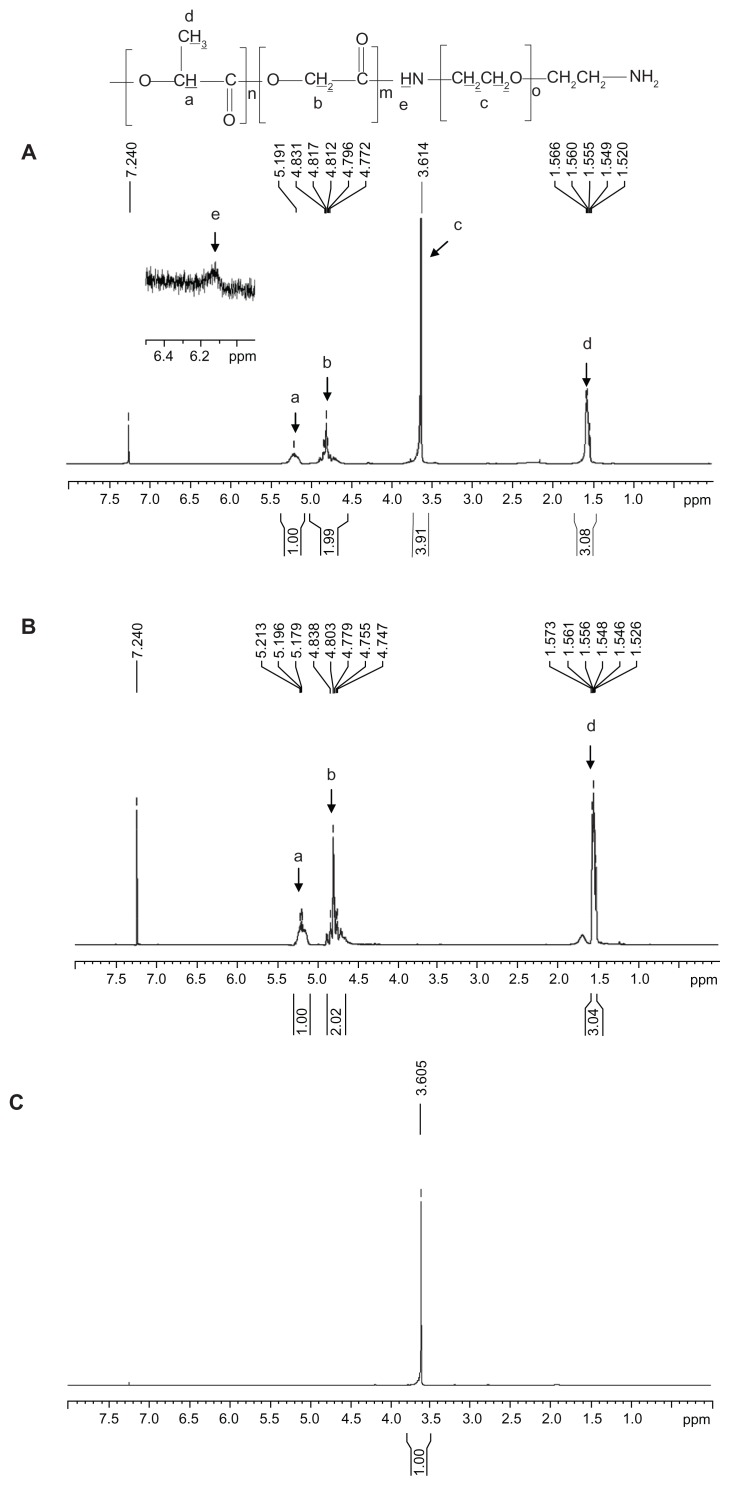
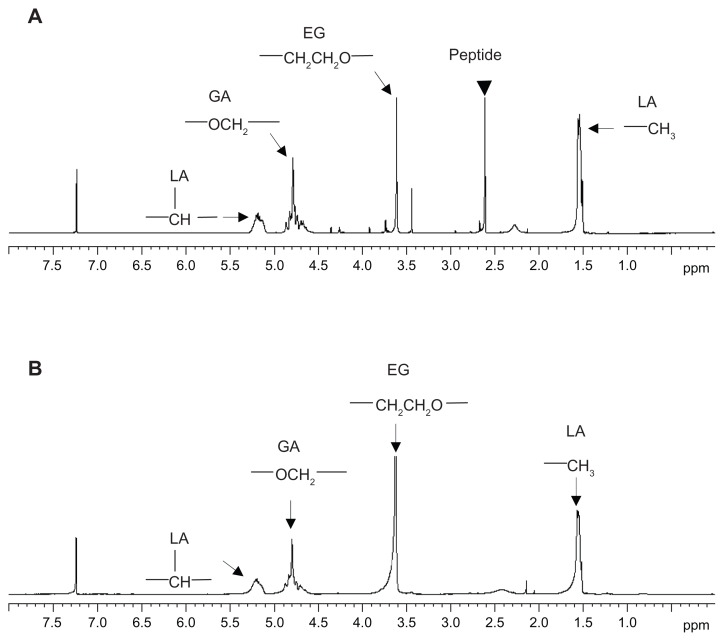
Figure 1 shows the 1H-NMR spectra for PLGA-PEG synthesized at a molar feed ratio of PLGA-NHS:PEG diamine 1:1. Four peaks were present, including peak a at δ 5.30–5.17 ppm for the CH proton of lactide, peak b at δ 4.90–4.56 ppm for the CH2 proton of glycolide, peak c at δ 3.61–3.55 ppm for the CH2 proton of ethylene glycol, and peak d at δ 1.62–1.45 ppm for the CH3 proton of lactide. There was a small peak at δ 6.10–6.20 ppm, indicating the presence of an amide bond between PLGA and PEG (Figure 1A inset).47 All of these lines of evidence confirmed the presence of both PLGA and PEG domains in the PLGA-PEG copolymer. The synthesis yield of PLGA-PEG was 94.1%, and the PEGylation efficiency was 97.0% in terms of 10.3% w/w PEG in the synthesized PLGA-PEG copolymer. The weight-average molecular weight (Mw), number-average molecular weight (Mn), and polydispersity of PLGA-PEG were 32.6 kDa, 17.6 kDa, and 1.8, respectively. The critical micelle concentration of the amphiphilic PLGA-PEG copolymer was determined using pyrene as a fluorescence probe. There were no micelles formed at a concentration below the critical micelle concentration. Above the critical micelle concentration, the fluorescence intensity abruptly increased and the slope markedly increased. The critical micelle concentration for PLGA-PEG was determined to be 3.0 × 10−8 mol/L. Lee et al demonstrated that PLA-PEG micelles with a critical micelle concentration of 8.1 × 10−7 mol/L had good stability in the bloodstream.48 The critical micelle concentration for our synthesized PLGA-PEG copolymer (3.0 × 10−8 mol/L) was lower than that of the reported value (8.1 × 10−7 mol/L). Our PLGA-PEG micelles had the potential to maintain their stable core-shell conformation in the circulation.
Figure 1.
1H-NMR spectra of (A) PLGA-PEG, (B) PLGA, and (C) PEG diamine.
Abbreviations: PEG, poly(ethylene glycol); PLGA, poly(D,L-lactide-co-glycolide).
Characterization of PLGA-PEG nanoparticles
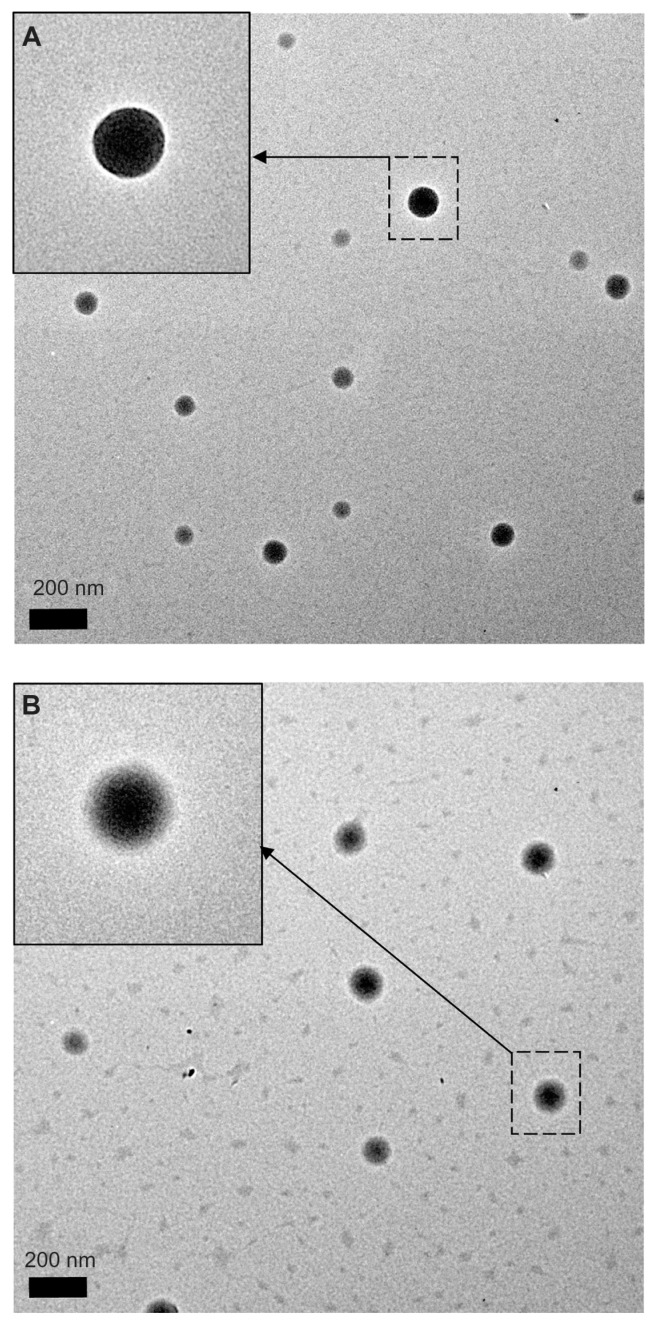
PLGA-PEG nanoparticles were prepared using the solvent evaporation method. The average size of the PLGA and PLGA-PEG nanoparticles was 105.3 ± 2.61 nm and 134.6 ± 5.57 nm, and the polydispersities were 0.023 ± 0.017 and 0.103 ± 0.085, indicating a narrow size distribution. The zeta potential of the PLGA and PLGA-PEG nanoparticles was −4.4 ± 0.85 mV and −13.3 ± 0.80 mV, respectively. Both nanoparticles were negatively charged, and thus able to avoid nonspecific interaction with the cell membrane by electrostatic interaction. Figure 2 shows the morphology of PLGA and PLGA-PEG nanoparticles observed by transmission electron microscopy. All of the nanoparticles had a spherical shape and uniform size without aggregation. The PLGA nanoparticles showed a clear and sharp margin around the surface (Figure 2A inset). However, the PLGA-PEG nanoparticles showed a rigid PLGA core with a fuzzy appearance around the surface (Figure 2B inset). The shielding effect of the PEG polymer chains around the PLGA-PEG nanoparticle surface accounted for this appearance. Covering of nanoparticle surfaces with hydrophilic PEG has been shown to prolong the circulation time of nanoparticles in vivo. The hydrophilic PEG polymer chains sterically stabilize nanoparticles against opsonization and subsequent phagocytosis. Kim et al reported that nanoparticles with a particle size less than 200 nm are preferred in a long circulating drug delivery system because they can evade uptake by the reticuloendothelial system in the spleen.49 Ioele et al demonstrated that extension of PEG polymer chains on the nanoparticle surface acts as a steric barrier to reduce clearance of the nanoparticles by macrophages in the liver and promote the enhanced permeation and retention effect.50 van Vlerken et al also found that PLGA nanoparticles surface-modified with PEG avoid uptake by the reticuloendothelial system, thereby improving the nanoparticle circulation time, and the nanoparticles are retained in the tumor tissue for prolonged periods of time.51 All of these observations supported our PEGylated PLGA nanoparticles, with a size less than 200 nm, having potential as a nanocarrier with long-circulating properties.
Figure 2.
Transmission electron microscopic images of (A) PLGA nanoparticles and (B) PLGA-PEG nanoparticles with magnification 9600×.
Note: The insets indicated by arrows have a magnification 19,000×.
Abbreviations: PEG, poly(ethylene glycol); PLGA, poly(D,L-lactide-co-glycolide).
Cytotoxicity of PLGA-PEG nanoparticles
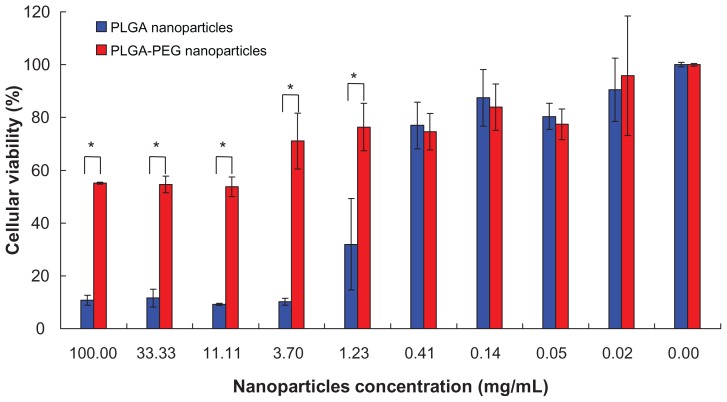
The cytotoxicity of the PLGA-PEG nanoparticles was evaluated using a L929 cell line of mouse connective tissue fibroblasts, as recommended by the American Society for Testing and Materials (ASTM F895, 2003).52 Figure 3 depicts cell viability with PLGA and the PLGA-PEG nanoparticles. The corresponding values at nanoparticle concentrations lower than 0.41 mg/mL were similar for these two types of nanoparticles and more than 75% of the cells were viable. However, cell viability was markedly decreased by PLGA nanoparticle concentrations higher than 1.23 mg/mL. Nevertheless, this did not occur for PLGA-PEG nanoparticles, even at a high concentration of 100 mg/mL, and 55.2% ± 0.4% of the cells were still viable. The calculated IC50 of the PLGA and PLGA-PEG nanoparticles was 1.02 ± 0.37 mg/mL and > 100 mg/mL, respectively. In other words, cell viability with PLGA-PEG nanoparticles was improved by more than 98-fold after PEGylation of 10.3% w/w. The shielding effect of PEG resulted in nanoparticles with better cell viability. The significantly higher IC50 of PLGA-PEG nanoparticles compared with PLGA nanoparticles indicated that the cytotoxicity of the nanoparticles was reduced and their biocompatibility was improved.
Figure 3.
L929 cellular viability of (
 ) PLGA and (
) PLGA and (
 ) PLGA-PEG nanoparticles (n = 3).
) PLGA-PEG nanoparticles (n = 3).
Notes: Percentage viability of L929 cell line was analyzed by MTT assay. The values are represented as the mean ± standard deviation, n = 3. Error bars indicate the standard deviation. *P < 0.05 by Student’s t-test.
Abbreviations: PEG, poly(ethylene glycol); PLGA, poly(D,L-lactide-co-glycolide).
Fluorescein conjugation of PLGA-PEG- FITC and PLGA-PEG NP-FITC
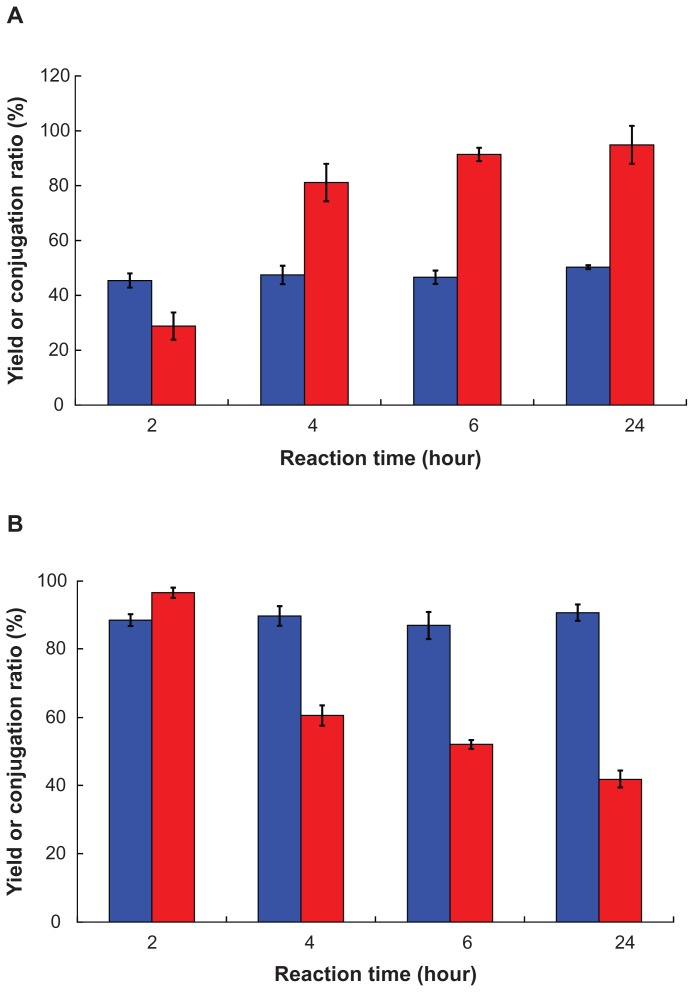
FITC-conjugated PLGA-PEG was prepared by two different methods. The first method involved conjugation of FITC with PLGA-PEG copolymer following nanoparticle preparation (Scheme 2A). The second method involved conjugation of FITC with previously prepared PLGA-PEG nanoparticles (Scheme 2B). With the first method, as the molar feed ratio of PLGA-PEG:FITC increased from 1:1 to 1:20 after reaction for 2 hours, FITC conjugation ratio of PLGA-PEG-FITC increased from 0.6 ± 0.3 mol% to 28.8 ± 4.9 mol%, but the yield decreased from 71.8% ± 2.1% to 42.5% ± 1.3%. This result shows that the FITC conjugation ratio was significantly improved by increasing the molar feed ratio of FITC. With the second method, the much higher FITC conjugation ratio and yield of PLGA-PEG NP=FITC were achieved as the molar feed ratio of PLGA-PEG NP: FITC 1:1 reaction for 2 hours, where the corresponding values were 96.5 ± 1.5 mol% and 88.5% ± 1.8%. The reaction time was further investigated at 2–24 hours and a molar feed ratio of PLGA-PEG:FITC 1:20 and PLGA-PEG NP:FITC 1:1, and the result is shown in Figure 4. The yields of PLGA-PEG-FITC copolymer were between 45.41% ± 2.57% and 50.28% ± 0.70% which increased slightly as the reaction time was increased from 2 to 24 hours (Figure 4A). In contrast, the FITC conjugation ratio was markedly increased from 28.80 ± 4.94 mol% to 81.13 ± 6.84 mol% as the reaction time increased from 2 to 4 hours. A further increase in reaction time to 6 hours and 24 hours only increased the conjugation ratios slightly to 91.37 ± 2.43 mol% and 94.87 ± 6.89 mol%, respectively. This result indicates that the 6-hour reaction period was long enough to achieve 90 mol% conjugation of FITC with PLGA-PEG- FITC. The effect of reaction time (2–24 hours) on the FITC conjugation molar ratio and yield of PLGA-PEG NP-FITC was investigated at a PLGA-PEG nanoparticle molar feed ratio of FITC 1:1, and the result is shown in Figure 4B. There was no prominent change in yield of PLGA-PEG NP-FITC within 24 hours of reaction time, and the corresponding values were in the range of 86.9% ± 4.0% and 90.6% ± 2.4%. However, the FITC conjugation ratio was markedly decreased from 96.5 ± 1.5 mol% to 41.9 ± 2.5 mol% when the reaction time was increased from 2 to 24 hours. All of these results indicate that conjugation of FITC with PLGA-PEG was preferred after formation of the micellar core-shell conformation. The amphiphilic properties of PLGA-PEG formed a core-shell conformation in the aqueous environment during the conjugation process, whereby the hydrophilic PEG chains extended outside of PLGA-PEG nanoparticles. This special feature enabled the amino group at the end of the PEG chain surrounding the surface of the PLGA-PEG nanoparticles to react efficiently with FITC. This resulted in a FITC conjugation ratio of PLGA-PEG NP-FITC that was higher than PLGA-PEG-FITC.
Figure 4.
Effect of reaction time on (
 ) yield and (
) yield and (
 ) FITC conjugation ratio of (A) PLGA-PEG-FITC reacted at a molar feed ratio of PLGA-PEG:FITC 1:20, and (B) PLGA-PEG NP-FITC reacted at a molar feed ratio of PLGA-PEG NP:FITC 1:1.
) FITC conjugation ratio of (A) PLGA-PEG-FITC reacted at a molar feed ratio of PLGA-PEG:FITC 1:20, and (B) PLGA-PEG NP-FITC reacted at a molar feed ratio of PLGA-PEG NP:FITC 1:1.
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol); FITC, fluorescein isothiocyanate; NP, nanoparticle.
Characterization of PLGA-PEG NP-FITC and PLGA-PEG NP-peptide-FITC
A peptide which mimicked the EGF receptor was used in this study as a ligand. The terminal amino group of PLGA-PEG was further conjugated with the carboxyl group of peptide-FITC as depicted in Scheme 2B(II), whereby the molar feed ratio of PLGA-PEG NP-peptide-FITC was 1:1 and the reaction time was 2 hours. The conjugation ratio and yield of the resulting PLGA-PEG NP-peptide-FITC were 71.1 ± 4.8 mol% and 89.6 ± 6.3%, respectively. The larger molecular weight of peptide-FITC (molecular weight 1276.49) than of FITC (molecular weight 389.4) might be an obstacle resulting conjugation ratio of PLGA-PEG NP-peptide- FITC (71.1 ± 4.8 mol%) lower than PLGA-PEG NP-FITC (96.5 ± 1.5 mol%). The particle size of the peptide- free and peptide-conjugated PLGA-PEG nanoparticles was 178.9 ± 4.47 nm and 179.7 ± 2.39 nm, respectively, with a polydispersity of 0.103 ± 0.087 and 0.111 ± 0.044, indicating a narrow distribution, and the corresponding zeta potential was −26.60 ± 5.06 mV and −16.70 ± 3.04 mV. All nanoparticles maintained a negatively charged surface, and the peptide-conjugated PLGA-PEG nanoparticles were less negative than for peptide-free PLGA-PEG nanoparticles due to the presence of two positively charged arginine amino acids in the conjugated peptide. Figure 5 shows the 1H-NMR spectra for the peptide-conjugated and peptide-free PLGA-PEG nanoparticles (400 mHz, CDCl3). Singh et al reported that the NMR chemical shifts in peptides represent a population-weighted average of rapidly interconverting peptide structures, and the chemical shifts of asparagine and arginine were reported to be 2.7 ppm and 2.9 ppm, respectively.53 There is a new peak at 2.6 ppm in Figure 5A of peptide-conjugated PLGA-PEG nanoparticles. Asparagine and arginine are two amino acids partially comprising the NR7 peptide. The presence of a 2.6 ppm peak confirmed that the NR7 peptide was successfully conjugated with the PLGA-PEG nanoparticles.
Figure 5.
1H-NMR spectra of (A) peptide-conjugated and (B) peptide-free PLGA-PEG nanoparticles (400 mHz, CDCl3).
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol).
Cell uptake
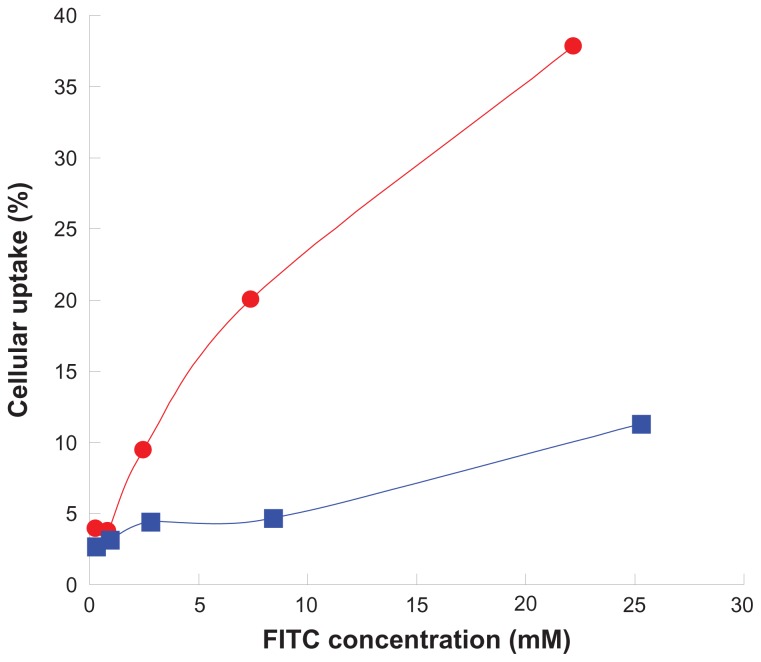
SKOV3 is a cell line with high expression of the EGF receptor and has been used to demonstrate the cellular uptake of peptide-free and peptide-conjugated PLGA-PEG nanoparticles. The result is shown in Figure 6. Cellular uptake of the nanoparticles increased with increasing nanoparticle concentration, and more enhancement was observed for peptide-conjugated nanoparticles than for peptide-free nanoparticles. After incubation with a given 22.2 μM fluorescein concentration of peptide-conjugated nanoparticles with SKOV3 cells for 30 minutes, cellular uptake was observed to be 37.9%, but was only 11.3% after incubation with a given 25.3 μM fluorescein concentration of peptide-free nanoparticles. The presence of the peptide enhanced nanoparticle uptake by more than three-fold in SKOV3 cells with high expression of the EGF receptor. The peptide on the surface of the PLGA-PEG nanoparticles enabled binding to the EGF receptor and was further internalized into SKOV3 cells.54 All of these results suggest that the terminal amino group of the amphiphilic PLGA-PEG copolymer was successfully conjugated with the NR7 peptide, and the resulting peptide-conjugated nanoparticles possessed active targeting ability to SKOV3 cells with high expression of the EGF receptor.
Figure 6.
Cellular uptake of (
 ) peptide-conjugated and (
) peptide-conjugated and (
 ) peptide-free PLGA-PEG nanoparticles in a SKOV3 cell line with high expression of the epithelial growth factor receptor at 37°C for 30 minutes.
) peptide-free PLGA-PEG nanoparticles in a SKOV3 cell line with high expression of the epithelial growth factor receptor at 37°C for 30 minutes.
Abbreviations: FITC, fluorescein isothiocyanate; PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol).
Characterization of doxorubicin-loaded nanoparticles
Doxorubicin is one of the most potent and widely used anticancer drugs. Its anticancer activity results from inhibition of nucleic acid synthesis in cancer cells.55 Doxorubicin-loaded nanoparticles were prepared and characterized. The yields of doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles were 76.5% ± 3.4% and 81.0% ± 0.8% and the corresponding particle size was 185.7 ± 16.7 nm and 178.5 ± 16.2 nm. The polydispersity index was 0.111 ± 0.044 and 0.029 ± 0.005, respectively, and the zeta potential was −15.0 ± 4.3 mV and 0.60 ± 5.04 mV. The particle size of both kinds of doxorubicin-loaded nanoparticles was less than 200 nm, with a narrow distribution, and the zeta potential of the peptide-conjugated nanoparticles was less negative than that of the peptide-free nanoparticles due to conjugation with positively charged peptides. The doxorubicin loading of the peptide-free and peptide-conjugated PLGA-PEG nanoparticles was 4.1% ± 1.4% and 4.8% ± 0.7%, and the encapsulation efficiency was 99.9% ± 5.8% and 97.7% ± 4.1%. The encapsulation of doxorubicin in the two types of nanoparticles was similar, regardless of the presence of the peptide.
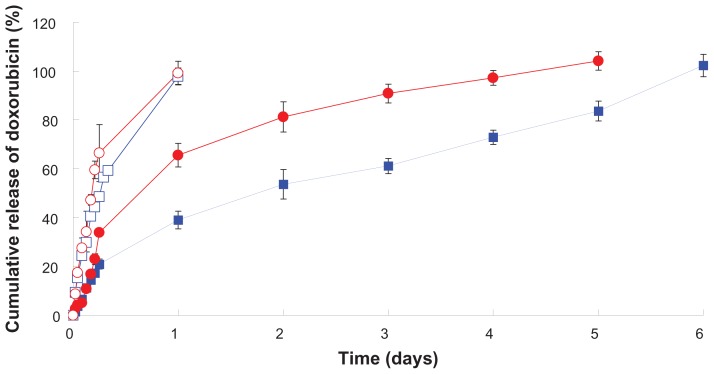
In vitro release of doxorubicin-loaded nanoparticles
Figure 7 shows the release of doxorubicin from peptide-free and peptide-conjugated PLGA-PEG nanoparticles at physiological pH (7.4) and simulated lysosomal pH (4.0, 500 U lipase) release media. The initial release of doxorubicin from peptide-free and peptide-conjugated nanoparticles was 21.0% ± 2.1% and 33.9% ± 0.6% at pH 7.4 in the first 6 hours. The surface to volume ratio of the nanoparticles was large, resulting in rapid drug release at or near the particle surface.44 There was 65.6% ± 4.9% and 99.2% ± 4.8% drug release from the peptide-conjugated nanoparticles in pH 7.4 and pH 4.0 (500 U lipase) during the first 24 hours, and the corresponding value released from the peptide-free nanoparticles was 39.1% ± 3.7% and 97.9% ± 3.4%. The doxorubicin release pattern was similar for both kinds of PLGA-PEG nanoparticles, whereby the drug was completely released at pH 4.0 (500 U lipase) within 24 hours, but release of doxorubicin was sustained for 5–6 days at pH 7.4. The release of doxorubicin from the peptide-conjugated nanoparticles was faster than from the peptide-free nanoparticles at pH 7.4. Khoee et al demonstrated that faster drug release from folate nanoparticles was due to the hydrophilicity of folate which facilitated uptake of water from nanoparticles and caused faster drug release.56 The NR7 peptide was hydrophilic, which might enhance uptake of water by nanoparticles and accelerate hydrolysis of the PLGA-PEG copolymer. All of these factors resulted in faster doxorubicin release from the peptide-conjugated nanoparticles. The release of doxorubicin from peptide-free and peptide-conjugated PLGA-PEG nanoparticles in pH 4.0 medium containing lipase was significantly faster than in pH 7.4 medium. Gillies et al demonstrated that the increase in aqueous solubility of doxorubicin and enhancing hydrolysis of the polymer in a mildly acidic medium (pH 5.0) resulted in faster release of doxorubicin.57 Unger et al indicated that PLGA was degraded by hydrolysis and that acidic conditions further catalyzed the hydrolysis process.58 Our results indicate that the release of doxorubicin was faster in pH 4.0 medium containing lipase than in pH 7.4 medium. The faster hydrolysis of the PLGA-PEG copolymer in acid buffer than in phosphate-buffered saline partially accounts for this result. In addition, the PLGA-PEG copolymer can be degraded by lipase, a type of esterase, to enhance doxorubicin release in a simulated lysosomal environment (pH 4.0 with lipase).59 The pH values of the intracellular compartments, including the endosomes and lysosomes, were reported to be in the range of 4.5–6.5. The average extracellular pH value of cancerous tissue is 6.8, which is caused by hypoxia that upregulates glycolysis following by production of lactate and protons in the extracellular microenvironment.60 Compared with normal tissue (pH 7.4), the existing pH of tumor tissue is considered to be an ideal trigger for selective release of doxorubicin from tumor tissues and within tumor cells. Overall, these results indicate that doxorubicin-loaded peptidefree and peptide-conjugated PLGA-PEG nanoparticles were stable at pH 7.4, whereby doxorubicin was not released from the nanoparticles during circulation in the blood and the undesirable toxicity of free doxorubicin in normal organs could be avoided. Both the doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles accumulated in cancerous tissue via the enhanced permeation and retention effect. Most peptide-conjugated nanoparticles were taken up by the cancer cells through receptor-mediated endocytosis, but only a few peptide-free nanoparticles entered the cancer cells. Doxorubicin would be released rapidly from peptide-conjugated nanoparticles in the lysosome. Free doxorubicin released from the nanoparticles in the cancer cells entered the nuclei and induced apoptosis of cancer cells without being toxic to normal cells. All of these results indicate that release of doxorubicin from PLGA-PEG nanoparticles was pH-dependent and enzyme-sensitive.
Figure 7.
In vitro release of doxorubicin from peptide-free and peptide-conjugated PLGA-PEG nanoparticles in pH 4.0 (500 U lipase) and pH 7.4 release media. (
 ) Peptide-conjugated nanoparticles, pH 4.0 (500 U lipase). (
) Peptide-conjugated nanoparticles, pH 4.0 (500 U lipase). (
 ) Peptide-free nanoparticles, pH 4.0 (500 U lipase). (
) Peptide-free nanoparticles, pH 4.0 (500 U lipase). (
 ) Peptide-conjugated nanoparticles, pH 7.4. (
) Peptide-conjugated nanoparticles, pH 7.4. (
 ) Peptide-free nanoparticles, pH 7.4.
) Peptide-free nanoparticles, pH 7.4.
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol).
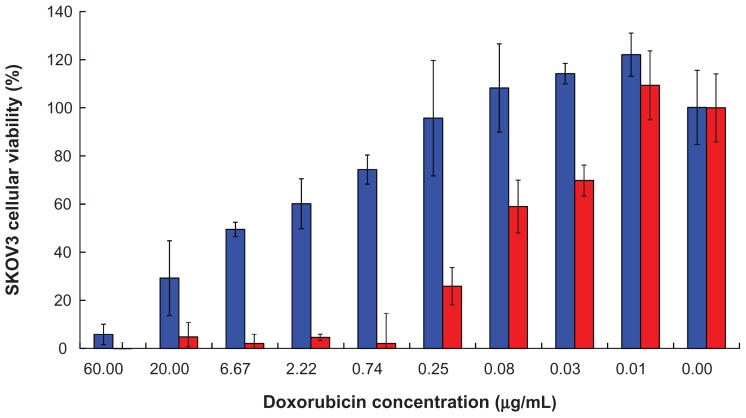
Inhibition of cell growth
Inhibition of SKOV3 cell growth by doxorubicin-loaded nanoparticles was investigated after 72 hours of incubation, and the result is shown in Figure 8. The IC50 for doxorubicin-loaded peptide-free and for peptide-conjugated PLGA-PEG nanoparticles was 3.12 ± 1.44 μg/mL and 0.05 ± 0.03 μg/mL, respectively (P < 0.01). In other words, the IC50 of doxorubicin-loaded peptide-conjugated nanoparticles in SKOV3 cells was 62.4-fold lower than that of doxorubicin-loaded peptide-free nanoparticles. These results indicate that the cytotoxicity of doxorubicin was significantly enhanced by peptide-conjugated nanoparticles. The IC50 of free doxorubicin has been reported to be 0.18 ± 0.02 μg/mL in SKOV3 cells after 72 hours of incubation.61 The IC50 of our doxorubicin-loaded peptide-conjugated PLGA-PEG nanoparticles was 0.05 ± 0.03 μg/mL, which was lower than that of free doxorubicin. In other words, inhibition of cell growth by free doxorubicin was also improved by peptide-conjugated PLGA-PEG nanoparticles. Enhancement of PLGA-PEG nanoparticle internalization via conjugation of a peptide that recognizes the EGF receptor results in more drug being transported into SKOV3 cells with high expression of the EGF receptor to achieve better inhibition of cell growth.
Figure 8.
SKOV3 cell growth inhibition by (
 ) doxorubicin-loaded peptide-free PLGA-PEG nanoparticles and (
) doxorubicin-loaded peptide-free PLGA-PEG nanoparticles and (
 ) doxorubicin-loaded peptide-conjugated nanoparticles after 72 hours of incubation.
) doxorubicin-loaded peptide-conjugated nanoparticles after 72 hours of incubation.
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol).
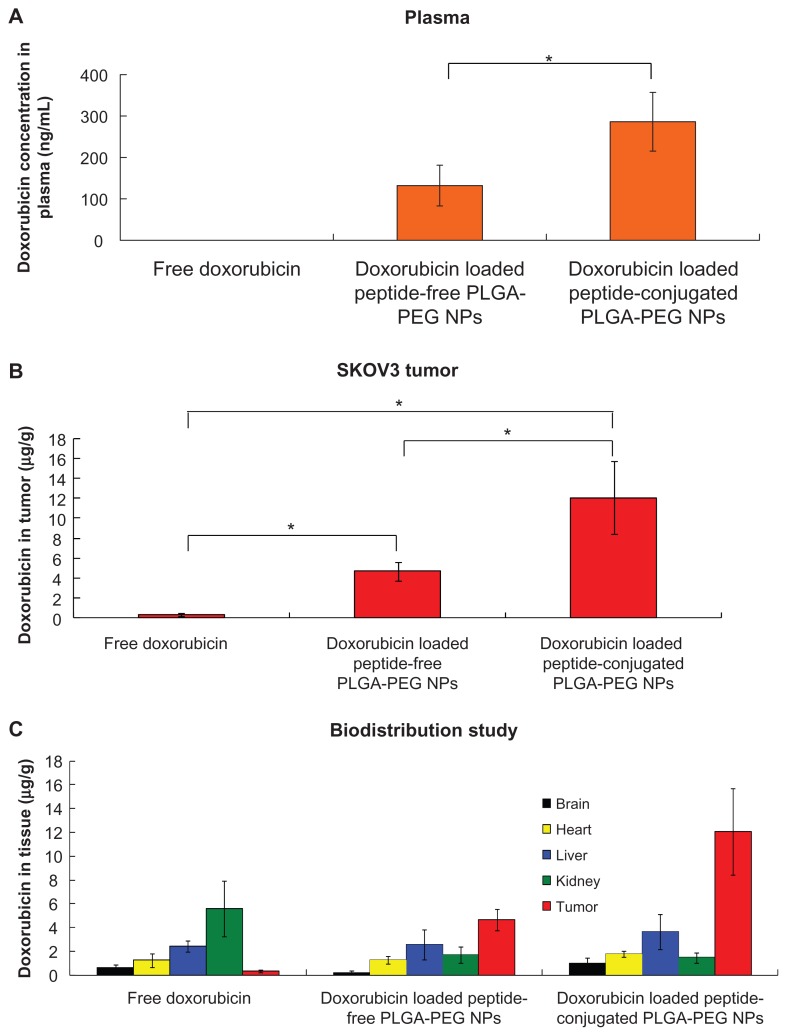
Biodistribution of doxorubicin in SKOV3 tumor-bearing mice
In order to investigate the passive and active targeting effects of doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles, the amounts of doxorubicin in the blood, tumor, brain, heart, liver, and kidney tissues of SKOV3 mice with tumor xenografts were determined. Figure 9A shows the plasma doxorubicin concentration 24 hours following administration of free doxorubicin, and peptide-free and peptide-conjugated PLGA-PEG nanoparticles. No doxorubicin was detected with free doxorubicin, which rapidly disappeared from the circulation within 24 hours of systemic administration due to its short half-life.61 For peptide-free and peptide-conjugated PLGA-PEG nanoparticles, the plasma doxorubicin levels were 132.06 ± 49.14 ng/mL and 286.14 ± 70.84 ng/mL, respectively. The superiority of doxorubicin-loaded nanoparticles in vivo as compared with free doxorubicin may be attributable to the sustained release and prolonged circulation time of the nanoparticles. In addition, the doxorubicin concentration achieved by peptide-conjugated PLGA-PEG nanoparticles was higher than that achieved by peptide-free PLGA-PEG nanoparticles. Figure 9B shows accumulation of doxorubicin in tumor tissue 24 hours following administration of free doxorubicin, and peptide-free and peptide-conjugated PLGA-PEG nanoparticles. For free doxorubicin, the doxorubicin level in the tumor was 0.34 ± 0.13 μg/g. In contrast, when peptide-free or peptide-conjugated PLGA-PEG nanoparticles were administered, a higher doxorubicin level was maintained, and the corresponding values were 4.69 ± 0.90 μg/g and 12.06 ± 3.65 μg/g, respectively. It can be seen that the peptide-free and peptide-conjugated PLGA-PEG nanoparticles accumulated at the tumor sites to a greater extent than did free doxorubicin. This can be attributed to the enhanced permeation and retention effect. Moreover, the peptide-conjugated PLGA-PEG nanoparticles were more readily taken up by tumor cells via receptor-mediated endocytosis. The combined effect of active targeting and enhanced cellular uptake of peptide-conjugated PLGA-PEG nanoparticles would be the main reason for the amount of doxorubicin accumulated in the tumor tissue to be 2.6-fold higher than for peptide-free PLGA-PEG nanoparticles. Figure 9C shows accumulation of doxorubicin in the brain, heart, liver, kidney, and tumor tissue. A significantly smaller amount of doxorubicin was detected in the kidney after administration of peptide-free PLGA-PEG nanoparticles (1.71 ± 0.66 μg/g) and peptide-conjugated PLGA-PEG nanoparticles (1.46 ± 0.41 μg/g) than with free doxorubicin (5.59 ± 2.32 μg/g) at 24 hours. The reduced amount of drug accumulation in the kidney could be attributable to selective localization of doxorubicin within the tumor via the enhanced permeation and retention effect. Biodistribution and accumulation of doxorubicin in the brain, heart, liver, and kidney were similar for both kinds of nanoparticles. However, the tumor targeting efficiency was markedly improved by peptide-conjugated PLGA-PEG nanoparticles as compared with peptide-free PLGA-PEG nanoparticles. This result indicates that the peptide acted as an active targeting ligand to enhance internalization of nanoparticles into tumor cells via receptor-mediated endocytosis. The doxorubicin accumulation ratio of tumor to liver was in the order of peptide-conjugated PLGA-PEG nanoparticles > peptide-free PLGA-PEG nanoparticles > doxorubicin solution. This result further implies that the lower liver uptake and higher tumor accumulation of doxorubicin was achieved by peptide-conjugated PLGA-PEG nanoparticles due to PEGylation and conjugation of a specific peptide ligand. The doxorubicin-loaded peptide-conjugated nanoparticles led to 30 times more drug being distributed in tumor tissue as compared with intravenous injection of free doxorubicin. This result demonstrates a way to maximize the chemotherapeutic effect of doxorubicin while minimizing its toxic side effects. Considering the results of cellular uptake, and in vitro release and biodistribution studies, doxorubicin was retained in the peptide-conjugated PLGA-PEG nanoparticles in a stable manner, which reduced their nonspecific toxicity to normal tissues during circulation in the blood after intravenous administration. The doxorubicin-loaded peptide-conjugated PLGA-PEG nanoparticles accumulated in tumor tissue via the enhanced permeation and retention effect and receptor-mediated endocytosis. When the peptide-conjugated nanoparticles arrived at lysosomes through the endolysosomal pathway, doxorubicin was released rapidly in the lysosomes of SKOV3 cells. The doxorubicin released inside the cancer cells entered the nuclei and induced apoptosis of these cells. Doxorubicin-loaded peptide-conjugated PLGA-PEG nanoparticles acted as an active targeting carrier that increased intracellular accumulation of doxorubicin in SKOV3 cells and reduced side effects in normal tissue.
Figure 9.
Accumulation and biodistribution of doxorubicin in (A) plasma, (B) tumor, and (C) tissues after intravenous administration of free doxorubicin, and doxorubicin-loaded peptide-free and peptide-conjugated PLGA-PEG nanoparticles in SKOV3 tumor-bearing mice at 24 hours.
Abbreviations: PLGA, poly(D,L-lactide-co-glycolide); PEG, poly(ethylene glycol); NPs, nanoparticles.
Conclusion
In this study, it was demonstrated that the amphiphilic PLGA-PEG copolymer provides an amino end group for peptide conjugation. The PLGA-PEG copolymers synthesized at a 1:1 molar feed ratio for PLGA-NHS:PEG diamine had a PEGylation efficiency of 97.0% and a low critical micelle concentration of 3.0 × 10−8 mol/L. The cytotoxicity of PLGA was markedly reduced by more than 98-fold after PEGylation. The FITC conjugation ratio of PLGA-PEG NP-FITC was significantly higher than of PLGA-PEG-FITC. The core-shell conformation provided an amino group at the end of the PEG chain surrounding the surface of the PLGA-PEG nanoparticles to react efficiently with FITC, which resulted in a higher FITC conjugation ratio of PLGA-PEG NP-FITC. Further conjugation of PLGA-PEG nanoparticles with the peptide recognizing the EGF receptor had a cellular uptake efficiency which was 3.3-fold superior to that of peptide-free PLGA-PEG nanoparticles. The doxorubicin-loaded nanoparticles were smaller than 200 nm with a narrow size distribution. The less negative zeta potential of peptide-conjugated nanoparticles compared to peptide-free nanoparticles was due to conjugation of the positively charged peptide with the former. In vitro release of doxorubicin from both kinds of nanoparticles was similar, but was pH-dependent and enzyme-sensitive. The IC50 of doxorubicin-loaded peptide-conjugated nanoparticles in SKOV3 cells expressing high levels of the EGF receptor was 0.05 ± 0.03 μg/mL, which was lower than for the doxorubicin- loaded peptide-free nanoparticles (3.12 ± 1.44 μg/mL). These results confirm that doxorubicin-loaded peptide- conjugated PLGA-PEG nanoparticles possessing active targeting ability not only enhance cellular uptake in SKOV3 cells expressing high levels of the EGF receptor but also have better cellular growth inhibition activity. This finding was further demonstrated in an in vivo biodistribution study in SKOV3 tumor-bearing mice, whereby higher tumor accumulation of doxorubicin was achieved by peptide-conjugated PLGA-PEG nanoparticles.
Acknowledgments
This work was supported by the National Science Council in Taiwan (NSC 99-2320-B-002-021-MY3). The authors appreciate the kind assistance of Professor Fu Hsiung Chang from the Graduate Institute of Biochemistry and Molecular Biology, College of Medicine, National Taiwan University in Taiwan, in the particle size analysis.
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Lu JM, Wang X, Marin-Muller C, et al. Current advances in research and clinical applications of PLGA-based nanotechnology. Expert Rev Mol Diagn. 2009;9:325–341. doi: 10.1586/erm.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semete B, Booysen L, Lemmer Y, et al. In vivo evaluation of the biodistribution and safety of PLGA nanoparticles as drug delivery systems. Nanomedicine. 2010;6:662–671. doi: 10.1016/j.nano.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Jain RA. The manufacturing techniques of various drug loaded biodegradable poly(lactide-co-glycolide) (PLGA) devices. Biomaterials. 2000;21:2475–2490. doi: 10.1016/s0142-9612(00)00115-0. [DOI] [PubMed] [Google Scholar]
- 4.Lim TY, Poh CK, Wang W. Poly (lactic-co-glycolic acid) as a controlled release delivery device. J Mater Sci Mater Med. 2009;20:1669–1675. doi: 10.1007/s10856-009-3727-z. [DOI] [PubMed] [Google Scholar]
- 5.Du L, Mei X, Wang C, Li X, Zhang F, Jin Y. In-vitro/in-vivo studies of the biodegradable poly-(D,L-lactide-co-glycolide) microspheres of a novel luteinizing hormone-releasing hormone antagonist for prostate cancer treatment. Anticancer Drugs. 2011;22:262–272. doi: 10.1097/cad.0b013e3283425c2a. [DOI] [PubMed] [Google Scholar]
- 6.Grabrucker AM, Garner CC, Boeckers TM, et al. Development of novel Zn loaded nanoparticles designed for cell-type targeted drug release in CNS neurons: in vitro evidences. PLoS One. 2011;6:e17851. doi: 10.1371/journal.pone.0017851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranganath SH, Fu Y, Arifin DY, et al. The use of submicron/nanoscale PLGA implants to deliver paclitaxel with enhanced pharmacokinetics and therapeutic efficacy in intracranial glioblastoma in mice. Biomaterials. 2010;31:5199–5207. doi: 10.1016/j.biomaterials.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Avgoustakis K. Pegylated poly(lactide) and poly(lactide-co-glycolide) nanoparticles: preparation, properties and possible applications in drug delivery. Curr Drug Deliv. 2004;1:321–333. doi: 10.2174/1567201043334605. [DOI] [PubMed] [Google Scholar]
- 9.Betancourt T, Byrne JD, Sunaryo N, et al. PEGylation strategies for active targeting of PLA/PLGA nanoparticles. J Biomed Mater Res A. 2009;91:263–276. doi: 10.1002/jbm.a.32247. [DOI] [PubMed] [Google Scholar]
- 10.Gref R, Luck M, Quellec P, et al. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 11.Ding H, Yong KT, Roy I, et al. Bioconjugated PLGA-4-arm-PEG branched polymeric nanoparticles as novel tumor targeting carriers. Nanotechnology. 2011;22:165101. doi: 10.1088/0957-4484/22/16/165101. [DOI] [PubMed] [Google Scholar]
- 12.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci U S A. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 14.Avgoustakis K, Beletsi A, Panagi Z, et al. Effect of copolymer composition on the physicochemical characteristics, in vitro stability, and biodistribution of PLGA-mPEG nanoparticles. Int J Pharm. 2003;259:115–127. doi: 10.1016/s0378-5173(03)00224-2. [DOI] [PubMed] [Google Scholar]
- 15.Sledge GW, Miller KD., Jr Exploiting the hallmarks of cancer: the future conquest of breast cancer. Eur J Cancer. 2003;39:1668–1675. doi: 10.1016/s0959-8049(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 16.Teicher BA. Molecular targets and cancer therapeutics: discovery, development and clinical validation. Drug Resist Updat. 2000;3:67–73. doi: 10.1054/drup.2000.0123. [DOI] [PubMed] [Google Scholar]
- 17.Maeda H. Tumor-selective delivery of macromolecular drugs via the EPR effect: background and future prospects. Bioconjug Chem. 2010;21:797–802. doi: 10.1021/bc100070g. [DOI] [PubMed] [Google Scholar]
- 18.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;6:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 20.Nakanishi T, Fukushima S, Okamoto K, et al. Development of the polymer micelle carrier system for doxorubicin. J Control Release. 2001;74:295–302. doi: 10.1016/s0168-3659(01)00341-8. [DOI] [PubMed] [Google Scholar]
- 21.Tobio M, Gref R, Sanchez A, Langer R, Alonso MJ. Stealth PLA-PEG nanoparticles as protein carriers for nasal administration. Pharm Res. 1998;15:270–275. doi: 10.1023/a:1011922819926. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Yung LY. Selectivity of folate conjugated polymer micelles against different tumor cells. Int J Pharm. 2008;349:256–268. doi: 10.1016/j.ijpharm.2007.07.040. [DOI] [PubMed] [Google Scholar]
- 23.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 24.Cheng J, Teply BA, Sherifi I, et al. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28:869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farokhzad OC, Cheng J, Teply BA, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esmaeili F, Ghahremani MH, Ostad SN, et al. Folate-receptor-targeted delivery of docetaxel nanoparticles prepared by PLGA-PEG-folate conjugate. J Drug Target. 2008;16:415–423. doi: 10.1080/10611860802088630. [DOI] [PubMed] [Google Scholar]
- 27.Lee H, Hu M, Reilly RM, Allen C. Apoptotic epidermal growth factor (EGF)-conjugated block copolymer micelles as a nanotechnology platform for targeted combination therapy. Mol Pharm. 2007;4:769–781. doi: 10.1021/mp0700559. [DOI] [PubMed] [Google Scholar]
- 28.Acharya S, Dilnawaz F, Sahoo SK. Targeted epidermal growth factor receptor nanoparticle bioconjugates for breast cancer therapy. Biomaterials. 2009;30:5737–5750. doi: 10.1016/j.biomaterials.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Chittasupho C, Duangrat C, Siahaan TJ, Berkland C. PLGA nanoparticle-peptide conjugate effectively targets intercellular cell-adhesion molecule-1. Bioconjug Chem. 2008;19:145–152. doi: 10.1021/bc700227z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriya T, Kanomata N, Kozuka Y, et al. Molecular morphological approach to the pathological study of development and advancement of human breast cancer. Med Mol Morphol. 2010;43:67–73. doi: 10.1007/s00795-010-0504-5. [DOI] [PubMed] [Google Scholar]
- 31.Alshenawy HA. Immunohistochemical expression of epidermal growth factor receptor, E-cadherin, and matrix metalloproteinase-9 in ovarian epithelial cancer and relation to patient deaths. Ann Diagn Pathol. 2010;14:387–395. doi: 10.1016/j.anndiagpath.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Simonetti S, Molina MA, Queralt C, et al. Detection of EGFR mutations with mutation-specific antibodies in stage IV non-small-cell lung cancer. Transl Med. 2010;8:135. doi: 10.1186/1479-5876-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal SK, Figlin RA, Reckamp K. Targeted therapies for non-small cell lung cancer: an evolving landscape. Mol Cancer Ther. 2010;9:1931–1944. doi: 10.1158/1535-7163.MCT-10-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolonin MG, Bover L, Sun J, et al. Ligand-directed surface profiling of human cancer cells with combinatorial peptide libraries. Cancer Res. 2006;66:34–40. doi: 10.1158/0008-5472.CAN-05-2748. [DOI] [PubMed] [Google Scholar]
- 35.Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp Cell Res. 2003;284:2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- 36.Tarasevich BJ, Gutowska A, Li XS, Jeong BM. The effect of polymer composition on the gelation behavior of PLGA-g-PEG biodegradable thermoreversible gels. J Biomed Mater Res A. 2009;89:248–254. doi: 10.1002/jbm.a.32025. [DOI] [PubMed] [Google Scholar]
- 37.Ghahremankhani AA, Dorkoosh F, Dinarvand R. PLGA-PEG-PLGA tri-block copolymers as in situ gel-forming peptide delivery system: effect of formulation properties on peptide release. Pharm Dev Technol. 2008;13:49–55. doi: 10.1080/10837450701702842. [DOI] [PubMed] [Google Scholar]
- 38.Goddard ED, Turro NJ, Kuo PL, Ananthapadmanabhan KP. Fluorescence probes for critical micelle concentration determination. Langmuir. 1985;1:352–355. doi: 10.1021/la00063a015. [DOI] [PubMed] [Google Scholar]
- 39.Lin WJ, Chen MH. Synthesis of multifunctional chitosan with galactose as a targeting ligand for glycoprotein receptor. Carbohydr Polym. 2007;67:474–480. [Google Scholar]
- 40.Yin Y, Chen D, Qiao M, Wei X, Hu H. Lectin-conjugated PLGA nanoparticles loaded with thymopentin: ex vivo bioadhesion and in vivo biodistribution. J Control Release. 2007;123:27–38. doi: 10.1016/j.jconrel.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Davaran S, Rashidi MR, Pourabbas B, Dadashzadeh M, Haghshenas NM. Adriamycin release from poly(lactide-coglycolide)-polyethylene glycol nanoparticles: synthesis, and in vitro characterization. Int J Nanomedicine. 2006;1:535–539. doi: 10.2147/nano.2006.1.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciapetti G, Cenni E, Pratelli L, Pizzoferrato A. In vitro evaluation of cell/biomaterial interaction by MTT assay. Biomaterials. 1993;14:359–364. doi: 10.1016/0142-9612(93)90055-7. [DOI] [PubMed] [Google Scholar]
- 43.Campos-Lara M, Mendoza-Espinoza JA. Cytotoxic evaluation of fluvastatin and rosuvastatin and effect of fluvastatin in the HeLa cell cycle. Afr J Pharm Pharmacol. 2011;5:322–326. [Google Scholar]
- 44.Chittasupho C, Xie SX, Baoum A, Yakovleva T, Siahaan TJ, Berkland CJ. ICAM-1 targeting of doxorubicin-loaded PLGA nanoparticles to lung epithelial cells. Eur J Pharm Sci. 2009;37:141–150. doi: 10.1016/j.ejps.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin C, Qian N, Zhao W, et al. Improved therapeutic effect of DOX-PLGA-PEG micelles decorated with bivalent fragment HAb18 F(ab’)(2) for hepatocellular carcinoma. Biomacromolecules. 2010;11:2422–2431. doi: 10.1021/bm1005992. [DOI] [PubMed] [Google Scholar]
- 46.Yadav AK, Mishra P, Mishra AK, Mishra P, Jain S, Agrawal GP. Development and characterization of hyaluronic acid-anchored PLGA nanoparticulate carriers of doxorubicin. Nanomedicine. 2007;3:246–257. doi: 10.1016/j.nano.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Buwalda SJ, Dijkstra PJ, Calucci L, Forte C, Feijen J. Influence of amide versus ester linkages on the properties of eight-armed PEG-PLA star block copolymer hydrogels. Biomacromolecules. 2010;11:224–232. doi: 10.1021/bm901080d. [DOI] [PubMed] [Google Scholar]
- 48.Lee ES, Oh KT, Kim D, Youn YS, Bae YH. Tumor pH-responsive flower-like micelles of poly(L-lactic acid)-b-poly(ethylene glycol)-b-poly( L-histidine) J Control Release. 2007;123:19–26. doi: 10.1016/j.jconrel.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SY, Shin IG, Lee YM, Cho CS, Sung YK. Methoxy poly(ethylene glycol) and epsilon-caprolactone amphiphilic block copolymeric micelle containing indomethacin. II. Micelle formation and drug release behaviours. J Control Release. 1998;51:13–22. doi: 10.1016/s0168-3659(97)00124-7. [DOI] [PubMed] [Google Scholar]
- 50.Ioele G, Cione E, Risoli A, Genchi G, Ragno G. Accelerated photostability study of tretinoin and isotretinoin in liposome formulations. Int J Pharm. 2005;293:251–260. doi: 10.1016/j.ijpharm.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 51.van Vlerken LE, Duan Z, Little SR, Seiden MV, Amiji MM. Biodistribution and pharmacokinetic analysis of paclitaxel and ceramide administered in multifunctional polymer-blend nanoparticles in drug resistant breast cancer model. Mol Pharm. 2008;5:516–526. doi: 10.1021/mp800030k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zange R, Li Y, Kissel T. Biocompatibility testing of ABA triblock copolymers consisting of poly(L-lactic-co-glycolic acid) A blocks attached to a central poly(ethylene oxide) B block under in vitro conditions using different L929 mouse fibroblasts cell culture models. J Control Release. 1998;56:249–258. doi: 10.1016/s0168-3659(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 53.Singh A, Wilczynski A, Holder JR, et al. Incorporation of a bioactive reverse-turn heterocycle into a peptide template using solid-phase synthesis to probe melanocortin receptor selectivity and ligand conformations by 2D 1H NMR. J Med Chem. 2011;54:1379–1390. doi: 10.1021/jm101425m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milane L, Duan Z, Amiji M. Development of EGFR-targeted polymer blend nanocarriers for combination paclitaxel/lonidamine delivery to treat multi-drug resistance in human breast and ovarian tumor cells. Mol Pharm. 2011;8:185–203. doi: 10.1021/mp1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousefpour P, Atyabi F, Vasheghani-Farahani E, Movahedi AA, Dinarvand R. Targeted delivery of doxorubicin-utilizing chitosan nanoparticles surface-functionalized with anti-Her2 trastuzumab. Int J Nanomedicine. 2011;6:1977–1990. doi: 10.2147/IJN.S21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khoee S, Rahmatolahzadeh R. Synthesis and characterization of pH-responsive and folated nanoparticles based on self-assembled brush-like PLGA/PEG/AEMA copolymer with targeted cancer therapy properties: a comprehensive kinetic study. Eur J Med Chem. 2012;50:416–427. doi: 10.1016/j.ejmech.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 57.Gillies ER, Fréchet JM. pH-responsive copolymer assemblies for controlled release of doxorubicin. Bioconjug Chem. 2005;16:361–368. doi: 10.1021/bc049851c. [DOI] [PubMed] [Google Scholar]
- 58.Unger F, Wittmar M, Morell F, Kissel T. Branched polyesters based on poly[vinyl-3-(dialkylamino)alkylcarbamate-co-vinyl acetate-co- vinyl alcohol]-graft-poly (D,L-lactide-co-glycolide): effects of polymer structure on in vitro degradation behaviour. Biomaterials. 2008;29:2007–2014. doi: 10.1016/j.biomaterials.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 59.Xue HY, Wong HL. Tailoring nanostructured solid-lipid carriers for time-controlled intracellular siRNA kinetics to sustain RNAi-mediated chemosensitization. Biomaterials. 2011;32:2662–2672. doi: 10.1016/j.biomaterials.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 60.Manchun S, Dass CR, Sriamornsak P. Targeted therapy for cancer using pH-responsive nanocarrier systems. Life Sci. 2012;90:381–387. doi: 10.1016/j.lfs.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 61.Han HD, Lee A, Hwang T, et al. Enhanced circulation time and antitumor activity of doxorubicin by comblike polymer-incorporated liposomes. J Control Release. 2007;120:161–168. doi: 10.1016/j.jconrel.2007.03.020. [DOI] [PubMed] [Google Scholar]