Abstract
Background
Enterococci are among the leading causes of hospital-acquired infections in the United States and Europe, with Enterococcus faecalis and Enterococcus faecium being the two most common species isolated from enterococcal infections. In the last decade, the proportion of enterococcal infections caused by E. faecium has steadily increased compared to other Enterococcus species. Although the underlying mechanism for the gradual replacement of E. faecalis by E. faecium in the hospital environment is not yet understood, many studies using genotyping and phylogenetic analysis have shown the emergence of a globally dispersed polyclonal subcluster of E. faecium strains in clinical environments. Systematic study of the molecular epidemiology and pathogenesis of E. faecium has been hindered by the lack of closed, complete E. faecium genomes that can be used as references.
Results
In this study, we report the complete genome sequence of the E. faecium strain TX16, also known as DO, which belongs to multilocus sequence type (ST) 18, and was the first E. faecium strain ever sequenced. Whole genome comparison of the TX16 genome with 21 E. faecium draft genomes confirmed that most clinical, outbreak, and hospital-associated (HA) strains (including STs 16, 17, 18, and 78), in addition to strains of non-hospital origin, group in the same clade (referred to as the HA clade) and are evolutionally considerably more closely related to each other by phylogenetic and gene content similarity analyses than to isolates in the community-associated (CA) clade with approximately a 3–4% average nucleotide sequence difference between the two clades at the core genome level. Our study also revealed that many genomic loci in the TX16 genome are unique to the HA clade. 380 ORFs in TX16 are HA-clade specific and antibiotic resistance genes are enriched in HA-clade strains. Mobile elements such as IS16 and transposons were also found almost exclusively in HA strains, as previously reported.
Conclusions
Our findings along with other studies show that HA clonal lineages harbor specific genetic elements as well as sequence differences in the core genome which may confer selection advantages over the more heterogeneous CA E. faecium isolates. Which of these differences are important for the success of specific E. faecium lineages in the hospital environment remain(s) to be determined.
Background
Enterococci are normal constituents of the gastro-intestinal flora of humans and other animals [1-3]. Although they only occasionally cause infections in healthy individuals, they are the third most commonly isolated gram positive organisms from hospital-associated (HA) infections in the United States and are increasingly reported in other countries [4,5]. Enterococcal infections are often difficult to treat due to the number of antibiotics to which these organisms are resistant. Some antibiotic resistances are intrinsic, such as resistances to cephalosporins, while other antibiotic resistances are acquired through mutations or horizontal gene transfer, most notably the van systems that encode vancomycin resistance [6-12]. Several recent studies also confirmed that enterococci can transfer their resistance to even more virulent organisms, such as Staphylococcus aureus[13].
Enterococcus faecalis is the most common enterococcal species recovered from infections. However, in the last decade, infections with Enterococcus faecium have been on the rise in the United States, Europe, and South America [2-5,14]. In the US, isolates of E. faecium now account for ca. 35% of nosocomial enterococcal isolates identified to the species level [4]. It is still not clear what has caused the ecological replacement of E. faecalis with E. faecium in the nosocomial setting, but it is speculated that the intense use of antibiotics in hospitals and the multiple antibiotic resistances of E. faecium have been major contributing factors [11,15]. A few genes have been suggested as being virulence determinants in E. faecium due to their enrichment in clinical isolates, such as the fms or hyl genes [16-22]. However, only three genes have been experimentally implicated to have an impact on virulence in animal models, namely esp, which has a role in biofilm, urinary tract infection, and endocarditis [23,24]; acm, encoding a collagen binding adhesin contributing to endocarditis [25,26]; and the ebpfm operon which encodes pili that are important in biofilm and urinary tract infection [27]. In addition, conjugative transfer of a plasmid with a hyl-like gene not only conferred increased resistance to vancomycin but also increased virulence in transconjugants in the mouse peritonitis model [28], and a different hyl-plasmid conferred colonization in the murine gut [29]. While the gene(s) responsible for this increase in virulence and colonization have yet to be determined, the deletion of the hyl gene did not cause attenuation in the peritonitis model [19].
Molecular epidemiological studies of outbreaks of E. faecium using MLST initially indicated that there was a specific lineage or genogroup of strains, designated clonal complex 17, that was predominant in the hospital environment [2,5,15,30]. Other studies using pyrosequencing and whole-genome microarray subsequently indicated that, while there appeared to be a globally dispersed clade containing the vast majority of epidemic and clinical isolates which harbor a large content of accessory genes specific to this clade [31,32], isolates associated with healthcare settings were not strictly clonally related to each other. In particular, while CC17 genogroup isolates are part of the HA subpopulation, not all HA isolates are considered part of the ST17 lineage [33]. Recent studies in our laboratory and others have shown large differences (~3–4%) in the sequence of the core genome, as well as differences in the 16-S rRNA, between two different clades which were named the hospital-associated clade (HA) and community-associated (CA) clade strains, (also known as clade A and B [34])[32,33]. The HA clade contains most clinical and HA-associated strains but also included strains from non-hospital origin [35,36].
Molecular studies and comprehensive comparative genomic studies of E. faecium have long been hindered by the lack of a complete genome sequence. The TX16 (DO) genome was initially sequenced at the Department of Energy’s Joint Genome Institute (JGI) in Walnut Creek, Ca. in 1999 in an effort to demonstrate capabilities of the sequencing technology at that time by sequencing the genome in only 1 day. However, the genome was far from closed and the past decade has been spent on annotation, final assembly, and analyses of this genome. Recently, while this manuscript was in review, a closed E. faecium genome was published by Lam et al. using the ST17 isolate Aus0004, which was isolated from the bloodstream of a patient in Melbourne, Australia [37].
In this study, we report the closed genome of the US E. faecium endocarditis isolate TX16 (DO), and a comparative analysis of this strain’s genome with 21 other available E. faecium draft genomes [32,38], as well as the recently published Aus0004 [37]. Due to the fact the TX16 genome has been used in multiple pathogenesis studies and is a part of the clonal group representing the majority of clinical strains globally [2,5,30,36], the complete genome sequence of E. faecium TX16 will facilitate future research by providing a critical starting point for genome-wide functional studies to determine the molecular basis of pathogenesis and to further understand the evolution and molecular epidemiology of E. faecium infective strains.
Results
E. faecium TX16 general genome features
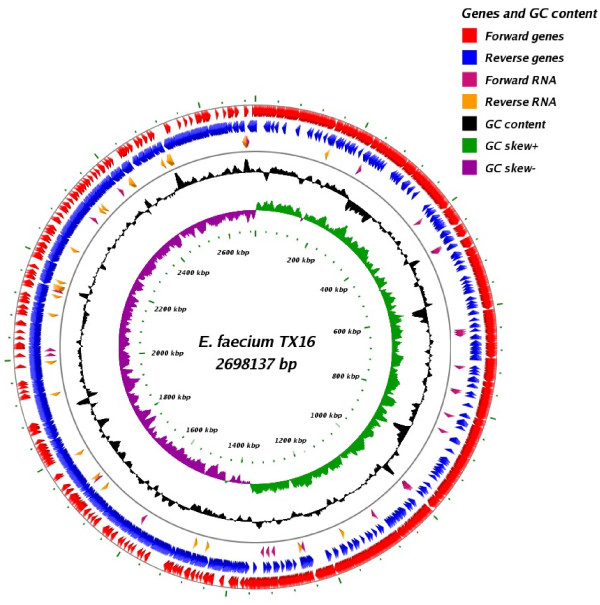
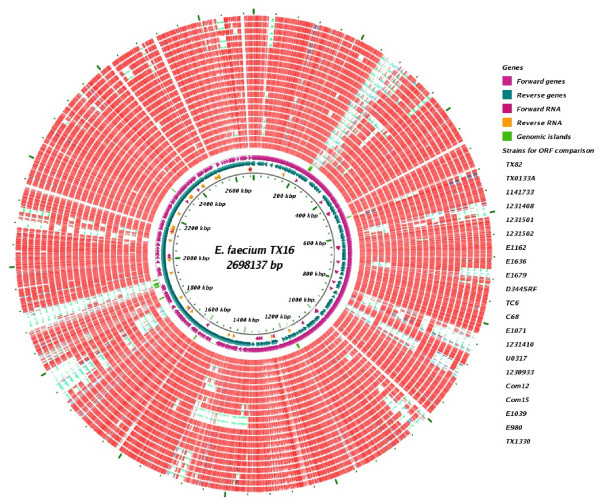
The E. faecium TX16 genome consists of one chromosome and three plasmids. The chromosome (Figure 1) contains 2,698,137 bp with 2,703 protein-coding ORFs, 62 tRNAs, 6 copies of ribosomal rRNA and 32 other non-coding RNAs (Table 1). The chromosome has a GC content of 38.15%, and it shows a clear GC skew at the origin of replication (Figure 1). The sizes of the three plasmids (pDO1, pDO2, and pDO3) are 36,262, 66,247 and 251,926 bp, encoding 43, 85, and 283 ORFs, respectively (Table 1).
Figure 1.
Circular map of theE. faeciumTX16 genome. Tracks from inside to outside are as follows: GC skew (G-C)/(G + C), GC content, forward and reverse RNA, reverse genes, and forward genes.
Table 1.
General features ofE. faeciumTX16 genome
| Features | Chromosome | Plasmid pDO1 | Plasmid pDO2 | Plasmid pDO3 |
|---|---|---|---|---|
|
Size (bp) |
2698137 |
36262 |
66247 |
251926 |
|
G + C % |
38.15 |
36.51 |
34.38 |
35.97 |
|
ORFs |
2703 |
43 |
85 |
283 |
|
rRNA operons |
6 |
0 |
0 |
0 |
|
tRNAs |
62 |
0 |
2 |
0 |
| ncRNAs | 32 | 1 | 0 | 0 |
To investigate the conservation of the gene order of E. faecium compared to its close relative E. faecalis, a BLASTP alignment of all the predicted proteins from the TX16 and V583 genomes was performed followed by ORF synteny analysis using DAGchainer [39]. The result showed that E. faecium TX16 gene order is very different from that of E. faecalis strain V583 (and therefore OG1RF, which has a very similar synteny to V583 [40,41]) and all ORF synteny blocks were relatively short (Additional file 1: Figure S1).
Interestingly, when comparing TX16 to the closed genome Aus0004, which was published while this paper was in review, Mauve genome alignment analysis resulted in 5 locally collinear blocks for both TX16 and Aus0004 ranging from 33,563–836,291 bp for TX16 and 32,326–905,025 bp for Aus0004 (Additional file 2: Figure S2). The two isolates had very similar synteny, although two regions found in TX16 were inverted in Aus0004. Two site-specific tyrosine family recombinases (EFAU004_01466 and EFAU004_02416) were found flanking these two inversions (Additional file 2: Figure S2).
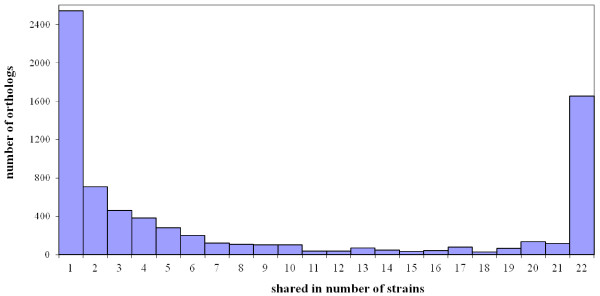
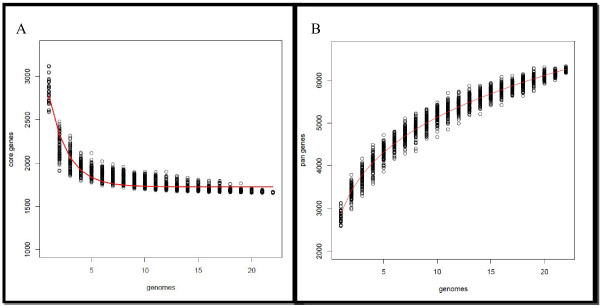
The genome size of the E. faecium strains vary substantially from 2.50 Mb (E1039) to 3.14 Mb (1,230,933), while the number of ORFs varies from 2,587 (E1039) to 3,118 (TX0133A). Ortholog analysis of TX16 compared to TX1330 and all the available but unfinished E. faecium genomes using BLASTP of predicted protein sequences and orthoMCL resulted in 3,169 distributed genes shared among some strains (Figure 2), 2,543 unique genes (Figure 2), and 1,652 core gene families, of which 1,608 genes are present in a single copy in all strains and 44 gene families are present in multiple copies. The number of core genes (including those in single and multiple copies) converged to 1,726 at the 22nd genome, while the number of pan genes reached 6,262 genes at the 22nd genome (Figure 3A and B). The extrapolated number of core genes is very close to the number of core genes (1,772 genes) Leavis et al. reported in their microarray-based study which used 97 isolates, yet the estimated number of pan genes is higher in the present analysis [31]. Furthermore, this study differs slightly from the analysis of van Schaik et al. which estimates the E. faecium core genome to be 2172 ± 20 CDS [32]. Our data do, however, concur with the conclusion that a sizeable fraction of the E. faecium genome is accessory and that the pan genome is considered to open.
Figure 2.
Distribution of orthologs in 22E. faeciumstrains. The orthologs were determined by orthoMCL as described in the Material and Methods. ORFs of the 3 plasmids in E. faecium TX16 were not included in the ortholog analysis.
Figure 3.
E. faeciumcore and pan genomes.A. E. faecium core genes. The number of shared genes is plotted as the function of number of strains (n) added sequentially. An open circle represents the number of shared genes for each permutation at a give number of strains (n). 1,608 single copy genes are shared by all 22 genomes. The red line represents the least-squares fit to the exponential decay function Fc = κc exp[−n/τc] + Ω (κc = 1871 ± 25, τc = 1.751 ± 0.027, Ω = 1726 ± 2). B. E. faecium pan-genes. The number of total genes is plotted as the function of strains (n). The open circle represents the number of total genes for each permutation at a give number of strains (n). The red line represents the least-squares fit to the power law function n = κ Nγ (κ = 2876 ± 7, γ = 0.2517 ± 0.009).
Phylogenetic, multi-locus sequence typing (MLST) and gene content similarity analysis
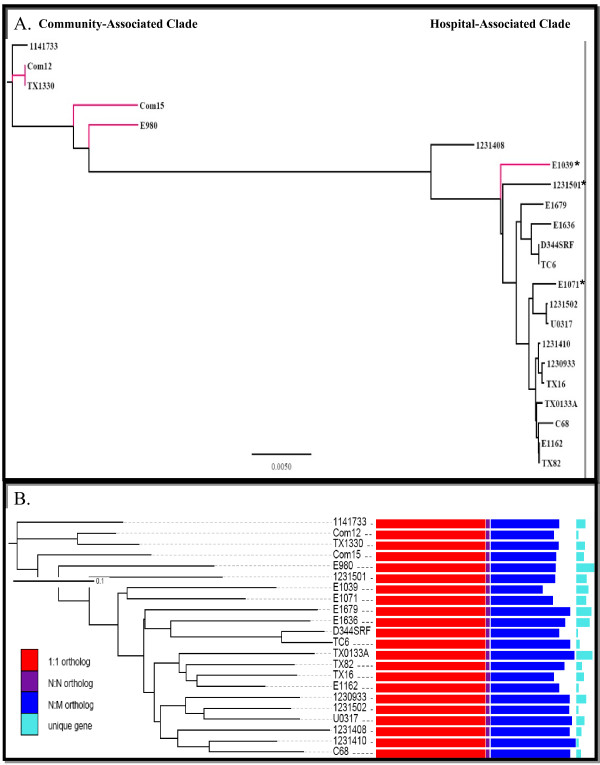
Analysis of the 22 E. faecium genomes (Table 2) showed that the isolates separate into two clades, one branch consisting mostly of CA isolates, with most HA isolates found in the other, as was noted in our previous study [33] (Figure 4A and B). When analyzing the phylogenetic distances among these 22 isolates using 628 single-copy ortholog genes of the same length (Figure 4A), similar clade patterns were observed for the E. faecium strains as seen in the 100 core gene analysis by Galloway-Pena et.al [33]. All isolates predicted to be part of the CC17 genogroup [2,5,30] cluster more closely together and branched more distantly than other HA-clade isolates (Figure 4A). The dendogram construction from the gene content dissimilarity represented by Jaccard distance (Figure 4B) also showed most hospital-isolated strains cluster together except hospital- isolated strain 1,141,733 which was shown genetically to belong to the CA clade. In addition, although E1039 is a community- isolated fecal strain, it is genetically closer to the HA strains. The phylogenetic and gene content dissimilarity analysis results all support the existence of two very distinct clades of E. faecium, which has been previously described using pyrosequencing, microarray, and the concatenation of a 100 core genes, estimated to have diverged anywhere from 300,000 to 3 million years ago [31-33].
Table 2.
The 22 sequencedEnterococcus faeciumgenomes
| Strain | ST | CC17 | Country | Year | Source | Reference |
|---|---|---|---|---|---|---|
| 1,231,408a |
582 |
Yes |
NAb |
NA |
Blood Culture of Hospitalized Patient |
[38] |
| 1,231,501 |
52 |
No |
NA |
NA |
Blood Culture of Hospitalized Patient |
[38] |
| Com15 |
583 |
No |
USA (MA) |
2006 |
Healthy Volunteer Feces |
[38] |
| 1,141,733 |
327 |
No |
NA |
NA |
Blood Culture of Hospitalized Patient |
[38] |
| 1,230,933 |
18 |
Yes |
NA |
NA |
Wound Swab of Hospitalized Patient |
[38] |
| 1,231,410 |
17 |
Yes |
NA |
NA |
Skin and Soft Tissue Infection |
[38] |
| 1,231,502 |
203 |
Yes |
NA |
NA |
Blood Culture of Hospitalized Patient |
[38] |
| Com12 |
107 |
No |
USA (MA) |
2006 |
Healthy Volunteer Feces |
[38] |
| E1039 |
42 |
No |
Netherlands |
1998 |
Healthy Volunteer Feces |
[32] |
| E1162 |
17 |
Yes |
France |
1997 |
Blood Culture of Hospitalized Patient |
[32] |
| E1071 |
32 |
No |
Netherlands |
2000 |
Hospitalized Patient Feces |
[32] |
| E1679 |
114 |
No |
Brazil |
1998 |
Swab of Vascular Catheter |
[32] |
| E1636 |
106 |
No |
Netherlands |
1961 |
Blood Culture of Hospitalized Patient |
[32] |
| E980 |
94 |
No |
Netherlands |
1998 |
Healthy Volunteer Feces |
[32] |
| U0317 |
78 |
Yes |
Netherlands |
2005 |
UTI of Hospitalized Patient |
[32] |
| D344SRFc |
21 |
No |
France |
1985 |
Clinical (Site not specified) |
[42] |
| TC6 |
21 |
No |
USA (OH) |
NA |
Transconjugant of C68 and D344SRF |
[29] |
| C68 |
16 |
Yes |
USA (OH) |
1998 |
Endocarditis Patient (Feces) |
[9] |
| TX0133 |
17 |
Yes |
USA (TX) |
2006 |
Endocarditis Patient (Blood) |
This study |
| TX82 |
17 |
Yes |
USA (TX) |
1999 |
Endocarditis Patient (Blood) |
[25] |
| TX16 |
18 |
Yes |
USA (TX) |
1992 |
Endocarditis Patient (Blood) |
[43] |
| TX1330 | 107 | No | USA (TX) | 1994 | Healthy Volunteer Feces | [17] |
aHybrid genome with ~1/3 of the core genes from the CA clade and 2/3 from the HA clade.
bIndicates this information was not available.
cA rifampin- and fusidic acid-resistant derivative of clinical strain E. faecium D344S in which the spontaneous loss of pbp5 and its surrounding region resulted in an ampicillin-susceptible phenotype.
Figure 4.
Enterococcus faeciumphylogenetics. 4A. A maximum-likelihood phylogenetic tree using 628 core genes. Distance bar indicates the sequence divergence. Strains isolated from the community are labeled with branches in red. An asterisk (*) indicates a strain within the HA clade lacking IS16. 4B. A hierarchical clustering using Jaccard distance of gene content by unweighted pair group method with arithmetic mean (UPGMA) (see Materials and Methods). The core, distributed and unique gene counts are also presented in the right panel. 1:1 ortholog, orthologs present with one copy in all strains; N:N ortholog, orthologs present with multiple copies in all strains; N:M ortholog, orthologs present in some strains.
Comparison of E. faecium TX16’s predicted proteins to predicted proteins from the other 21 E. faecium genomes using BLASTP revealed a mosaic-like structure, as previously described [16,33], and many highly variable regions. Some of the TX16 variable regions are HA clade specific (Figure 5). Notably, regions from 27 to 38 kb, from 581 to 606 kb, from 702 to 717 kb, from 997 to 1,042 kb, from 1,737 to 1,802 kb and from 2,629 to 2,642 kb on the TX16 genome are missing or have low identity in the CA strains. Interestingly, region 1737 to 1802 kb encodes 4 surface proteins (HMPREF0351_11775, HMPREF0351_11776, and HMPREF0351_11777 which are the 3-gene pilus cluster, fms11-fms19-fms16 and HMPREF0351_11828 which is fms18, also known as EcbA, a collagen and fibrinogen binding MSCRAMM). Another notable region with low ORF identity hits or missing in strain D344SRF and TC6 is a ~145-kb region from 1,364 to 1,509 kb on the TX16 genome. Containing the pilus subunit protein EbpCfm (fms9) and other 2 pilus subunit proteins (EbpAfm and EbpBfm)(Figure 5).
Figure 5.
ORF comparisons of the 22E. faeciumgenomes. A circular map of BLASTP identity of predicted proteins from TX16 against the predicted proteins from other 21 E. faecium strains. Tracks from inside to outside: forward and reverse RNAs, reverse genes, foward genes, and genomic islands. In outer strain circles from inside to outside are the BLASTP precent identity of TX16 against ORFs from TX82, TX0133A, 1,141,733, 1,231,408, 1,231,501, 1,231,502, E1162, E1636, E1679, D344SRF, TC6, C68, E1071, 1,231,410, U0317, 1,230,933, Com12, Com15, E1039, E980, and TX1330. Red is 90–100% identity, purple is 60–89% identity, green is 0–59% identity.
Assessment of genomic rearrangements among E. faecium strains was more difficult because other genomes are not complete. We further investigated the genes that are unique to the HA-clade based on clade assignment of the strains in the phylogenetic analysis, and identified 378 ORFs (14% of TX16 ORFs) that are unique to the HA clade (shared at least between 2 HA clade isolates) (Additional file 3: Table S1). Of the 378 ORFs, 282 ORFs are conserved in at least half of the HA clade strains including 61 ORFs which are shared among all HA-clade isolates. Most of the HA clade unique genes are transposon-related genes, transporters, and prophage genes. Interestingly, a Cna B-type gene, the enterocin A operon, and two fms genes (see MSCRAMMs below) are among the HA-clade specific genes. Strain 1,231,408 was excluded from the HA unique gene analysis because it was previously shown to be a hybrid strain that contained both HA (~2/3) and CA (~1/3) alleles based on our 100 core gene analysis [33].
Mobile genetic elements
E. faecium isolates from patients typically have many mobile genetic elements which often contain antibiotic resistance genes that are easily transferable between strains. Bacteriophage-mediated transduction can transfer antibiotic resistance between enterococci [44,45] and many bacteriophages have also been identified in E. faecium[44]. To identify phage genes on the TX16 genome, Prophinder and Prophage Finder were used to search for prophage loci [46,47]. Both programs identified that two chromosomal regions (821–858 kb and 2,073–2,088 kb) with a total size of about 62 kb contain phage-related genes. Sixty-one and twenty one phage-related genes were identified in these regions, respectively (Additional file 4: Table S2). All CA strains have low identity ORF hits or missing ORFs in the predicted prophage locus from 821 to 857 kb, while most HA strains have similar ORFs in this locus. All CA strains and most HA strains lack similar ORFs in the other predicted prophage locus from 2,073 to 2,087 kb (Figure 5 and Additional file 3: Table S1). In addition to these two main regions, small numbers of phage-related genes were also identified throughout the chromosome, but these were not further analyzed.
IS elements and transposases are major mobile genetic elements in E. faecium and about 180 IS element and transposase-related genes were identified in the TX16 genome (Additional file 5: Table S3). About half of these IS elements and transposases are present on the three plasmids. Considering the sizes of the chromosome and three plasmids (chromosome, 2,698,137 bp; plasmid 1, 36,262 bp; plasmid 2; 66,247 bp; plasmid 3, 251,926 bp), plasmid DNAs appear to be more susceptible to IS element/transposase insertions. Some IS elements/transposases exist as multiple copies in specific locations on the chromosome or plasmids. Four copies of ISEnfa3 sequence (HMPREF0351_10172, HMPREF0351_10364, HMPREF0351_11866, and HMPREF0351_11868) were identified in the chromosome but not in the 3 TX16 plasmids whereas the sequences of IS1216 (HMPREF0351_12707, _12726, _12729, _12749, _12763, _12794, _12807, _12813, _12818), IS1297 (HMPREF0351_12910, _12920, _12891, _12875), and ISEfa4 (HMPREF0351_13111) were identified in the three plasmids but not in the chromosome. IS elements and transposases were found more frequently in HA strains than in CA strains. Previously, IS16 was suggested as a molecular screening marker to predict E. faecium pathogenicity because of its presence in clinical E. faecium isolates [31,48]. We performed a BLAST search of the 22 E. faecium genomes to identify the IS/transposase elements showing the same presence or absence patterns of IS16 (HMPREF0351_11812, _11855, _12352, and _12809). Many IS/transposase elements were found to have the same pattern of presence/absence in different strains as IS16; including ISEnfa3 (IS3/IS911 transposase: HMPREF0351_10172, _10364, _11866, and _11868), IS116/IS110/IS902 family transposases (HMPREF0351_11035, _11528, _12768, and _13088), IS66 transposases (HMPREF0351_10928, _11787, _11933, _12004, _12887, and_12948), and transposases (HMPREF0351_10878, _10880, _10927, _11934, and _12005). Therefore, all these IS elements and transposases (in addition to IS16) have potential as molecular markers to identify clinical E. faecium. However, these IS elements and transposases are not found in all HA-clade strains as 1,231,501; E1039; and E1071 do not have these IS elements and transposases, although they are present in all of the isolates considered to be part of the CC17 genogroup (Figure 4A).
Genomic islands
A pathogenicity island containing the esp gene has previously been reported in E. faecium[32,49]. The esp gene is not present in the TX16 genome but a search for other possible genomic islands (GIs) in TX16 using GI prediction programs including IslandPath-DIMOB [50], SIGI-HMM [51], and IslandPick [52,53], identified a total of 9 regions totaling 62,290 bp predicted as GIs. The GIs are shown in Figure 5, and the genes encoded by GIs are listed in Additional file 4: Table S2 and Additional file 6: Table S4. GIs 6, 7 and 8 might be a single GI, since they are located very close together. GIs 6 and 7 are separated by only 2 ORFs and 7 ORFs are present between GIs 7 and 8. The 9 predicted GIs have hypothetical proteins and transposon-related proteins in common. Among these putative GIs, islands 2, 3, 4, and 5 were frequently present in E. faecium of HA origin (data not shown). Island 2 contains 9 genes (6 genes encoding hypothetical proteins, and a predicted transposase and two transcriptional regulators). Island 3 contains 12 genes including 4 hypothetical proteins, 3 predicted ABC transport genes, a transposase, a Mg-dependent DNase, a LysM family protein, a cell wall protein, and a predicted fosfomycin resistance protein. Island 4 and 5 are composed of 7 and 9 genes, respectively. Island 4 contains 5 hypothetical proteins, a putative membrane protein, and a putative transposase. Four hypothetical proteins and 5 transposase related proteins were present in Island 5. The presence of a transposase in each island supports that these islands were acquired through horizontal gene transfer. While a potential role in pathogenesis has been suggested, there are many hypothetical proteins in each island and no genetic or experimental evidence to indicate such a role. However, island 3 which contains a predicted fosfomycin resistance protein might be important in promoting E. faecium colonization because of the selective advantage conferred when this antibiotic is used. The remaining GIs 1, 6, 7, 8, and 9 exist only in the TX16 genome or in a limited number of E. faecium strains.
We also searched for previously reported GIs [17,49] and pathogenicity islands [32] in the 22 E. faecium genomes. As reported [32], a pathogenicity island including the esp gene was observed in E1162; E1679; and U0317. In addition to these three strains, an island with a partial esp gene was also found in 1,231,502; C68; 1,231,410; TX0133A; and 1,230,933 strains when we performed a BLAST search. The esp gene could possibly be intact in these strains but interrupted in the draft assemblies, possibly as a consequence of the next-generation sequencing technology problems. A GI previously found to be specific to CC17 [49] was also observed in the HA clade strains TX0133A; TX82; C68; 1,231,410; 1,230,933; E1162; TX16; 1,231,502; U0317; and E1679. Intrestingly, 1,231,408, which is the mosaic strain [33], lacked this GI. The presence of a putative three-gene pilus-encoding cluster, fms11-fms19-fms16, previously proposed as a small GI [17], is described within the subsequent section on MSCRAMM-like proteins.
Genetic loci in E. faecium TX16 predicted to be involved in biosynthesis of surface polysaccharides
Our analysis of the E. faecium TX16 genome did not identify close homologs of the cpsC-K cluster of E. faecalis. Homologs of the two genes, cpsA and cpsB, were found and well conserved in TX16, but were recently reported to not be sufficient for capsule production in E. faecalis[54]. Similarly, homologs of cpsA-cpsB but not of cpsC-K were found in the 21 other E. faecium draft genomes.
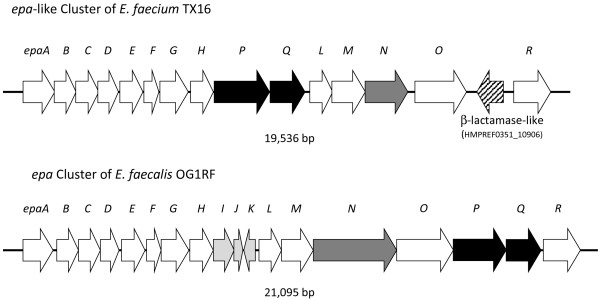
In contrast, a locus homologous to the epa locus, which was shown to produce a rhamnose, glucose, galactose, N-acetylgalactosamine and N-acetylglucosamine-containing antigenic cell wall polysaccharide in E. faecalis OG1RF[55,56], was found in the TX16 genome (Figure 6). However, identities of the encoded Epa-like proteins vary widely between orthologs of TX16 and OG1RF (ranging from 31% (EpaQ) to 92% (EpaE)). In addition, gene composition and order of the epa-like locus are partially different in these two organisms; the homologs of the three genes in the middle of the E. faecalis epa cluster, epaI, epaJ and epaK, are not present in TX16, while two other epa-like genes, epaP and epaQ are located at this site. All 15 epa-like genes of TX16 were found to be present, highly conserved and similarly organized in all 21 available E. faecium draft genomes (aa identities of the encoded proteins range from 88% to 100%), indicating that they are part of the core genome of this species. However, the absence of three epa genes in E. faecium, one encoding a glycosyl hydrolase (epaI), suggests the Epa polysaccharides of the two species have different sugar compositions.
Figure 6.
Comparison of the homologousepa-like loci ofE. faeciumTX16 andE. faecalisOG1RF. Orthologs of epaP and epaQ, located at different positions in the E. faecium and E. faecalis genomes, are indicated by black arrows. Genes epaI, epaJ and epaK, present only in E. faecalis, are indicated by light grey arrows. The epaN homolog of E. faecium, which is shorter than epaN of E. faecalis, is shown by a dark grey arrow. The TX16 ORF (HMPREF0351_10906) with relatively low similarity to the β-lactamase superfamily is shown by a hatched arrow. The epaA to epaR region of E. faecium TX16 corresponds to locus tags HMPREF0351_10891 to HMPREF0351_10907.
Genes encoding proteins predicted to be an initiating transferase of polysaccharide biosynthesis (undecaprenylphosphate sugar phosphotransferase), glycosyl transferases, acetyl transferases, sugar phosphate transferases and repeat unit polymerases are typically clustered together in loci that mediate polysaccharide synthesis in gram-positive bacteria. Our search for these features in the TX16 genome identified two additional regions that might be involved in polysaccharide production.
The first of these regions found in TX16 (Locus 4) is a downstream extension of the epa-like region (HMPREF0351_10908 - HMPREF0351_10923), immediately preceded by an undecaprenyl-phosphate galactose-phosphotransferase (encoded by epaR) (Additional file 7: Figure S3). Unlike the epa region, however, the extension (HMPREF0351_10908 - HMPREF0351_10923; Locus 4) is present in only 5 of the other E. faecium draft genomes; all except one of these strains (E980) belong to the HA clade . This Locus was also observed in these strains by Palmer et al. [34]. TX16 and these 5 draft genomes also have an additional ORF (HMPREF0351_10906 in TX16), encoding a putative member of the large beta-lactamase-like superfamily (Pfam PF00144, e = 9.4 × 10−17) between epaO and epaR on the upstream side of this region (Figure 6) and a transposase (HMPREF0351_10924) in 5 of the 6 genomes on its downstream side.
Analysis of the remaining 16 draft genomes for a corresponding region revealed a predicted polysaccharide-encoding gene cluster downstream of the epa region in all of them, (Locus 1, 2, and 3 also described by Palmer et al. [34]), although these regions have only low similarities to those of TX16 and the 5 genomes above and extensive sequence variation among each other (Additional file 7: Figure S3). Locus 3 (HMPREFD9522_ 02513–02504) was found in only HA clade strains, while Locus 1 (EFWG_01379-01370) and Locus 2 (HMPREF0352_0048-0457), although found in some HA-clade strains, were only found in non-CC17 isolates as well as in four of the five CA-clade isolates, indicating some specificity of polysaccharide biosynthesis genes for certain lineages or niches. Of note, none of Locus 2 strains have IS16, only two of the Locus 1 strains have IS16, while all that had Locus 3 or 4 have IS16.
The second region found in TX16 that appears likely to be involved in polysaccharide biosynthesis (HMPREF0351_11938 - HMPREF0351_11970) is largely unique to this genome, with only the first four ORFs present in 20 of the genomes and the whole region completely absent in one of the genomes (E1039). However, each of the 20 other genomes has additional genes predicted to be involved in polysaccharide biosynthesis at this location.
Distribution of genes encoding MSCRAMM-like proteins, putative virulence genes, antibiotic resistance determinants, and CRISPRs
Previous studies of E. faecium TX16 identified 15 genes encoding LPXTG family cell-wall anchored proteins with MSCRAMM-like features, such as immunoglobulin-like folding; 11 of these were found in four gene clusters, each predicted/demonstrated to encode a different pilus, and four were found as individual MSCRAMM-encoding genes [18,21,22]. Our search for these genes in 21 unique E. faecium draft genomes in this study found all of the MSCRAMM-encoding genes to be widely distributed except fms18 (ecbA) and fms15 which were only in HA-clade isolates (although some are present as variants or pseudogenes within the HA-clade) (Additional file 8: Table S5). Moreover, our analysis revealed that ebpA-ebpB-ebpCfmfms14-fms17-fms13fms20, scm, and fms18 (the latter present in only HA isolates) all have sequence variants in some of the 21 strains, with identities of the encoded variant proteins ranging from 39% (fms20 homolog) to 94% (ebpC) versus their counterparts in TX16 (Additional file 8: Table S5). In general, most of the MSCRAMMS followed the CA/HA clade groupings with a variant representing each clade. Variant 1 of the fms11-fms19-fms16 locus was strictly found in the HA-clade, and variant 2 in the CA-clade except for 1,231,501 which only had one of the three proteins (fms16) as a CA-variant, suggesting recombination by this isolate. Variant 1 of fms14-fms17-fms13 was found in all but one HA clade isolate (1,231,408, a hybrid of HA and CA clades, has variant 2) and variant 2 in all 5 CA-clade strains. Variant 1 of scm was found to be exclusively carried by all 16 HA clade strains and variant 2 by 4 of the 5 CA clade strains. Although the differences between these MSCRAMMs in CA- vs. HA-clade strains are generally greater (ranging from 2 to 27% with an average of 10%) than the differences (3–4%) previously reported for the clade-specific differences in a set of core genes that excluded predicted surface proteins, they are comparable to the differences seen in several other surface proteins that have been studied [33,57].
Interestingly, the majority of HA clade strains (12/16, including TX16) were found to have variant 1 of the ebp pilus operon, while variant 2 was exclusively found in the 5 CA-clade strains in addition to variant 1 in three of the five isolates. In contrast, variation within fms20 was restricted to the HA clade; all CA clade isolates carried fms20 variant 1, but the percent identity between these two variants is much smaller (39%), possibly indicating the need for a new gene name. Also of note was the acm gene, which is present as a pseudogene in all of the CA-clade isolates except 1,141,733 which is the only CA-clade isolate that is from a hospitalized patient; acm pseudogenes were also found in non-CC17 HA-clade isolates.
Of note, our search for MSCRAMMs and potential pilus proteins also found one to three new individually encoded CnaB domain-containing MSCRAMMs in five of the E. faecium draft genomes and a new pilus encoding gene cluster in strain E1071; the latter consists of three genes one of which is a relatively distant homolog of bee1 (35% aa identity) and two are identical or highly homologous to bee2 or bee3 (100% and 98%, respectively) of a plasmid-encoded bee pilus gene cluster found in a small percentage of E. faecalis isolates [58].
To identify possible virulence genes in the E. faecium genomes, the enterococcal virulence factors listed in the Virulence Factors Database (VFDB) [59] were aligned to the ORF protein sequences using BLASTP and filtered with 50% identity and 50% match length. The homologs of efaA, EF0954 (a homolog of BopD which is a transcriptional regulator involved in biofilm production of E. faecalis[42,60] ), cpsA and cpsB genes are present in all E. faecium strains (see surface polysaccharides above for cpsA and cpsB), and espEfm and hylEfm are exclusively present in some HA clade strains while the homolog of EF0818 (a putative hyaluronidase and annotated as a Family 8 polysaccharide lyase, also similar to the LPXTG protein EF3023) is exclusively present in the CA-clade strains (except strain 1,141,733). Homologs of other E. faecalis virulence factors listed in the VFDB were not found in TX16 genome.
We also searched the 22 E. faecium isolates for the presence and absence of 13 resistance genes. Our data correspond to previously published data for some of the isolates [32,61]. We observed that there is a clear distinction between the isolates of the genetically defined CA clade and those of the HA clade with none of the CA clade isolates having any of the antibiotic resistance determinants analyzed (Table 3). On the other hand, all of the HA-clade isolates have multiple resistance determinants, including the pbp5-R allele that confers ampicillin resistance previously reported by Galloway-Pena et al. [57], except for strains 1,231,501 and E1039. 1,231,501, which is in the HA-clade but lacks all antibiotic resistances including pbp5-R, may have lost the allele via recombination and acquired pbp5-S or may even represent a more ancestral isolate. Indeed, 1,231,501 was shown to be a hybrid of HA and CA genomes by Palmer, et al., with the replacement (hybrid) region including pbp5-S, which could explain the origin of pbp5-S in this strain [34]. E1039, which has the pbp5-R allele but none of the other resistance genes, is genetically defined as a HA-clade isolate, but came from a healthy volunteer, perhaps explaining its lack of other antibiotic resistances. Interestingly, neither of these strains has IS16. D344SRF is the only other HA-clade isolate that lacks the pbp5-R allele; however, this strain is known to have spontaneously lost pbp5 and the surrounding region and contains many other resistances [62]. Of note, E1636 only has two of the 13 resistances analyzed (tetM and pbp5-R); however, this could possibly be explained by its early isolation in 1961. This again suggests that these isolates are more distantly related to the other strains within the HA-clade.
Table 3.
Antibiotic resistance gene profiles of the 21E. faeciumstrains
|
Gene |
cat |
ermA |
ermB |
aad6 |
aad9 |
aadE |
aacA- aphD |
tetL |
tetM |
vanA |
gyrAb |
parCc |
pbp5-Rd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistance | CHL | ERY | ERY | SPC/ STR | SPC/ STR | SPC/ STR | GEN | TET | TET | VAN | CIP | CIP | AMP |
| Strains |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1,141,733 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Com12 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Com15 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| E980 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| TX1330 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1,230,933 |
|
|
X |
X |
|
X |
X |
|
X |
X |
X |
X |
X |
| 1,231,408 |
|
|
X |
X |
|
X |
X |
|
|
|
X |
X |
X |
| 1,231,410 |
|
|
X |
X |
|
X |
|
|
|
X |
X |
|
X |
| 1,231,501 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1,231,502 |
|
|
X |
X |
|
X |
X |
|
|
X |
X |
X |
X |
| C68 |
|
|
X |
X |
|
X |
X |
|
X |
|
X |
X |
X |
| D344SRFa |
|
|
X |
X |
|
X |
|
X |
X |
|
|
|
|
| TX16 |
X |
|
X |
X |
|
X |
|
X |
X |
|
|
|
X |
| E1039 |
|
|
|
|
|
|
|
|
|
|
|
|
X |
| E1071 |
X |
|
X |
X |
X |
X |
|
X |
|
X |
|
|
X |
| E1162 |
|
|
|
|
|
|
|
X |
X |
|
|
|
X |
| E1636 |
|
|
|
|
|
|
|
|
X |
|
|
|
X |
| E1679 |
|
X |
X |
X |
X |
|
X |
|
|
X |
X |
X |
X |
| TX82 |
|
|
X |
X |
|
X |
|
|
X |
X |
X |
X |
X |
| TX0133A |
X |
|
X |
X |
|
X |
X |
|
X |
|
X |
X |
X |
| U0317 | X | X | X | X | X | X | X |
aA rifampin- and fusidic acid-resistant derivative of clinical strain E. faecium D344S in which the spontaneous loss of pbp5 and its surrounding region resulted in an ampicillin-susceptible phenotype.
bAmino acid change (E to K/G) in residue 87 or (S to R/Y/I) in residue 83 of GyrA.
cAmino acid change (E to K) in residue 86 or (S to R/I) in residue 82 of ParC.
dConsensus sequence of the pbp5-R allele encoding the low affinity Pbp5-R.
eTC6 was not included in this analysis as it is a transconjugant of C68 and D344SRF, so therefore is not a unique genome.
Two groups have previously analyzed CRISPR-associated genes within E. faecalis and E. faecium genomes [32,61]. Partial CRISPR-like loci were previously described in E1071, E1679, and U0317; however, these loci were within a gene and were considered non-functional [32]. In addition, Palmer et al. identified CRISPR-cas predicted proteins in the Broad Institute strains Com12; 1,141,733; and 1,231,408 [61]. Similarly, we only found a CRISPR-cas locus in strain TX1330 (Additional file 9: Table S6) out of the 6 strains not previously studied (TX1330; TX16; TX0082; TX0133A; D344SRF; and C68). In summary, out of the 22 available genomes, only one of the HA-clade isolates contained CRISP-loci, namely the hybrid strain 1,231,408. The three other strains containing CRISPR-loci of the CA-clade (Com12; 1,141,733; and TX1330) all lacked antibiotic resistance determinants. Therefore, our data coincide with the previous observation that members of the recently emerged high-risk enterococcal lineages lack CRISPR-loci and the inverse relationship between the presence of a CRISPR-cas locus and acquired antibiotic resistance [61].
Metabolic pathway
Metabolic pathways of E. faecium might have contributed to the recently increased incidence of E. faecium colonization and infection. To help understand E. faecium metabolism, the KEGG pathway (with EC number) and KAAS (with amino acid sequences) databases were used. Both databases predicted more than 100 pathways using TX16 genomic information. E. faecium exhibits major genomic differences in the genes involved in energy metabolism compared to that of other facultative anaerobic bacteria. However, like other species in the Lactobacillaceae order, genes for typical aerobic energy (ATP) generation through the TCA cycle and electron transport chain do not exist, i.e., genes encoding complex I (NADH dehydrogenase), II (succinate dehydrogenase,), III (cytochrome bc1 complex), and IV (cytochrome c oxidase).
When we compared the metabolic pathways of TX16 to those of E. faecalis V583 using the KEGG database, all 82 metabolic pathways of E. faecalis were also predicted in TX16. Indeed, more diverse metabolic activities were observed in TX16 (Additional file 10: Table S7 and Additional file 11: Table S8). Additional files 10: Table S7 and Additional files 11: Table S8 show lists of enzymes that only exist in E. faecium TX16 or E. faecalis V583 when KEGG enzymes from both strains were compared. Many of these enzymes were also described by van Schaik et al. who compared 7 European strains (also included in this study) to E. faecalis V583. They found 70 COGs present in their E. faecium genomes lacking in V583, whereas we found 176 predicted enzymes present in TX16 lacking in E. faecalis V583 according to KEGG analysis. Additionally, they found 140 COGs specific for E. faecalis V583, compared to the European strains, whereas we found only 112 enzymes specific to V583 when compared to TX16 according to KEGG analysis [32].
Plasmids
Alignment of ORFs from the three plasmids of TX16 to the ORFs from the other 21 E. faecium genomes by BLASTP showed that all strains shared some ORFs that are similar to the ORFs of the three E. faecium TX16 plasmids (pDO1, pDO2 and pDO3), but none of them have more than 90% of the ORFs from any of the plasmids. It is likely that some strains may have similar but not identical plasmids as TX16, but identification of plasmids in other strains is difficult since those genomes are draft sequences. Alignment of ORFs of the three TX16 plasmids to 22 complete E. faecium plasmid sequences available in NCBI using TBLASTN with 90% identity and 50% match length cutoffs showed that pDO1 is most similar to plasmid pM7M2, a 19.5 kb plasmid which shared 27 ORFs of the 43 ORFs (62.8%) from pDO1, and that pDO2 is somewhat similar to plasmids pRUM and pS177 with 44.7% and 41.2% match to pDO2 ORFs respectively. TX16 plasmid pDO3 does not seem to be similar to any completely sequenced E. faecium plasmids but has similarity to the partially sequenced E. faecium large plasmid pLG1, Both pDO3 and pLG1plasmids harbor the hyaluronidase gene (hylEfm), The hylEfm gene was also found in HA strains 1,230,933, 1,231,410, 1,231,502, C68, TC6 and U0317.
Discussion
TX16 was the first E. faecium strain sequenced and has been used in various studies since [26,28,63,64]. The TX16 genome is characterized by numerous hyper variant loci and a large number of IS elements and transposons. Ortholog analysis as well as core and pan-genome analysis of TX16 and the other 21 sequenced strains revealed that E. faecium genomes are highly heterogeneous in gene content and possess a large number of dispensable genes. Similar to the findings by van Schaik et al. [32], pan and core genome analysis predict the pan genome to be open. Phylogenetic analysis using single-copy orthologs of the same length and gene content dissimilarity analysis in addition to recent studies [33,57] looking at core genes, SNPs and 16S rRNA, all indicate a large divergence between CA-clade isolates and HA-clade isolates. Furthermore, our previous analysis [33,57] and analyses within this study show that CC17 genogroup isolates cluster more closely together and further away from the CA-clade isolates than the other non-CC17 HA-clade isolates, indicating the CC17 genogroup is a more recently evolved genogroup.
Genomic island analysis by codon usage bias and composition variation showed that TX16 has 9 GIs, although TX16 also possesses a large number of hyper variant loci, suggesting that most of the genomic variable loci in TX16 were acquired through lateral gene transfer, possibly through mobile elements such as transposons. In general, strains in the HA clade harbored more transposons than the CA strains and certain IS elements such as IS16. These findings are consistent with a previous study using whole genome microarray [31].
Although IS16 presence has been proposed as an indicator of hospital-associated strains such as those apart of the CC17 genogroup [48], IS16 was not found in all HA-clade strains. Of note, however, all HA-clade strains contained the pbp5-R allele (except for 1,231,501 and D344SRF which is a spontaneous deletion mutant of pbp5) which may indicate that this is a reliable marker for hospital-associated isolates. Indeed, the pbp5-R allele is also found in animal and community isolates that are considered within the HA-clade, but not considered clinically associated [35,36]. The exception, 1,231,501 is interesting in that it is the HA-clade isolate from the blood of a hospitalized patient with no resistance genes, possibly supporting the concept that the genomic content of a strain, not just antibiotic resistance, adds to the survival in the hospital environment. In the 100 gene analysis by Galloway-Pena et al., it was found that 5 of the 92 genes of this strain studied grouped with the community clade, indicating it is a hybrid strain [33] as also reported in a recent study [34].
Capsular and other cell envelope polysaccharides of several gram-positive bacteria are known to have important roles in virulence and protective immunity [65-67]. Although the majority of studies on enterococcal surface polysaccharides have focused on E. faecalis, similar molecules have also been identified in E. faecium and suggested as targets for opsonic antibodies and as potential vaccine candidates [43,68], and also implicated in resistance of TX16 to phagocytosis in normal human serum [63]. Two such gene clusters, cps and epa, have been identified in E. faecalis[55,56,69,70]. Although a 7-9-gene cps region (cpsC to cpsK) was recently determined necessary for the production of an E. faecalis capsular polysaccharide [54] and shown to contribute to pathogenesis and evasion of the host innate immune response [67,69], TX16 only contains two homologs of the genes in this locus (cpsA-cpsB)[54]. In contrast, 15 of the 18 E. faecalis epa polysaccharide genes have homologs in TX16 and the other 21 E. faecium genomes, although their sequences vary between the two species. Therefore, it is likely that E. faecalis and E. faecium produce compositionally related, but not identical, Epa surface polysaccharides.
The hyper variable nature of the two polysaccharide loci found in TX16 raises the possibility that they are involved in biosynthesis of antigenically diverse surface polysaccharides which could help protect E. faecium against host immune responses. Similar to other gram-positive bacteria, various MSCRAMM-like cell wall anchored proteins have been previously identified in E. faecium; these include the collagen adhesin Acm and biofilm-associated Ebp pili, shown to be important for endocarditis and UTI in animal models [26,71], respectively, as well as two other collagen-binding MSCRAMMs, Scm and Fms18 (EcbA) [21,72]. Our comparison of 15 previously described MSCRAMM and pilus encoding genes of TX16 [17,18,21] with those of 21 E. faecium draft genomes found them to be common among these strains and the majority of them (12/15) to be enriched among HA clade strains or have a sequence variant mostly/exclusively carried by CA clade strains. Thus, these findings agree with previous hybridization results [14,16,17,22] and with the presence of two distinct subpopulations of E. faecium. Furthermore, one of these genes, acm, was previously found to be expressed more often by clinical versus non-clinical isolates, whereas a pseudogene was often found in isolates from the community [26,64]. Taken together, these data indicate a clear difference in the MSCRAMM and pilus gene profiles of the HA and CA clades, suggesting that these genes may have favored the emergence of HA-clade E. faecium in nosocomial infections.
When we combined our finding with previously published results, four of the 21 E. faecium genomes contain the CRISPR-cas locus. Three of these strains are within the CA clade and lack all antibiotic resistances analyzed in this study. One of the strains, 1,231,408, is a unique strain in which its genome is a hybrid of CA and HA genes. However, it does have 8 antibiotic resistance associated genes, showing there is not always an inverse relation between the number of antibiotic resistance determinants and the presence of CRISPR loci. More strains containing CRISPR-loci will need to be studied in order to determine if 1,231,408 is just an exception to the rule, or if the highly recombinant nature of E. faecium makes it different from E. faecalis with respect to the presence of CRISPR-loci in relation to antibiotic resistance determinants.
Overall, there seem to be some patterns that point to specific evolutionary events throughout E. faecium’s history as a species. First and foremost, there is a large ancestral split between the CA- and HA-clade strains which are separated by at least a 3–4% difference in their core genome [33]. The CA-clade isolates, except one, do not have either polysaccharide synthesis Locus 3 or 4 downstream of the epa region, antibiotic resistance genes, certain genomic islands, or IS elements. After the HA-clade diverged from CA-clade there was further evolution within the HA clade and some HA-clade strains studied here may represent phylogenetic transitional lineages (Figure 4B and C). Like the CA-clade strains, these transitional lineages are characterized by a lack of IS16 (E1039; 1,231,501; and E1071) and have neither Locus 3 nor 4 (E1039; 1,231,501; E1071; E1636; E1679) in the epa extension. Although the data are limited, one scenario that could explain these observations is if Locus 1 replaced Locus 2 in a HA-clade ancestral strain, after the split from the CA clade, which later acquired IS16 and then, subsequently, Locus 3 or 4 replaced Locus 1 in the epa extension region. Even if this is not the case, it seems clear that only strains further along in the phylogenetic trees, indicating a division within the HA-clade (Figure 4A and B), acquired IS16 and the polysaccharide biosynthesis Loci 3 and 4. The exception is E980, a strain previously shown to have 8 of 92 genes from the HA-clade, which could have gained Locus 4 via recombination. Also of note, three of the four strains that have Locus 1 downstream of the epa locus lack the ebp genes, possibly suggesting there may have been some kind of gain and loss through homologous recombination.
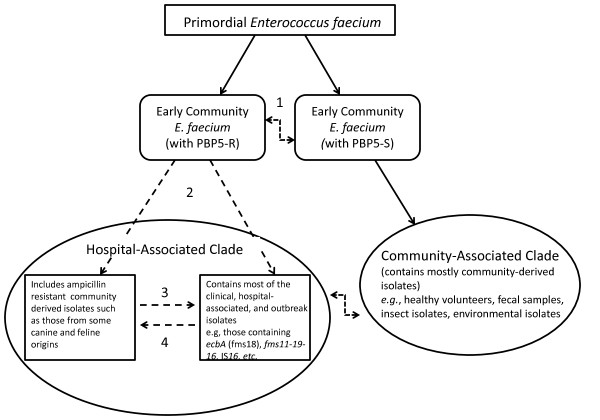
Figure 7 shows the projected scenarios for the evolution of the two clades of E. faecium as can be envisioned using our data as well as other previous publications [31,33,34,57]. The hypothesis is that there was a primordial type of E. faecium which split many millinea ago and evolved into two early community groups which had homologous genes e.g. the pbp5-S or pbp5-R alleles, the latter representing community sources of ARE (ampicillin resistant E. faecium). These lineages could recombine with each other resulting in hybrid strains (i.e. 1,231,408 and 1,231,501) (scenario 1). The divergence between the two community groups eventually reached a core genomic difference of approximately 3–4%, creating a HA clade, which includes both ampicillin- resistant, community-based isolates, such as those from some canine and feline origins, as well as most of the clinical-, hospital- and outbreak- associated isolates and a CA clade, which consists mostly of community derived isolates. Most likely, community and hospital ARE isolates split from the same ancestor, as represented by scenario two. However, it is also possible that ARE clones evolved from the animal reservoir (scenario 3), or that animal ARE isolates represent evolutionary descendants of hospital ARE transferred from humans to their pets (scenario 4).
Figure 7.
The projected evolution of the two clades ofE. faecium. A figure addressing the possible scenarios which may have occurred in the evolution of Enterococcus faecium resulting in the HA-clade and CA-clade. Specifically, a primordial type of Enterococcus faecium split into early community isolates which had homologous core genomes with significant sequence differences (e.g., the pbp5-S or pbp5-R allele). These early community groups further segmented into a hospital-associated clade and the community clade. Scenario one depicts that these lineages could recombine with each other (represented by the bent dashed arrow) resulting in hybrid strains, scenario two depicts community and hospital ARE isolates splitting from the same ancestor, scenario three depicts ARE clones evolving from the animal reservoir, and scenario four depicts animal ARE isolates representing descendants of hospital ARE transferred from humans to their pets.
Conclusions
In conclusion, the completion of the TX16 genome has provided insight into the intricate genomic features of E. faecium, and will surely serve as an important reference for those studying E. faecium genomics in the future. By studying TX16, an endocarditis isolate belonging to CC17, and comparing the TX16 genome to the other 21 draft genomes, we have been able to confirm the high genomic plasticity of this organism. The HA-clade isolates contain a number of unique IS elements, transposons, phages, plasmids, genomic islands, and inherent and acquired antibiotic resistance determinants, most likely contributing to the emergence of this organism in the hospital environment that has occurred in the last 30 years.
Methods
Bacterial strains and DNA sequencing
The E. faecium strain TX16 (DO) was isolated from the blood of a patient with endocarditis [63] and E. faecium TX1330 was isolated from the stool of a healthy volunteer [18,73]. Routine bacterial growth was on BHI agar or broth, and genomic DNA was isolated from overnight culture using the method previously described [74].
Both E. faecium TX16 and TX1330 were sequenced, assembled and annotated as part of the reference genome project in the Human Microbiome Project (HMP). E. faecium TX16 was initially sequenced by traditional Sanger sequencing technology to 15.6x read sequence coverage, and subsequently by 454 GS20 technology to 11x read sequence coverage of fragment reads, 7.5x sequence coverage of 2 kb insert paired end reads, and by 454 FLX platform to 73x sequence coverage of 8 kb insert paired-end reads. Both Sanger and 454 reads were assembled using 454 Newbler assembler. The gaps between contigs in scaffolds were closed using the unassembled mate paired reads or by PCR sequencing of the DNA products amplified from the primers flanking the gaps. The assembly and gap closure of TX16 was difficult due to large number of repetitive sequences in the genome. The addition of the large insert 8 kb library with deep clone coverage was able to facilitate the assembly and scaffolding to generate high quality contigs and scaffolds in the de novo assembly. E. faecium strain TX1330 was sequenced by 454 GS20 technology to 6x sequence coverage for fragment reads and by 454 FLX to 69.8x sequence coverage for paired end reads, respectively. TX1330 was also assembled using 454 Newbler assembler.
Plasmids were identified by circularization of DNA sequences by paired end reads, and were also experimentally verified by PFGE analysis of SmaI and ApaI digested genomic DNA followed by hybridization with PCR-generated probes complementary to 5′ and 3′ ends of plasmid contigs. PFGE hybridization profiles were then compared to identify neighboring plasmid contigs.
The gene prediction for both E. faecium TX16 and TX1330 was accomplished by Glimmer 3 [75] and GeneMark [76]. tRNAScan [77] was used for tRNA prediction, RNAmmer [78] for rRNA prediction, and RFAM/infernal for other non-coding RNA genes [79]. Manual annotation was facilitated by Genboree genome browser (http://www.genboree.org). Conserved protein domains were searched using Pfam [80], COG [81], and InterProScan [82]. Other tools such as PsortB [83,84], ExPASy ENZYME [85], and the Transport Classification Database [86] were also used to facilitate the annotation. For manual annotation, each entry was annotated by two annotators independently and the differences were reconciliated at the end of the annotation.
Genomic sequences and annotations for 20 other draft E. faecium strains, including 1,141,733; 1,230,933; 1,231,408; 1,231,410; 1,231,501; 1,231,502; C68; Com12; Com15; D344SRF; E1039; E1071; E1162; E1636; E1679; E980; TC6; TX82; TX0133A; U0317, were obtained from NCBI. A complete list of the strains and their clinical sources is provided in Table 2.
Genome characterization
DNA and protein sequence alignments were performed using BLASTN and BLASTP [87], respectively, unless otherwise stated. Prophage loci were identified using both Prophinder program [47] and Prophage Finder [46]. Prophinder uses BLASTP to search phage proteins in the ACLAME database while Prophage Finder uses BLASTX to search input DNA sequence to an NCBI database of phage genomes. Possible prophage loci were also reviewed manually. IslandViewer [52] server was used to analyze possible genomic islands on the chromosome. IslandViewer integrated sequence composition based genomic island prediction programs including IslandPath-DIMOB [50] and SIGI-HMM [51] as well as comparative genome based program IslandPick [53] for genomic island prediction. Genes and DNA sequence in the identified genomic regions were used to perform the BLAST search against the other 21 E. faecium genomes to investigate the presence or absence of clade specific genomic islands. Repeat sequences were identified by RepeatScout [88]. Circular genome maps were generated using the CGView program [89].
BLASTN and BLASTX as well as ISfinder server [90] were used to identify IS sequences and transposons in the TX16 chromosome and plasmids. Genomic regions with homology to IS and transposon sequences from both BLAST analyses were verified with the gene annotation of TX16. Both BLAST searches identified many small regions as a part of IS elements and transposons. Regions with shorter than 60% match length to reference sequences were excluded from further analysis. Identified genes/regions by analyses above were also used to perform the BLAST search against the other 21 E. faecium genomes to investigate whether there are clade specific presences or absences.
Chromosomal DNA sequences of TX16 and Aus0004 were aligned using Mauve 2.3.1 and performed a comparative genomic analysis [91,92]. Junction sites of 5 locally collinear blocks (LCB) of Mauve alignment were further investigated with genome annotation to identify possible reasons of two inversions and DNA insertions.
Six genomes that had yet to be studied for CRISPR-loci were analyzed for CRISPR loci (TX1330, TX16, TX82, TX0133A, D344SRF, and C68). We searched for CRISPR loci in the six genomes by performing BLAST using the sequences from the ORFs previously described for CRISPR-loci in E. faecium EFVG_01551 to EFVG_01555 [61], as well as using CRISPRfinder (http://crispr.u-psud.fr/Server/CRISPRfinder.php) and the CRT program [93] to detect prophage CRISPR palindromic repeats in TX16.
Conserved gene orders between E. faecium TX16, E. faecalis V583 [41] and E. faecalis OG1RF genomes [40] were identified using BLASTP with E value of 1e-3 and DAGchainer with default parameters [39].
The extrapolation of core-genome and pan-genome was performed as described previously [94,95]. ORF protein sequences were aligned using BLASTP, and a gene pair was considered present in two strains if the alignment covered at least 50% length of the shorter gene with at least 70% sequence identity. Due to the large number of possible combinations of 22 strains, only 100 permutations were performed for each nth genome.
Metabolic pathways of the TX16 genome were analyzed with enzyme commission (EC) numbers as well as with the predicted amino acid sequences of all TX16 ORFs. 528 unique EC numbers of TX16 genome are analyzed at the KEGG server (http://www.genome.jp/kegg/pathway.html) to predict the metabolic pathway. Also, KEGG automatic annotation server (http://www.genome.ad.jp/kaas-bin/kaas_main) was used for functional annotation of the TX16 ORFs. Metabolic pathways and enzymes identified from TX16 were compared to that of E. faecalis V583 (KEGG genome T00123) in KEGG pathway database.
Ortholog, phylogenetic and multi-locus sequence typing (MLST) analysis
Protein ortholog groups of E. faecium genomes were identified using OrthoMCL program [96] using BLASTP E value of 1e-5 and default MCL inflation parameter of 1.5 with 80% sequence identity and 60% match length cutoffs. The match length percentage was set relatively low because all the genomes except TX16 are draft sequences. The dissimilarity in gene content among the E. faecium genomes was calculated using Jaccard distance (1- Jaccard coefficient) as described previously [97], and the Jaccard distance matrix was used for hierarchical clustering using the unweighted pair group method with arithmetic mean (UPGMA). Single-copy orthologs with the same length in all strains were chosen for phylogenetic analysis after removing genes that may have undergone recombination detected by PHI program [98]. Multiple sequence alignments were performed by MAFFT program [99] and the topology of the phylogenetic tree was inferred by maximum-likelihood algorithm using PhyML [100] with bootstrap value of 100. 16S rRNA phylogenetic analysis was performed in another manuscript [33]. iTOL program [101] was used for phylogenetic tree visualization.
The in silico multi-locus sequence types were determined either by extracting the allele types of adkatpAddlgdhgydpstS, and purK from the genomic sequence, or using the allele numbers previously obtained through experimentation [57]. The allele numbers and sequence types were used to construct an UPGMA dendogram using S.T.A.R.T.2 software (http://pubmlst.org/).
Identification of putative virulence-associated genes and antibiotic resistance determinants
Putative virulence genes were identified by BLASTP of E. faecium ORF protein sequences to the enterococcal virulence factors in the Virulence Factors Database (VFDB) [59], and hits were manually inspected.
To identify antibiotic resistance genes, BLASTN was performed using the nucleotide sequences of 13 antibiotic resistance genes including cat (chloramphenicol O-acetyltransferase) using the EfmE1071_2206 sequence which is an ortholog to the cat gene found on the E. faecium plasmid pRUM [102]ermA (rRNA adenine N-6-methyltransferase) using the EfmE1679_0214 sequence and located on Tn554 [103]; ermB (rRNA adenine N-6-methyltransferase) using the EfmE1071_2296 sequence, an ortholog to the ermB gene found on the E. faecalis plasmids pRE25 and pSL1[104]; aad6 (aminoglycoside 6-adenylyltransferase) using the EfmE1071_1021 sequence an ortholog to the genes found on the E. faecalis plasmid pEF418 (Genbank:AF408195); aad9 (streptomycin 3″-adenylyltransferase) using EfmE1679_0213 sequence and located on Tn554[103]; aadE (aminoglycoside 6-adenylyltransferase) using EfmU0317_2169 sequence an ortholog to the gene found on the E. faecalis plasmid pRE25 [104]; aacA-aphD (bifunctional aminoglycoside modifying enzyme) using the EfmU0317_2161 sequence; tetL using the EfmE1071_1017 sequence [105]; tetM using the EfmE1162_0404 sequence [105]; vanA using the EfmE1071_0104 to EfmE1071_0110 sequence which is identical to the vanA gene cluster found on Tn1546[106]; gyrA using EfmE1679_2520 to determine amino acid changes of E87K/G or S83R/Y/I [107]; parC using EfmE1679_0369 to determine amino acid changes of E86K or S82R/I [107]; and pbp5 (GenBank accession no. ZP_00603984) to search for the low-affinity pbp5 consensus sequence [57,108].
Database submission
The genome sequences, plasmid sequences, and the gene annotation of E. faecium TX16, pDO1, pDO2, and pDO3, were submitted to GenBank with the accession numbers of CP003583, CP003584, CP003585, and CP003586 respectively. The draft sequence of TX1330 was submitted to GenBank with the accession number ACHL01000000.
Authors’ contributions
XQ carried out the annotations, genome characterization, genome analyses, closure of the genome and drafting of the manuscript. JGP carried out annotations, phylogenetic, antibiotic resistance, and CRISPR analyses, and writing /submission of the manuscript. JS carried out the annotations, genome, MSCRAMM, virulence genes, and polysaccharide biosynthesis analyses, and drafting of the manuscript. JHR carried out metabolic pathway, genomic island, and mobile element analyses and drafting of the manuscript. The rest of the authors contributed though annotating or sequencing of the genome. GMW and BEM contributed their study design, overseeing the study, and editing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1.Gene order synteny ofE. faeciumTX16 compared toE. faecalisV583. A figure ploting the synteny blocks between TX16 and V583 with the coordinates of each genome.
Figure S2.Genome alignment of TX16 and Aus0004. A figure comparing the two closed E. faecium genomes sequences available using Mauve genome alignment analysis.
Table S1.Hospital-associated clade unique genes. A table listing the genes and their corresponding ORF in TX16 that are unique to the hospital clade and how many of the HA clade strains the gene is present in.
Table S2.Prophage loci and genes onE. faeciumTX16 genome. A table listing the two prophage loci, the predicted gene products within these two loci, and the corresponding ORFs in TX16.
Table S3.Mobile elements in theE. faeciumTX16 genome. A table listing all of the predicted mobile elements and their corresponding locus tags in TX16.
Table S4.E. faeciumTX16 genomic islands and genes. A table listing the nine genomic islands, the genes and predicted products within those islands, and the corresponding ORFs and coordinates within TX16.
Figure S3.ORF composition of the downstream extension of theepagene cluster in the 22E. faeciumgenomes (HMPREF0351_10908 - HMPREF0351_10923 in TX16). A figure depicting the predicted polysaccharide-encoding gene clusters found in the E. faecium genomes.
Table S5.Presence of genes encoding MSCRAMMs and pilins among 21E. faeciumgenomes. A table listing the different MSCRAMM and pilin variants present in each of the 22 genomes.
Table S6.Summary of CRISPRs found inE. faeciumsequenced strains. A table listing in what strains CRISPRs were found, the locus tag, and the functional assignment.
Table S7.Specific enzymes present in TX16 but not inE. faecalisV583. A table listing enzymes, KEGG information, and locus tags specific to TX16.
Table S8.Specific enzymes present inE. faecalis V583 but not in TX16. A table listing the enzymes and locus tags specific to V583.
Contributor Information
Xiang Qin, Email: xqin@bcm.edu.
Jessica R Galloway-Peña, Email: Jessica.Galloway-Pena@uth.tmc.edu.
Jouko Sillanpaa, Email: Jouko.Sillanpaa@uth.tmc.edu.
Jung Hyeob Roh, Email: Jung.H.Roh@uth.tmc.edu.
Sreedhar R Nallapareddy, Email: Sreedhar.R.Nallapareddy@uth.tmc.edu.
Shahreen Chowdhury, Email: shahreenachowdhury@gmail.com.
Agathe Bourgogne, Email: agathe.bourgogne@gmail.com.
Tina Choudhury, Email: Tina_Choudhury16@yahoo.com.
Donna M Muzny, Email: donnam@bcm.edu.
Christian J Buhay, Email: cbuhay@bcm.edu.
Yan Ding, Email: yding@bcm.edu.
Shannon Dugan-Rocha, Email: sdugan@bcm.edu.
Wen Liu, Email: wliu@bcm.edu.
Christie Kovar, Email: ckovar@bcm.edu.
Erica Sodergren, Email: esodergr@wustl.edu.
Sarah Highlander, Email: sarahh@bcm.edu.
Joseph F Petrosino, Email: jpetrosi@bcm.edu.
Kim C Worley, Email: kworley@bcm.edu.
Richard A Gibbs, Email: agibbs@bcm.edu.
George M Weinstock, Email: geowei@mac.com.
Barbara E Murray, Email: bem.asst@uth.tmc.edu.
Acknowledgments
This work was partially supported by NIH/NHGRI grant 1U54HG004973-0 and NIH/NIAID grants R01 AI42399 and R01 AI067861. JGP was supported by T32 AI55449 and is currently supported by F31 AI092891.
References
- Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009;4(9):1125–1135. doi: 10.2217/fmb.09.82. [DOI] [PubMed] [Google Scholar]
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;9(5):454–460. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Boyd DA, Cabral T, Van Caeseele P, Wylie J, Mulvey MR. Molecular characterization of the vanE gene cluster in vancomycin-resistant Enterococcus faecalis N00-410 isolated in Canada. Antimicrob Agents Chemother. 2002;46(6):1977–1979. doi: 10.1128/AAC.46.6.1977-1979.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DA, Du T, Hizon R, Kaplen B, Murphy T, Tyler S, Brown S, Jamieson F, Weiss K, Mulvey MR. VanG-type vancomycin-resistant Enterococcus faecalis strains isolated in Canada. Antimicrob Agents Chemother. 2006;50(6):2217–2221. doi: 10.1128/AAC.01541-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother. 2008;52(7):2667–2672. doi: 10.1128/AAC.01516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carias LL, Rudin SD, Donskey CJ, Rice LB. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180(17):4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42(Suppl 1):S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- Goossens H. Spread of vancomycin-resistant enterococci: differences between the United States and Europe. Infect Control Hosp Epidemiol. 1998;19(8):546–551. doi: 10.1086/647871. [DOI] [PubMed] [Google Scholar]
- Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH. et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13(47) pii: 19046 3. [PubMed] [Google Scholar]
- de Niederhausern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, Anacarso I. Vancomycin-resistance transferability from vanA enterococci to Staphylococcus aureus. Curr Microbiol. 2011;62(5):1363–1367. doi: 10.1007/s00284-011-9868-6. [DOI] [PubMed] [Google Scholar]
- Panesso D, Reyes J, Rincon S, Diaz L, Galloway-Pena J, Zurita J, Carrillo C, Merentes A, Guzman M, Adachi JA. et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J Clin Microbiol. 2010;48(5):1562–1569. doi: 10.1128/JCM.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top J, Willems R, Blok H, de Regt M, Jalink K, Troelstra A, Goorhuis B, Bonten M. Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin Microbiol Infect. 2007;13(3):316–319. doi: 10.1111/j.1469-0691.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- Galloway-Pena JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009;200(10):1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189(22):8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47(6):1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- Panesso D, Montealegre MC, Rincon S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiol. 2011;11(1):20. doi: 10.1186/1471-2180-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187(3):508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- Sillanpaa J, Nallapareddy SR, Prakash VP, Qin X, Hook M, Weinstock GM, Murray BE. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology. 2008;154(Pt 10):3199–3211. doi: 10.1099/mic.0.2008/017319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. Distribution of genes encoding MSCRAMMs and Pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol. 2009;47(4):896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E, Bonten MJ, Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189(22):8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E, Singh KV, Jacques-Palaz KD, van Luit-Asbroek M, Oostdijk EA, Bonten MJ, Murray BE, Willems RJ. Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microbes Infect. 2011;13(14–15):1185–1190. doi: 10.1016/j.micinf.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72(1):334–345. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76(9):4120–4128. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun. 2011;79(7):2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;53(10):4240–4246. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Lakticova V, Carias LL, Rudin S, Hutton R, Marshall SH. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J Infect Dis. 2009;199(3):342–349. doi: 10.1086/595986. [DOI] [PubMed] [Google Scholar]
- Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008;52(3):297–308. doi: 10.1111/j.1574-695X.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- Leavis HL, Willems RJ, van Wamel WJ, Schuren FH, Caspers MP, Bonten MJ. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007;3(1):e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H. et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway-Pena J, Roh JH, Latorre M, Qin X, Murray BE. Genomic and SNP Analyses Demonstrate a Distant Separation of the Hospital and Community-Associated Clades of Enterococcus faecium. PLoS One. 2012;7(1):e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J. et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio. 2012;3(1):e00318–00311. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborg P, Top J, Hendrickx AP, Dawson S, Willems RJ, Guardabassi L. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl Environ Microbiol. 2009;75(8):2360–2365. doi: 10.1128/AEM.02035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Regt MJ, van Schaik W, van Luit-Asbroek M, Dekker HA, van Duijkeren E, Koning CJ, Bonten MJ, Willems RJ. Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One. 2012;7(2):e30319. doi: 10.1371/journal.pone.0030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD. et al. Comparative Analysis of the First Complete Enterococcus faecium Genome. J Bacteriol. 2012;194(9):2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, Zeng Q, Gevers D, Feldgarden M, Birren B. et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol. 2010;192(9):2469–2470. doi: 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20(18):3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H. et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9(7):R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417(6890):746–750. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188(8):2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakita RM, Quan VC, Jacques-Palaz K, Singh KV, Arduino RC, Mee M, Murray BE. Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol Med Microbiol. 2000;28(4):291–299. doi: 10.1111/j.1574-695X.2000.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. Novel Bacteriophages in Enterococcus spp. Curr Microbiol. 2010;60(6):400–406. doi: 10.1007/s00284-009-9555-z. [DOI] [PubMed] [Google Scholar]
- Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett Appl Microbiol. 2011;52(6):559–564. doi: 10.1111/j.1472-765X.2011.03043.x. [DOI] [PubMed] [Google Scholar]
- Bose M, Barber RD. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In silico biology. 2006;6(3):223–227. [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24(6):863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- Werner G, Fleige C, Geringer U, van Schaik W, Klare I, Witte W. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect Dis. 2011;11:80. doi: 10.1186/1471-2334-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E, van Schaik W, Leavis HL, Bonten MJ, Willems RJ. Identification of a novel genomic island specific to hospital-acquired clonal complex 17 Enterococcus faecium isolates. Appl Environ Microbiol. 2008;74(22):7094–7097. doi: 10.1128/AEM.01378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao WW, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FS. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1(5):e62. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, Surovcik K, Meinicke P, Merkl R. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinforma. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25(5):664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Hsiao WW, Brinkman FS. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinforma. 2008;9:329. doi: 10.1186/1471-2105-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Hancock LE. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J Bacteriol. 2009;191(20):6203–6210. doi: 10.1128/JB.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77(9):3759–3767. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Murray BE, Weinstock GM. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66(9):4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway-Pena JR, Rice LB, Murray BE. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob Agents Chemother. 2011;55(7):3272–3277. doi: 10.1128/AAC.00099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Sillanpaa J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, Sullam PM, Murray BE. Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun. 2011;79(7):2911–2920. doi: 10.1128/IAI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33((Database issue)):D325–328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creti R, Koch S, Fabretti F, Baldassarri L, Huebner J. Enterococcal colonization of the gastro-intestinal tract: role of biofilm and environmental oligosaccharides. BMC Microbiol. 2006;6:60. doi: 10.1186/1471-2180-6-60. pii: e00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4) doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias LL, Hutton-Thomas R, Sifaoui F, Gutmann L, Rudin SD. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2001;45(5):1480–1486. doi: 10.1128/AAC.45.5.1480-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino RC, Jacques-Palaz K, Murray BE, Rakita RM. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect Immun. 1994;62(12):5587–5594. doi: 10.1128/iai.62.12.5587-5594.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun. 2008;76(9):4110–4119. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ada G. Vaccines and vaccination. N Engl J Med. 2001;345(14):1042–1053. doi: 10.1056/NEJMra011223. [DOI] [PubMed] [Google Scholar]
- Teng F, Jacques-Palaz KD, Weinstock GM, Murray BE. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect Immun. 2002;70(4):2010–2015. doi: 10.1128/IAI.70.4.2010-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Fleming SD, Hancock LE. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect Immun. 2009;77(12):5551–5557. doi: 10.1128/IAI.00576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner J, Wang Y, Krueger WA, Madoff LC, Martirosian G, Boisot S, Goldmann DA, Kasper DL, Tzianabos AO, Pier GB. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect Immun. 1999;67(3):1213–1219. doi: 10.1128/iai.67.3.1213-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock LE, Gilmore MS. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc Natl Acad Sci U S A. 2002;99(3):1574–1579. doi: 10.1073/pnas.032448299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Singh KV, Qin X, Murray BE, Weinstock GM. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immun. 2000;68(2):815–823. doi: 10.1128/iai.68.2.815-823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence. 2010;1(4):236–246. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, van Luit-Asbroek M, Schapendonk CM, van Wamel WJ, Braat JC, Wijnands LM, Bonten MJ, Willems RJ. SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect Immun. 2009;77(11):5097–5106. doi: 10.1128/IAI.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque TM, Tomayko JF, Ricke SC, Okhyusen PC, Murray BE. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40(11):2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. Preparation of Genomic DNA from Bacteria. Green Publishing Associates, Brooklyn, N.Y.; 1994. [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27(19):3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33((Database issue)):D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. et al. The Pfam protein families database. Nucleic Acids Res. 2010;38((Database issue)):D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21(5):617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ. et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34((Database issue)):D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21(4):537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12(3):R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinforma. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11(5):472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Lefebure T, Hubisz MJ, Pavinski Bitar P, Lang P, Siepel A, Stanhope MJ. Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution. Genome Biol Evol. 2011;3:168–185. doi: 10.1093/gbe/evr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172(4):2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol. 2003;47(5):1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- Murphy E, Huwyler L. de Freire Bastos Mdo C: Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985;4(12):3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz FV, Perreten V, Teuber M. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid. 2001;46(3):170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- Burdett V, Inamine J, Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175(1):117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis HL, Willems RJ, Top J, Bonten MJ. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J Clin Microbiol. 2006;44(3):1059–1064. doi: 10.1128/JCM.44.3.1059-1064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Bellais S, Carias LL, Hutton-Thomas R, Bonomo RA, Caspers P, Page MG, Gutmann L. Impact of specific pbp5 mutations on expression of beta-lactam resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2004;48(8):3028–3032. doi: 10.1128/AAC.48.8.3028-3032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.Gene order synteny ofE. faeciumTX16 compared toE. faecalisV583. A figure ploting the synteny blocks between TX16 and V583 with the coordinates of each genome.
Figure S2.Genome alignment of TX16 and Aus0004. A figure comparing the two closed E. faecium genomes sequences available using Mauve genome alignment analysis.
Table S1.Hospital-associated clade unique genes. A table listing the genes and their corresponding ORF in TX16 that are unique to the hospital clade and how many of the HA clade strains the gene is present in.
Table S2.Prophage loci and genes onE. faeciumTX16 genome. A table listing the two prophage loci, the predicted gene products within these two loci, and the corresponding ORFs in TX16.
Table S3.Mobile elements in theE. faeciumTX16 genome. A table listing all of the predicted mobile elements and their corresponding locus tags in TX16.
Table S4.E. faeciumTX16 genomic islands and genes. A table listing the nine genomic islands, the genes and predicted products within those islands, and the corresponding ORFs and coordinates within TX16.
Figure S3.ORF composition of the downstream extension of theepagene cluster in the 22E. faeciumgenomes (HMPREF0351_10908 - HMPREF0351_10923 in TX16). A figure depicting the predicted polysaccharide-encoding gene clusters found in the E. faecium genomes.
Table S5.Presence of genes encoding MSCRAMMs and pilins among 21E. faeciumgenomes. A table listing the different MSCRAMM and pilin variants present in each of the 22 genomes.
Table S6.Summary of CRISPRs found inE. faeciumsequenced strains. A table listing in what strains CRISPRs were found, the locus tag, and the functional assignment.
Table S7.Specific enzymes present in TX16 but not inE. faecalisV583. A table listing enzymes, KEGG information, and locus tags specific to TX16.
Table S8.Specific enzymes present inE. faecalis V583 but not in TX16. A table listing the enzymes and locus tags specific to V583.