Abstract
Background
Huntington disease (HD) is caused by an expanded CAG repeat in the HD gene. Although the length of the CAG repeat strongly correlates with the age-at-onset (AAO), AAO in HD individuals may differ dramatically in spite of similar expanded CAG repeat lengths. Additional genetic or environmental factors are thought to influence the disease onset. Several modifier genes have been discovered so far but they do not fully explain the variability of AAO in HD. To potentially identify a novel genetic modifier, we analyzed single nucleotide polymorphisms (SNPs) in the kalirin (KALRN) gene. Kalirin is a protein crucially involved in spine plasticity and its interaction with huntingtin-associated protein-1 (HAP-1) and a potential protein dysfunction might contribute to spine pathogenesis in HD.
Methods
The selected SNPs were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and association of SNPs with AAO was investigated with the framework of linear models in an analysis of variance and covariance.
Results
Eleven SNPs in the kalirin gene were examined in an association study in European HD patients. The ten coding SNPs under investigation were monomorphic, whereas SNP rs10934657 in the promoter region showed a minor allele frequency >1%. An analysis of covariance together with the influence of the expanded HD allele was applied in 680 HD patients. SNP rs10934657 did not affect the AAO of the examined HD population.
Conclusions
The results did not reveal an association between the analyzed kalirin polymorphisms and the AAO in HD. However, it does not exclude other SNPs of the kalirin gene as susceptible genetic modifiers.
Background
Huntington disease (HD) is one of the most common autosomal-dominant neurodegenerative disorders caused by an expansion of an instable CAG repeat in the HD gene resulting in a polyglutamine tract near the amino terminus of the huntingtin protein (htt) [1]. The mutation leads to the selective loss of vulnerable neurons, notably medium spiny neurons in the caudate nucleus, which results in motor and cognitive impairment, personality changes and psychiatric illness [2]. The number of CAG repeats in the HD gene is the primary determinant of disease onset, however it only accounts for approximately 42-73% of the variance in age-at-onset (AAO) in HD [3,4]. Similar to several other neurodegenerative disorders, the detrimental gene alone does not fully determine the AAO in the course of the disease. Environmental factors, although not specifically defined yet, may contribute to different disease manifestation. The remaining variation of AAO may be due to modifier genes and seems to be strongly heritable [5]. Several genetic modifiers of HD have been identified so far, including huntingtin-associated protein-1 (HAP1) [6], ubiquitin C-terminal hydrolase 1 (UCHL1) [7,8], GluR6 subunit of kainate receptor (GRIK2) [9], the adenosinergic A2A receptor (ADORA2A)[10,11], autophagy-related protein 7 (Atg7) [12] and the peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) [13-15]. A recent review on HD modifiers was provided by Arning and Epplen [16]. These modifiers with their respective functions contribute to different aspects of pathogenesis in HD.
Cell death in the striatum and aggregation of the mutant huntingtin protein are pathological hallmarks of HD [17]. Morphological alterations of dendrites and spines are also found in HD patients and animal models [18]. Ferrante et al. showed truncated dendritic arbors, focal swellings on dendrites and spine loss in patients with a severe grade of HD [19]. Similar dendritic degeneration has been observed in R6/2 mice expressing mutant huntingtin exon 1 with 150 CAG repeats and transgenic mice expressing full length huntingtin cDNA [20,21]. Spine morphogenesis and plasticity are modulated by actin dynamics, which is regulated by Rho-like small GTPases (Rac, RhoA and Cdc42) and their GDP/GTP exchange factors (GEFs) [22]. Kalirin is a brain-specific, multifunctional Rho GEF encoded by the KALRN gene on chromosome 3q21.2 that generates several isoforms by alternative splicing [23]. Rat kalirin-7, corresponding to isoform 2 of the human kalirin gene, is undetectable at birth and increases during synaptogenesis [23,24]. It contains a SEC14 domain, a spectrin-like domain, a RhoGEF domain and a Pleckstrin homology domain (PH) [25] which controls multiple functions of the protein. Kalirin-7 activates Rac1 and regulates dendritic spine morphogenesis, plasticity and development [26,27]. Another probable link between kalirin and HD pathogenesis is huntingtin-associated protein-1 (HAP-1), a HD genetic modifier which interacts with the kalirin protein [6,28].
Although several susceptibility modifier genes for HD have been identified, they are not sufficient to explain the rest of the variance in AAO in HD. The goal of the present study was to investigate if SNPs in the kalirin gene also have a modifier effect on the AAO in HD. We specifically focused on the isoform-2 of the kalirin gene since it is the major splice variant in the adult brain, which integrates various signaling inputs and modulates dendritic spine maturation, plasticity and dynamics. In this regard, we analyzed one SNP in the promoter region and ten non-synonymous SNPs (D451E, Q520R, Q585E, G654W, T727S, R837Q, X1112E, D1326E, N1389H, E1588G) in the translated region of isoform-2 of the kalirin gene.
Methods
Subjects
A total of 680 unrelated European HD patients were analyzed. Among them, 320 patients were of German descent and 171 patients were of Italian descent. The remaining 189 patients were from other European countries [6,12,15]. AAO was determined by neurologists specialized in HD, usually as the time point when motor symptoms were first noticed. The mean AAO was 44.0 years (SD 13.0, age range 5–80 years). CAG repeat lengths in the HD gene had been tested in all patients and CAG numbers had been standardized in a reference laboratory. The number of the expanded CAG repeats ranged from 39 to 90. All healthy individuals are samples of the Centre d’Etude du Polymorphisme Human cohort (CEPH).
Ethics
All participating individuals gave informed consent according to the Declaration of Helsinki. An ethics proposal was approved by the ethical review committee of the Medical Department of the University of Tuebingen (39/2003).
Genotyping
The selected SNPs were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The target sequences were amplified with mismatch forward or reverse primers and digested with specific restriction enzymes. The mismatch primers were generated using dCAPs Finder 2.0 software (http://helix.wustl.edu/dcaps/dcaps.html) and optimized by Primer 3 program (http://frodo.wi.mit.edu/primer3/). Primer sequences for PCR amplification are shown in Table 1. PCR was performed in a final volume of 25 μl using 5 μl DNA, 1 μl of each primer, 1x buffer, 0.2 mM dNTP and 1.5U DNA Taq polymerase (BioTherm™). The cycling profile was as follows: 95°C 5′; [95°C 30″; respective annealing temperature for each SNP is indicated in Table 1, 30″; 72°C 1′] for 35 cycles; 72°C 5′ and stored at 10°C. Five μl of the PCR product were electrophoresed on 2% agarose gels and only samples with positive signals were used, yielding the total of 680 HD samples. The PCR products were incubated with 3U AluI (rs10934657, rs111472457, rs61746078, rs2289838, rs2289838), 2.5U BtgI (rs35057827), 1.9U MscI (rs13074913), 3U BamHI (rs61745397), 3U SpeI (rs112304715), 3U NdeI (rs2289838) or 3U SacI (rs1062749) according to the manufacturer’s instructions (New England Biolabs, Inc., Beverly, MA, USA).
Table 1.
Primer design for fragment length analysis
| SNP | Sequence (5′ → 3′) | PCR product (bp) | Annealing temperature (°C) | Restriction enzyme |
|---|---|---|---|---|
| rs10934657 |
TGGCAAGAGGGAGAGGAG |
139 |
55.2 |
AluI |
| |
CTTCCTCCTCTGTAAACCAGAGAGA |
|
|
|
| rs111472457 |
CATCCGAGATGCAAGACCTAGA |
156 |
58.5 |
AluI |
| |
CCGTGAGGGATTCGGAGT |
|
|
|
| rs35057827 |
GCATGAGGTGTTACATCACCAGCCAC |
123 |
60.9 |
BtgI |
| |
CAATCCAGTCCAACACCTGCT |
|
|
|
| rs61746078 |
CAGCAGGATGTACAGCAGGT |
150 |
61.2 |
AluI |
| |
GTACGTATTCTGAGCCACAG |
|
|
|
| rs13074913 |
TCTACAAGGCAGCTCGACAC |
136 |
58 |
MscI |
| |
AGGTCTTCCATCCATGGCC |
|
|
|
| rs61745397 |
GCCAGGGACTCGGCTGGA |
154 |
60.6 |
BamHI |
| |
TCACCTCGATGGTGTACTGC |
|
|
|
| rs112304715 |
CAGCAGGGACAGGATCTGCACT |
170 |
62.5 |
SpeI |
| |
AGCCGCTTATGAGTCTGCTCT |
|
|
|
| rs77832285 |
TCCTGAGTGAGCTCCTGCATAT |
110 |
59 |
NdeI |
| |
GCTCGAACACCACATATTGC |
|
|
|
| rs2289838 |
AGCCCGGAAGAAAGAATTTA |
168 |
58.2 |
AluI |
| |
TGGATGTTGCCAAAGATGATAAG |
|
|
|
| rs74389479 |
AAGTACGAGCAACTGCCTGAG |
138 |
59.5 |
AluI |
| |
ATCAAAGAAGGTGCCCGCAA |
|
|
|
| rs1062749 |
CTGCAAATTCGCCTTGTGGT |
162 |
55.2 |
SacI |
| GCTGAAGTGGCTCCTTTAGAGCT |
Mismatch positions are highlighted in italics.
Statistical analysis
Statistical analyses were performed as in our previous studies [12,15]. To determine allele and genotype frequencies and Hardy-Weinberg distribution of the tested genotypes GENEPOP software version 4.0.10 (http://www.genepop.curtin.edu.au/) was used. With the framework of linear models in an analysis of variance and covariance (JMP® Version 8.0.2 SAS institute, Inc., Cory, NC, USA) we investigated the modifying role of the polymorphic SNP rs10934657 in the kalirin gene on the AAO of HD. First, we applied a model of analysis of covariance with rs10934657 and the expanded HD allele as independent variables and the AAO as a dependent variable. The goodness-of-fit was assessed by the proportion of variation in the AAO that is explained by the coefficient of determination (R²). We obtained the best fit of our data and a minimization of the residuals by logarithmic transformation of the AAO and the CAG repeat number in the HD gene. To determine the effect of SNP rs10934657 on AAO by an analysis of variance and covariance, the effect of the expanded HD allele (HD CAG) was calculated alone, as well as with SNP rs10934657. When factor rs10934657 is added to the effect of the expanded HD allele (ΔR²), a change of R² would indicate a relative improvement of the model. This method would identify the percentage of the variance that is attributable to the candidate modifier loci when there is a significant P-value (P≤0.05).
Results
To explore the potential modifying effects of the kalirin gene on the AAO in HD, we chose eleven single nucleotide polymorphisms (SNPs) that were published in the NCBI SNP database at the start of this study. According to our hypothesis, via interaction with HAP-1 or due to protein dysfunction, the abnormal kalirin protein may contribute to spine pathogenesis in HD. Therefore, in this study we focused on SNPs that could potentially influence protein function or expression based on their positions in functional domains or regulatory regions. Accordingly, ten of the selected SNPs are non-synonymous and are located in spectrin-like domains (rs111472457 in exon 8, rs35057827 in exon 9, rs61746078 in exon 10, rs13074913 in exon 11, rs61745397 in exon 13, rs112304715 in exon 14, rs77832285 in exon 20), the Rho GEF domain (rs2289838 in exon 25, rs74389479 in exon 27) and downstream of the pleckstrin homology domain (rs1062749 in exon 32), respectively (Figure 1). SNP rs10934657 is in the 5′ untranslated region (5′UTR), a predicted promoter region. Accession numbers and alleles of the analyzed SNPs are shown in Table 2. We first screened these SNPs in 60 control samples (CEPH) to monitor the allele frequencies of each polymorphism. SNP rs10934657 in the 5′UTR region (C>T) was polymorphic with a minor allele frequency≥1% in controls whereas the ten coding SNPs were monomorphic in our cohort (Table 2). Therefore, SNP rs10934657 was selected for further genotyping of HD patients.
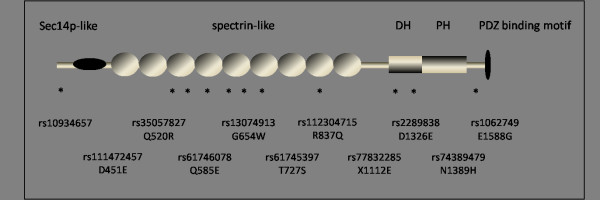
Figure 1 .
Domain structure of the kalirin gene, isoform 2. Dbl-homology (DH) and pleckstrin homology (PH) domains are responsible for the GEF activity of kalirin. The C terminus contains a unique 20 amino acid sequence with a PDZ domain-binding motif (STYV) that is specific for isoform 2. The localization of all SNPs examined in this study and rs numbers and changes on the protein level are indicated.
Table 2.
Overview of the SNPs studied
|
SNP ID |
Chromosome position chromosome 3 |
Allelesa(1/2) |
Gene location |
SNP type |
Protein level |
Controls (CEPH) |
HD patients |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
Allele frequency |
Genotype frequency |
Allele frequency |
||||
| 1 | 2 | 1-1 | 1-2 | 2-2 | 1 | 2 | ||||||
| rs10934657 |
123812836 |
C / T |
5′-UTR |
Non-coding |
|
0.8 |
0.2 |
487 |
174 |
19 |
0.844 |
0.156 |
| rs111472457 |
124048782 |
G / T |
exon 8 |
non-synonymous |
D451E |
1 |
0 |
- |
- |
- |
- |
- |
| rs35057827 |
124053260 |
G / A |
exon 9 |
non-synonymous |
Q520R |
1 |
0 |
- |
- |
- |
- |
- |
| rs61746078 |
124066099 |
G / C |
exon 10 |
non-synonymous |
Q585E |
1 |
0 |
- |
- |
- |
- |
- |
| rs13074913 |
124113985 |
T / G |
exon 11 |
non-synonymous |
G654W |
1 |
0 |
- |
- |
- |
- |
- |
| rs61745397 |
124117557 |
T / A |
exon 13 |
non-synonymous |
T727S |
1 |
0 |
- |
- |
- |
- |
- |
| rs112304715 |
124132486 |
A / G |
exon 14 |
non-synonymous |
R837Q |
1 |
0 |
- |
- |
- |
- |
- |
| rs77832285 |
124165034 |
G / T |
exon 20 |
non-synonymous |
X1112E |
1 |
0 |
- |
- |
- |
- |
- |
| rs2289838 |
124181433 |
G / T |
exon 25 |
non-synonymous |
D1326E |
1 |
0 |
- |
- |
- |
- |
- |
| rs74389479 |
124196161 |
C / A |
exon 27 |
non-synonymous |
N1389H |
1 |
0 |
- |
- |
- |
- |
- |
| rs1062749 | 124211666 | G / A | exon 32 | non-synonymous | E1588G | 1 | 0 | - | - | - | - | - |
Chromosome positions were obtained from NCBI single nucleotide polymorphism (SNP) browser (http://www.ncbi.nlm.nih.gov/SNP/).
Reference sequence for coding SNPs: NM_003947.4.
The studied genotype distributions were consistent with Hardy-Weinberg distribution P=0.4716.
a Alleles are described as 1 (wild type allele) or 2 (variant allele).
To examine the effect of the polymorphic SNP rs10934657 on disease onset, the respective genotypes were determined in a total of 680 HD patients. Genotyped allele frequencies are listed in Table 2. The allele frequencies of SNP rs10934657 in our European population were consistent with the HapMap-CEU population studies reported in the International HapMap project (http://www.hapmap.org) (C: 0.8; T: 0.2). In order to identify a possible modifying effect of SNP rs10934657 on the AAO of the analysed HD patients, an analysis of covariance together with the influence of the expanded HD allele was applied. Analysing the effect of the expanded CAG repeat in the HD gene itself, R² in the statistical model reaches a value of 0.5394 (Table 3). This indicates that the expanded HD alleles accounts for about 53% of the variance in the AAO, which is in good accordance to other studies [3,4]. However, the inclusion of SNP rs10934657 as covariant did not improve our model. This SNP did therefore not affect the AAO of the disease in the examined HD population (P=0.9713). Furthermore, there is no significant effect of the non-expanded CAG repeat on the AAO in our analysed population (data not shown).
Table 3.
Effect of SNP rs10934657 on AAO in HD (Analysis of covariance)
| Variable | R² | ΔR² | p-value | Least significant number of patients |
|---|---|---|---|---|
|
HD CAG |
0.5394 |
|
<0.0001 |
7 |
| HD CAG + SNP rs10934657 | 0.5394 | 0 | 0.9713 | 70065 |
Discussion
To our knowledge, this is the first study that examines an association of the kalirin gene with the AAO in HD. Previous studies have established a connection between kalirin variants and susceptibility to schizophrenia, Alzheimer disease, adult attention deficit hyperactivity disorder (ADHD), coronary artery disease and ischemic stroke [29-34]. In the present study, we hypothesized that isoform 2 of the kalirin gene, corresponding to rat kalirin-7, the predominant kalirin isoform in adult brain, may also contribute as a novel genetic modifier for HD based on its role in spine plasticity and its interaction with HAP-1.
Although the length of the expanded CAG tract in the HD gene is the main determinant of the HD phenotype, the manifestation of the disease is also modified by other risks, such as environmental or genetic factors. To date, two strategies have been applied for identifying genetic modifiers, the genome-wide approach and the candidate gene approach. The genome-wide studies are based on genetic linkage to search for specific chromosome regions, which might be associated with an alteration of age at neurological onset, including the HD-MAPS project which identified 6q23-24 as an association region [34] and the Venezuela pedigrees study [35]. The identified genomic regions in both studies were relatively large and it is difficult to detect specific modifiers, which are now analyzed with a combination of densely spaced SNPs and copy number probes. On the other hand, association studies that investigate candidate genes that are speculated to be involved in HD pathogenesis provide a straight forward option to identify these modifiers, although they are not comprehensive. Recent studies have demonstrated several genetic modifiers related to various mechanisms implicated in HD pathology, such as metabolic impairment, transcription dysregulation, oxidative stress and excitotoxicity [6,8,36,37]. Among them, the polymorphism T441M of HAP-1 showed an 8-year delay in AAO due to a tighter interaction of HAP-1 with mutant huntingtin (htt) protein and thus ameliorated htt-mediated toxicity [6]. HAP-1 is associated with huntingtin, dynactin p150/kinesin light chain (KLC), endosomal organelles and BDNF, suggesting its role in intracellular trafficking and endocytosis.
Here we examined polymorphisms in the kalirin gene due to its crucial role in spine plasticity and its interaction with HAP-1. Recently, several genome-wide association studies (GWAS) had revealed genetic associations of the kalirin gene with several diseases. An intronic SNP rs9289231 was associated with early onset coronary artery disease in an American white population [29], while intronic SNPs rs11712039, rs17286604 and rs11712619 were associated with ischemic stroke in a small Portuguese population [33]. A recent GWAS in a Japanese population showed that a missense mutation in the kalirin gene, P2255T (ss250607859), may be a genetic risk factor for schizophrenia [32]. In the present study we did not include any intronic SNPs, as we drew our attention to potential functional changes on the protein level. Also SNP ss250607859 was excluded from our analyses, as it affects another isoform of the kalirin gene.
Conclusions
Among the eleven SNPs that were screened in our study, we found only SNP rs10934657 to be polymorphic in our European control cohort. However, in an analysis of covariance, there was no significant effect (P=0.9713) of this SNP on the AAO in our cohort of European HD patients. Also testing for additional factors, such as sex or German or Italian ancestry, did not reveal a significant effect either (data not shown). Further power analysis was performed to determine the population size that would be required to show potentially significant effects of rs10934657. Our calculations revealed a minimum of 70065 samples, indicating that this SNP is very unlikely to have a big impact on AAO in HD.
In summary, although the genetic variations of the kalirin gene investigated here showed no effect on the AAO in HD, this does not exclude other SNPs of the kalirin gene as susceptible genetic modifiers.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YCT carried out the molecular genetic analyses, participated in the statistical analysis of the data and drafted the manuscript. SM performed the statistical analysis. OR participated in the study design and reviewed the manuscript. AS and HPN conceived, designed and coordinated the study, interpretated the data and participated in writing of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Yu-Chun Tsai, Email: yu-chun.tsai@med.uni-tuebingen.de.
Silke Metzger, Email: simetzger@yahoo.de.
Olaf Riess, Email: olaf.riess@med.uni-tuebingen.de.
Anne S Soehn, Email: anne.soehn@med.uni-tuebingen.de.
Huu Phuc Nguyen, Email: hoa.nguyen@med.uni-tuebingen.de.
Acknowledgement
We thank Adriana Redensek for careful proofreading of this manuscript. This work is supported by DFG grant (682/10-1) and a fellowship from the European Commission to Yu-Chun Tsai (MEST-CT-2005-020235, “NeuroTrain”).
References
- Group THsDCR. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- White J, Auerbach W, Duyao M, Vonsattel J, Gusella J, Joyner A, MacDonald M. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Brinkman R, Mezei M, Theilmann J, Almqvist E, Hayden M. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–1210. [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Pleasant N, Franz ML, Abbott MH, Folstein SE, Ross CA. Correlation between the onset age of Huntington’s disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet. 1993;2:1547–1549. doi: 10.1093/hmg/2.10.1547. [DOI] [PubMed] [Google Scholar]
- Wexler N, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts S, Gayan J. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci U S A. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Rong J, Nguyen H-P, Cape A, Tomiuk J, Soehn AS, Propping P, Freudenberg-Hua Y, Freudenberg J, Tong L. et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum Mol Genet. 2008;17:1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- Naze P, Vuillaume I, Destee A, Pasquier F, Sablonniere B. Mutation analysis and association studies of the ubiquitin carboxy-terminal hydrolase L1 gene in Huntington’s disease. Neurosci Lett. 2002;328:1–4. doi: 10.1016/S0304-3940(02)00231-8. [DOI] [PubMed] [Google Scholar]
- Metzger S, Bauer P, Riess MSO. JTFLSD CGPSHWLHW-S GKWBMVHLB SWLAJZAS DH-ZANBNEJZ VKMPPRLK MKBHFWFKMD. The S18Y polymorphism in the UCHL1 gene is a genetic modifier in Huntington’s disease. Neurogenetics. 2006;7:27–30. doi: 10.1007/s10048-005-0023-z. [DOI] [PubMed] [Google Scholar]
- Zeng W, Gillis T, Hakky M, Djousse L, Myers R, MacDonald M, Gusella J. Genetic analysis of the GRIK2 modifier effect in Huntington’s disease. BMC Neurosci. 2006;7:62. doi: 10.1186/1471-2202-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaenens C, Burnouf S, Simonin C, Van Brussel E, Duhamel A, Defebvre L, Duru C, Vuillaume I, Cazeneuve C, Charles P. A genetic variation in the ADORA2A gene modifies age at onset in Huntington’s disease. Neurobiol Dis. 2009;35:474–476. doi: 10.1016/j.nbd.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Wieczorek S, Epplen J, Arning L. Age at onset in Huntington’s disease: replication study on the associations of ADORA2A, HAP1 and OGG1. Neurogenetics. 2010;11:435–439. doi: 10.1007/s10048-010-0248-3. [DOI] [PubMed] [Google Scholar]
- Metzger S, Saukko M, Van Che H, Tong L, Puder Y, Riess O, Nguyen H. Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet. 2010;128:453–459. doi: 10.1007/s00439-010-0873-9. [DOI] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L. PGC-1alpha as modifier of onset age in Huntington disease. Mol Neurodegener. 2009;4:10. doi: 10.1186/1750-1326-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydt P, Soyal S, Gellera C, DiDonato S, Weidinger C, Oberkofler H, Landwehrmeyer B, Patsch W. The gene coding for PGC-1alpha modifies age at onset in Huntington’s Disease. Mol Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che HV, Metzger S, Portal E, Deyle C, Riess O, Nguyen H. Localization of sequence variations in PGC-1alpha influence their modifying effect in Huntington disease. Mol Neurodegener. 2011;6:1. doi: 10.1186/1750-1326-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arning L, Epplen JT. Genetic modifiers of Huntington’s disease: beyond CAG. Futur Neurol. 2011;7:93–109. [Google Scholar]
- Imarisio S, Carmichael J, Korolchuk V, Chen C-W, Saiki S, Rose C, Krishna G, Davies JE, Ttofi E, Underwood BR, Rubinsztein DC. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- Spires TL, Grote HE, Garry S, Cordery PM, Van Dellen A, Blakemore C, Hannan AJ. Dendritic spine pathology and deficits in experience-dependent dendritic plasticity in R6/1 Huntington’s disease transgenic mice. Eur J Neurosci. 2004;19:2799–2807. doi: 10.1111/j.0953-816X.2004.03374.x. [DOI] [PubMed] [Google Scholar]
- Ferrante R, Kowall N, Richardson E. Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidetti P, Charles V, Chen E-Y, Reddy PH, Kordower JH, Whetsell WO, Schwarcz R, Tagle DA. Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001;169:340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- Klapstein GJ, Fisher RS, Zanjani H, Cepeda C, Jokel ES, Chesselet M-F, Levine MS. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington’s disease transgenic mice. J Neurophysiol. 2001;86:2667–2677. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- Penzes P, Jones K. Dendritic spine dynamics - a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Penzes P, Eipper B, Mains R. Isoforms of kalirin, a neuronal Dbl family member, generated through use of different 5′- and 3′-ends along with an internal translational initiation site. J Biol Chem. 2000;275:19324–19333. doi: 10.1074/jbc.M000676200. [DOI] [PubMed] [Google Scholar]
- Cingolani L, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Alam M, Johnson R, Darlington D, Hand T, Mains R, Eipper B. Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine alpha-amidating monooxygenase, an integral membrane peptide-processing enzyme. J Biol Chem. 1997;272:12667–12675. doi: 10.1074/jbc.272.19.12667. [DOI] [PubMed] [Google Scholar]
- Xie Z, Srivastava D, Photowala H, Kai L, Cahill M, Woolfrey K, Shum C, Surmeier D, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Rapid induction of dendritic spine morphogenesis by trans-synaptic EphrinB-EphB receptor activation of the Rho-GEF Kalirin. Neuron. 2003;37:263–274. doi: 10.1016/S0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Colomer V, Engelender S, Sharp AH, Duan K, Cooper JK, Lanahan A, Lyford G, Worley P, Ross CA. Huntingtin-associated protein 1 (HAP1) binds to a Trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum Mol Genet. 1997;6:1519–1525. doi: 10.1093/hmg/6.9.1519. [DOI] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, Harris M, Nelson S, Hale AB, Granger CB. et al. Peakwide mapping on chromosome 3q13 identifies the Kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn H, Ji I, Ji HP, Markesbery WR, Ji TH. Under-expression of Kalirin-7 Increases iNOS activity in cultured cells and correlates to elevated iNOS activity in Alzheimer’s disease hippocampus. J Alzheimers Dis. 2007;12:271–281. doi: 10.3233/jad-2007-12309. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J. et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP. et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug T, Manso H, Gouveia L, Sobral J, Xavier J, Albergaria I, Gaspar G, Correia M, Viana-Baptista M, Simões R. et al. Kalirin: a novel genetic risk factor for ischemic stroke. Hum Genet. 2010;127:513–523. doi: 10.1007/s00439-010-0790-y. [DOI] [PubMed] [Google Scholar]
- Li J-L, Hayden M, Warby S, Durr A, Morrison P, Nance M, Ross C, Margolis R, Rosenblatt A, Squitieri F. et al. Genome-wide significance for a modifier of age at neurological onset in Huntington’s Disease at 6q23-24: the HD MAPS study. BMC Med Genet. 2006;7:71. doi: 10.1186/1471-2350-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayán J, Brocklebank D, Andresen JM, Alkorta-Aranburu G, Zameel Cader M, Roberts SA, Cherny SS, Wexler NS, Cardon LR, Housman DE. Genomewide linkage scan reveals novel loci modifying age of onset of Huntington’s disease in the Venezuelan HD kindreds. Genet Epidemiol. 2008;32:445–453. doi: 10.1002/gepi.20317. [DOI] [PubMed] [Google Scholar]
- Arning L, Saft C, Wieczorek S, Andrich J, Kraus P, Epplen J. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease in a sex-specific manner. Hum Genet. 2007;122:175–182. doi: 10.1007/s00439-007-0393-4. [DOI] [PubMed] [Google Scholar]
- Arning L, Monté D, Hansen W, Wieczorek S, Jagiello P, Akkad DA, Andrich J, Kraus PH, Saft C, Epplen JT. ASK1 and MAP2K6 as modifiers of age at onset in Huntington’s disease. J Mol Med. 2008;86:485–490. doi: 10.1007/s00109-007-0299-6. [DOI] [PubMed] [Google Scholar]