Abstract
Through genome-wide association meta-analyses of up to 133,010 individuals of European ancestry without diabetes, including individuals newly genotyped using the Metabochip, we have raised the number of confirmed loci influencing glycemic traits to 53, of which 33 also increase type 2 diabetes risk (q < 0.05). Loci influencing fasting insulin showed association with lipid levels and fat distribution, suggesting impact on insulin resistance. Gene-based analyses identified further biologically plausible loci, suggesting that additional loci beyond those reaching genome-wide significance are likely to represent real associations. This conclusion is supported by an excess of directionally consistent and nominally significant signals between discovery and follow-up studies. Functional follow-up of these newly discovered loci will further improve our understanding of glycemic control.
The Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) previously undertook meta-analyses of genome-wide association studies (GWAS) of glycemic traits in non-diabetic individuals, leading to the discovery of multiple associated loci: 16 for fasting glucose concentrations, two for fasting insulin concentrations and five for post-challenge glucose concentrations (2hGlu)1-3. These and subsequent studies highlighted important biological pathways implicated in glucose and insulin regulation4,5. They also demonstrated that some, but not all, loci associated with glycemic traits in non-diabetic individuals also affect the risk of type 2 diabetes (T2D)1,6. Despite the success of these efforts, the identification of new loci was limited by de novo genotyping capacity and cost, such that only a limited number of promising loci from discovery analyses were taken forward to follow-up analyses (often those reaching a threshold of ~P < 10−5 in discovery). Therefore, it is likely that many additional associations with common, low penetrance variants remain to be found among SNPs not previously selected for replication7,8.
The Illumina CardioMetabochip (Metabochip) is a custom Illumina iSELECT array of 196,725 SNPs developed to support cost-effective large-scale follow-up studies of putative association signals for a range cardiovascular and metabolic traits (~66,000 SNPs) and to fine-map established loci (~120,000 SNPs) (Supplementary Fig. 1)9. The ~66,000 follow-up SNPs were selected to enable genotyping of the most significant association signals for each of 23 metabolic traits contributed by a range of consortia. MAGIC contributed ~5,000 top ranking SNPs for fasting glucose, and ~1,000 each for fasting insulin and 2hGlu that had shown nominal association in discovery analyses (Pdiscovery < 0.02)1,2.
In the present study, we combined newly available samples with genotype data for these 66,000 follow-up SNPs with previous discovery meta-analyses to discover new association signals with glycemic traits. This approach identified 41 glycemic associations not previously described1,2: 20 for fasting glucose, 17 for fasting insulin and four for 2hGlu. This raises the number of associated loci to 36 for fasting glucose, 19 for fasting insulin and 9 for 2hGlu, explaining 4.8%, 1.2% and 1.7% of the variance in these traits, respectively. Of these 53 non-overlapping loci, 33 were also associated with T2D (P < 0.05), which while supporting the previous assertion of an imperfect correlation between these traits, also implicates new loci in the etiology of T2D and increases the overlap between glycemic and T2D loci.
RESULTS
Approaches to identify loci associated with glycemic traits
To follow up loci showing evidence of association (Pstage one < 0.02) in discovery GWAS, we investigated the 66,000 Metabochip follow-up SNPs for association with fasting glucose, fasting insulin and 2hGlu. We combined in meta-analysis data from up to 133,010 (fasting glucose), 108,557 (fasting insulin) and 42,854 (2hGlu) non-diabetic individuals of European ancestry, including individuals from the previous meta-analyses1,2, individuals from new GWAS and individuals newly genotyped on the Metabochip array (Supplementary Fig. 2). All study characteristics are shown in Supplementary Table 1. Genome-wide association data for Filipino women were available (Supplementary Table 1), for which we report the effect directions and allele frequencies in Supplementary Tables 2a,b. Genome-wide significant (P < 5 × 10−8) association signals located more than 500 kb from, and not in LD (Hapmap CEU: r2 < 0.05) with, any variant already known to be associated with the trait were considered novel. Associated loci are referred to by the name of the nearest gene, unless a more biologically plausible gene was nearby, or a nearby gene was previously associated with another trait. In such cases, we list the nearest genes in Supplementary Tables 2a-d. As body mass index (BMI) is a major risk factor for T2D and is correlated with glycemic traits, we also performed analyses adjusted for BMI.
Though not the main focus of this effort, given the increased variant density available on the Metabochip for established glycemic loci, we investigated whether these data would enable fine-mapping of underlying functional variants1-3. In these analyses, we included data from up to 53,622 individuals for fasting glucose, 42,384 for fasting insulin and 27,602 for 2hGlu from studies with Metabochip genotypes only. However, given the lack of samples from different ancestries and the absence of full conditional analyses, for the most part these analyses did not improve the resolution of association signals.
Beyond individual SNP investigations for each glycemic trait, we also tested the hypothesis that gene-based analyses using VEGAS10 would identify genes that harbor multiple association signals, which individually did not reach genome-wide significance. Among the ~66,000 SNPs, we used VEGAS to pool the results for all SNPs within each gene (± 50 kb) to identify genes with more evidence of association than expected by chance (given gene size and linkage disequilibrium structure) by simulation, and significant after Bonferroni-correction for multiple testing (P < 5 × 10−6).
Fasting glucose
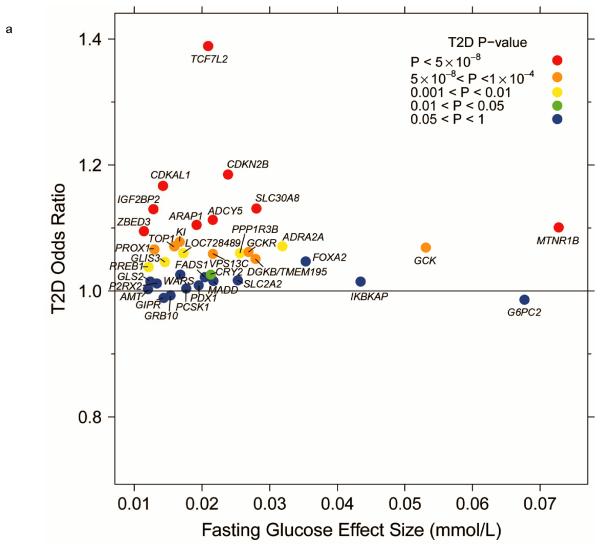
In analyses of up to 133,010 individuals, we identified 20 loci with genome-wide significant associations to fasting glucose (P < 5 × 10−8) (Table 1 and Supplementary Figs. 3 and 4) and confirmed previously established loci1 (Supplementary Table 2e). Of these 20 loci, nine (in or near IBKAP, LOC728489, WARS, KL, TOP1, P2RX2, AMT, RREB1 and GLS2) had not previously been associated with other metabolic traits (Box 1). Among these, KL (Klotho) is of particular interest. In addition to being associated with fasting glucose (but not fasting insulin), the glucose-raising allele is also associated with an increased risk of T2D (OR = 1.08 (1.04-1.11), P = 1.1 × 10−5) (Fig. 1). KL was first identified as a gene related to suppression of aging: its reduced expression was associated with reduced lifespan, as well as hypoglycemia11. Despite further animal studies supporting a role for KL in glucose metabolism12 and insulin sensitivity13, human studies have generally been small and inconclusive14,15.
Table 1.
SNPs associated with fasting glucose, fasting insulin and 2 hour-glucose at genome-wide significance level in Europeans
| Primary trait |
FI (BMI- adjusted) |
2hGlu | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary Trait |
SNP | Chr | Position | Gene | Alleles (effect/ other) |
Freq effect allele |
Effect (SE) | Global analysis P value |
Global analysis n |
I2 estimate (P value) |
Effect (SE) |
Global analysis P value |
Global analysi s n |
Effect (SE) |
Global analysis P value |
Global analysi s n |
| rs10811661 | 9 | 22124094 | CDKN2B | T/C | 0.82 | 0.0238 (0.003) |
5.6 × 10−18 | 128,488 | 0.00 (1.00) |
−0.0065 (0.003) |
0.019 | 98,880 | 0.0567 (0.014) |
8.8 × 10−5 | 42,801 | |
| rs4869272 | 5 | 95565204 | PCSK1* | T/C | 0.69 | 0.0177 (0.002) |
1.0 × 10−15 | 131,872 | 0.00 (1.00) |
0.0016 (0.002) |
0.469 | 103,493 | −0.0322 (0.012) |
0.006 | 42,848 | |
| rs11619319 | 13 | 27385599 | PDX1 | G/A | 0.23 | 0.0195 (0.002) |
1.3 × 10−15 | 132,996 | 0.00 (1.00) |
0.0001 (0.002) |
0.977 | 103,492 | 0.0185 (0.013) |
0.156 | 42,848 | |
| FG | rs983309 | 8 | 9215142 | PPP1R3B* | T/G | 0.12 | 0.0256 (0.003) |
6.3 × 10−15 | 127,470 | 0.14 (0.32) |
0.0223 (0.003) |
1.2 × 10−12 | 99,024 | −0.0548 (0.016) |
0.001 | 42,846 |
| rs6943153 | 7 | 50759073 | GRB10 | T/C | 0.34 | 0.0154 (0.002) |
1.6 × 10−12 | 131,795 | 0.00 (1.00) |
0.0091 (0.002) |
2.3 × 10−5 | 103,447 | 0.0110 (0.011) |
0.333 | 42,794 | |
| rs11603334 | 11 | 72110633 | ARAP1 | G/A | 0.83 | 0.0192 (0.003) |
1.1 × 10−11 | 128,139 | 0.00 (1.00) |
−0.0046 (0.003) |
0.086 | 99,026 | 0.0294 (0.014) |
0.037 | 42,839 | |
| rs6113722 | 20 | 22505099 | FOXA2 | G/A | 0.96 | 0.0353 (0.005) |
2.5 × 10−11 | 123,665 | 0.04 (0.78) |
−0.0095 (0.005) |
0.064 | 103,471 | 0.0493 (0.030) |
0.101 | 41,416 | |
| rs16913693 | 9 | 110720180 | IKBKAP | T/G | 0.97 | 0.0434 (0.007) |
3.5 × 10−11 | 125,115 | 0.00 (1.00) |
−0.0018 (0.007) |
0.785 | 96,357 | 0.0639 (0.034) |
0.062 | 40,522 | |
| rs3829109 | 9 | 138376587 | LOC728489 | G/A | 0.71 | 0.0172 (0.003) |
1.1 × 10−10 | 115,310 | 0.25 (0.07) |
−0.0002 (0.003) |
0.948 | 94,964 | 0.0343 (0.014) |
0.013 | 36,803 | |
| rs3783347 | 14 | 99909014 | WARS | G/T | 0.79 | 0.0168 (0.003) |
1.3 × 10−10 | 132,544 | 0.02 (0.89) |
0.0017 (0.003) |
0.515 | 103,339 | 0.0274 (0.014) |
0.044 | 42,850 | |
| rs2302593 | 19 | 50888474 | GIPR | C/G | 0.50 | 0.0144 (0.002) |
9.3 × 10−10 | 116,141 | 0.27 (0.05 |
0.0025 (0.002) |
0.265 | 96,976 | −0.0322 (0.012) |
0.006 | 40,781 | |
| rs9368222 | 6 | 20794975 | CDKAL1 | A/C | 0.28 | 0.0143 (0.002) |
1.0 × 10−9 | 128,453 | 0.09 (0.50) |
−0.0047 (0.002) |
0.037 | 98,894 | 0.0279 (0.012) |
0.023 | 42,825 | |
| rs10747083 | 12 | 131551691 | P2RX2 | A/G | 0.66 | 0.0133 (0.002) |
7.6 × 10−9 | 127,111 | 0.00 (1.00) |
−0.0006 (0.002) |
0.785 | 99,895 | 0.0269 (0.012) |
0.026 | 42,790 | |
| rs6072275 | 20 | 39177319 | TOP1 | A/G | 0.16 | 0.0159 (0.003) |
1.7 × 10−8 | 128,616 | 0.00 (1.00) |
0.0038 (0.003) |
0.169 | 99,018 | −0.0110 (0.014) |
0.435 | 42,853 | |
| rs7651090 | 3 | 186996086 | IGF2BP2 | G/A | 0.31 | 0.0128 (0.002) |
1.75 × 10−8 | 128,548 | 0.02 (0.86) |
0.0003 (0.002) |
0.900 | 98,924 | 0.0583 (0.012) |
1.05 × 10−6 | 42,814 | |
| rs576674 | 13 | 32452302 | KL | G/A | 0.15 | 0.0167 (0.003) |
2.3 × 10−8 | 131,856 | 0.00 (1.00) |
−0.0001 (0.003) |
0.983 | 103,472 | 0.0308 (0.016) |
0.060 | 42,849 | |
| rs11715915 | 3 | 49430334 | AMT | C/T | 0.68 | 0.0120 (0.002) |
4.9 × 10−8 | 131,523 | 0.30 (0.02) |
0.0059 (0.002) |
0.006 | 103,398 | 0.0273 (0.012) |
0.018 | 42,851 | |
|
|
||||||||||||||||
| FG (BMI-adjusted) | rs17762454 | 6 | 7158199 | RREB1 | T/C | 0.26 | 0.0140 (0.002) |
9.6 × 10−9 | 123,247 | 0.00 (1.00) |
−0.0002 (0.002) |
0.919 | 103,470 | 0.0007 (0.013) |
0.953 | 42,848 |
| rs7708285 | 5 | 76461623 | ZBED3 | G/A | 0.27 | 0.0150 (0.003) |
1.2 × 10−8 | 117,931 | 0.00 (1.00) |
0.0027 (0.002) |
0.265 | 98,341 | 0.0349 (0.013) |
0.008 | 42,803 | |
| rs2657879 | 12 | 55151605 | GLS2 | G/A | 0.18 | 0.0157 (0.003) |
3.9 × 10−8 | 121,596 | 0.39 (0.03) |
−0.0024 (0.003) |
0.366 | 102,175 | 0.0200 (0.014) |
0.164 | 42,670 | |
|
|
||||||||||||||||
|
Primary
trait |
FG | 2hGlu | ||||||||||||||
| rs1421085 | 16 | 52358455 | FTO | C/T | 0.42 | 0.0200 (0.003) |
1.9 × 10−15 | 104,062 | 0.00 (1.00) |
0.0074 (0.002) |
0.001 | 128,597 | 0.0122 (0.011) |
0.278 | 42,849 | |
| FI | rs983309 | 8 | 9215142 | PPP1R3B* | T/G | 0.12 | 0.0287 (0.004) |
3.8 × 10−14 | 103,030 | 0.04 (0.77) |
0.0256 (0.003) |
6.3 × 10−15 | 127,470 | −0.0548 (0.016) |
0.001 | 42,846 |
| rs9884482 | 4 | 106301085 | TET2 | C/T | 0.39 | 0.0165 (0.002) |
1.4 × 10−11 | 108,420 | 0.00 (1.00) |
0.0001 (0.002) |
0.946 | 132,869 | 0.0004 (0.011) |
0.973 | 42,745 | |
| rs7903146 | 10 | 114748339 | TCF7L2 | C/T | 0.72 | 0.0181 (0.003) |
6.1 × 10−11 | 103,037 | 0.31 (0.02) |
−0.0220 (0.002) |
2.7 × 10−20 | 127,477 | −0.0885 (0.013) |
5.6 × 10−12 | 42,851 | |
| rs10195252 | 2 | 165221337 | GRB14* | T/C | 0.59 | 0.0159 (0.003) |
4.9 × 10−10 | 99,126 | 0.00 (1.00) |
0.0053 (0.002) |
0.014 | 127,005 | 0.0361 (0.011) |
0.001 | 42,846 | |
| rs1167800 | 7 | 75014132 | HIP1 | A/G | 0.54 | 0.0156 (0.003) |
2.6 × 10−9 | 91,416 | 0.00 (1.00) |
0.0016 (0.002) |
0.470 | 118,536 | −0.0133 (0.012) |
0.272 | 38,884 | |
| rs2820436 | 1 | 217707303 | LYPLAL1 | C/A | 0.67 | 0.0153 (0.003) |
4.4 × 10−9 | 104,044 | 0.01 (0.97) |
0.0077 (0.002) |
0.001 | 128,580 | −0.0041 (0.012) |
0.723 | 42,843 | |
| rs2745353 | 6 | 127494628 | RSPO3 | T/C | 0.51 | 0.0143 (0.002) |
5.5 × 10−9 | 104,075 | 0.06 (0.67) |
−0.0009 (0.002) |
0.677 | 128,615 | −0.0005 (0.011) |
0.962 | 42,853 | |
| rs731839 | 19 | 38590905 | PEPD | G/A | 0.34 | 0.0145 (0.003) |
1.7 × 10−8 | 104,636 | 0.13 (0.38) |
0.0046 (0.002) |
0.038 | 132,528 | 0.0142 (0.012) |
0.220 | 42,846 | |
| rs4865796 | 5 | 53308421 | ARL15 | A/G | 0.67 | 0.0146 (0.003) |
2.1 × 10−8 | 100,001 | 0.03 (0.81) |
0.0043 (0.002) |
0.052 | 127,784 | 0.0337 (0.012) |
0.004 | 42,852 | |
| rs2972143 | 2 | 226824609 | IRS1 | G/A | 0.62 | 0.0142 (0.003) |
3.2 × 10−8 | 99,566 | 0.00 (1.00) |
0.0035 (0.002) |
0.107 | 127,473 | 0.0195 (0.011) |
0.082 | 42,853 | |
| rs1530559 | 2 | 135472099 | YSK4 | A/G | 0.52 | 0.0145 (0.003) |
3.4 × 10−8 | 107,281 | 0.19 (0.18) |
0.0037 (0.002) |
0.100 | 129,880 | 0.0200 (0.011) |
0.077 | 42,849 | |
|
| ||||||||||||||||
| rs2943645 | 2 | 226807424 | IRS1 | T/C | 0.63 | 0.0193 (0.002) |
2.3 × 10−19 | 99,023 | 0.00 (1.00) |
0.0034 (0.002) |
0.112 | 127475 | 0.0210 (0.011) |
0.061 | 42,846 | |
| rs10195252 | 2 | 165221337 | GRB14* | T/C | 0.60 | 0.0174 (0.002) |
1.3 × 10−16 | 98,997 | 0.00 (1.00) |
0.0053 (0.002) |
0.014 | 127005 | 0.0361 (0.011) |
0.001 | 42,846 | |
| rs2126259 | 8 | 9222556 | PPP1R3B | T/C | 0.11 | 0.0238 (0.003) |
3.3 × 10−13 | 99,021 | 0.14 (0.51) |
0.0213 (0.003) |
5.4 × 10−10 | 127480 | −0.0877 (0.017) |
1.8 × 10−7 | 42,849 | |
| FI (BMI-adjusted) | rs4865796 | 5 | 53308421 | ARL15 | A/G | 0.67 | 0.0154 (0.002) |
2.2 × 10−12 | 98,314 | 0.48 (0.01) |
0.0043 (0.002) |
0.052 | 127784 | 0.0337 (0.012) |
0.004 | 42,852 |
| rs17036328 | 3 | 12365484 | PPARG | T/C | 0.86 | 0.0212 (0.003) |
3.6 × 10−12 | 98,984 | 0.21 (0.31) |
0.0051 (0.003) |
0.103 | 128567 | 0.0335 (0.016) |
0.031 | 42,843 | |
| rs731839 | 19 | 38590905 | PEPD | G/A | 0.34 | 0.0148 (0.002) |
5.1 × 10−12 | 103,252 | 0.13 (0.55) |
0.0046 (0.002) |
0.038 | 132528 | 0.0142 (0.012) |
0.220 | 42,847 | |
| rs974801 | 4 | 106290513 | TET2 | G/A | 0.38 | 0.0139 (0.002) |
3.3 × 10−11 | 103,489 | 0.09 (0.67) |
0.0012 (0.002) |
0.582 | 131866 | 0.0052 (0.011) |
0.643 | 42,849 | |
| rs459193 | 5 | 55842508 |
ANKRD55/M
AP3K1 |
G/A | 0.73 | 0.0147 (0.002) |
1.12 × 10−10 | 103,378 | 0.27 (0.17) |
0.0111 (0.002) |
1.6 × 10−6 | 132989 | 0.0276 (0.012) |
0.023 | 42,849 | |
| rs6822892 | 4 | 157954125 | PDGFC | A/G | 0.68 | 0.0138 (0.002) |
2.6 × 10−10 | 103,432 | 0.00 (1.00) |
0.0010 (0.002) |
0.636 | 132951 | 0.0256 (0.012) |
0.031 | 42,836 | |
| rs4846565 | 1 | 217788727 | LYPLAL1 | G/A | 0.67 | 0.0132 (0.002) |
1.8 × 10−9 | 99,014 | 0.00 (1.00) |
0.0066 (0.002) |
0.003 | 127468 | 0.0132 (0.012) |
0.254 | 42,853 | |
| rs3822072 | 4 | 89960292 | FAM13A1 | A/G | 0.48 | 0.0116 (0.002) |
1.8 × 10−8 | 99,977 | 0.00 (1.00) |
0.0025 (0.002) |
0.236 | 129432 | 0.0161 (0.011) |
0.143 | 42,850 | |
| rs6912327 | 6 | 34872900 | UHRF1BP1 | T/C | 0.80 | 0.0165 (0.003) |
2.3 × 10−8 | 80,010 | 0.04 (0.91) |
0.0074 (0.003) |
0.011 | 103826 | 0.0139 | 0.391 | 34,761 | |
|
Primary
trait |
FG |
FI
(BMI- adjuste d) |
||||||||||||||
|
|
||||||||||||||||
| 2hGlu | rs6975024 | 7 | 44198411 | GCK | C/T | 0.15 | 0.1026 (0.016) |
5.2 × 10−11 | 42,842 | 0.00 (1.00) |
0.0605 (0.003) |
2.9 × 10−99 | 103,517 | 0.0063 (0.003) |
0.030 | 98,458 |
| rs11782386 | 8 | 9239197 | PPP1R3B* | C/T | 0.87 | 0.0985 (0.017) |
2.2 × 10−9 | 42,852 | 0.00 (1.00) |
−0.0167 (0.003) |
5.5 × 10−7 | 100,595 | −0.0164 (0.003) |
6.9 × 10−7 | 95,565 | |
| rs1019503 | 5 | 96280573 | ERAP2 | A/G | 0.48 | 0.0628 (0.011) |
8.9 × 10−9 | 42,851 | 19.6 (0.42) |
−0.0061 (0.002) |
0.003 | 108,113 | 0.0004 (0.002) |
0.851 | 103,448 | |
|
|
||||||||||||||||
|
2hGlu
(BMI- adjusted) |
rs7651090 | 3 | 186996086 | IGF2BP2 | G/A | 0.30 | 0.064 (0.012) |
4.5 × 10−8 | 42,792 | 63.4 (0.01) |
0.0128 (0.002) |
1.8 × 10−8 | 104,019 | 0.0003 (0.002) |
0.900 | 98,924 |
Genome-wide loci for fasting glucose (FG), fasting insulin (FI), FI (adjusted for BMI) and 2hGlu are shown along with results for the other traits aligned to the trait-raising allele for the primary trait. “Non-MAGIC” SNPs (identified in other consortia and selected for the Metabochip to follow-up on other non-MAGIC traits) are indicated in bold. Freq denotes the allele frequency of the primary trait-raising allele. Per allele effect (SE) for FI represents changes of natural-log transformed levels of FI. N represents sample size. Heterogeneity was assessed using the I2 index86. The gene shown is the nearest gene to the lead SNP, other than those loci marked with an asterisk. For these loci, the nearest gene is also listed in Supplementary Tables 2a-d.
Box 1.
Fasting glucose
IKBKAP (inhibitor of kappa light polypeptide gene enhancer in β-cells, kinase complex-associated protein) encodes a scaffold protein that binds IKKs and NF-κB-inducing kinase (NIK), assembling them into different active complexes. Splicing mutations in this gene lead to familial dysautonomia56. Also mapping to this region are C9orf4, C9orf5 and C9orf6, MIR32 (microRNA 32, unknown function), as well as ACTL7A (actin-like 7A) and ACTL7B (actin-like 7B).
WARS (tryptophanyl-tRNA synthetase) catalyzes the aminoacylation of tRNA(Trp) with tryptophan. The intronic SNP rs3783347 is associated with WARS expression in liver: the glucose-raising allele associated with lower mRNA expression (age- and sex-adjusted P = 4.19 × 10−5) and is in perfect LD (r2 = 1, D’ = 1) with a 3′UTR SNP in SLC25A47 (rs3736952) and in modest LD (r2 = 0.3, D’ = 1) with non-synonymous Arg135Leu (qualified as tolerated by SIFT and probably damaging by Polyphen). Nearby YY1 (YY1 transcription factor) codes for a zinc-finger transcription factor involved in regulating a broad set of promoters. It has been suggested that YY1-regulated transcription is linked to glucose metabolism via O-GlcNAcylation57.
KL (klotho) encodes a type-I membrane protein related to β-glucosidases. rs576674 lies ~36 kb upstream of KL. Variation in KL has been associated with insulin regulation, insulin resistance phenotypes and cardiovascular disease in some studies14,15,58,59 but KL variants were not associated with diabetes risk60. The various SNPs in these studies are all in weak LD with rs576674 (r2 < 0.125). Variation in KL is also associated with bone metabolism and may play a role in associations of energy metabolism with bone metabolism61,62.
TOP1 (topoisomerase (DNA) I). rs6072275 is intronic in TOP1 and lies in a large region of high LD in Europeans, which includes the plausible biological candidate LPIN3 (lipin 3). In mice, a related homolog Lpin1 is associated with fatty liver dystrophy63, a phenotype similar to human lipodystrophy (loss of body fat, fatty liver, hypertriglyceridemia, and insulin resistance). Lpin1 mRNA is expressed at high levels in adipose tissue and induced during differentiation of preadipocytes, suggesting that lipin is required for normal adipose tissue development, while LPIN2 has been suggested to be associated with T2D and glucose metabolism64. rs6072275 lies in the middle of a large CNV that extends from within the 3′ end of TOP1 to the 5′ end of PLCG1 (phospholipase C, gamma 1).
P2RX2 (purinergic receptor P2X, ligand-gated ion channel, 2). rs10747083 lies in a small CNV about 150 kb upstream of five protein-coding genes, including P2RX2, encoding one of a family of purinoceptors for ATP; GALNT9 (UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9), encoding a member of the UDP-N-acetyl-alpha-D-galactosamine polypeptide N-acetylgalactosaminyltransferase (GalNAc-T) family of enzymes and expressed specifically in the brain; FBRSL1 (fibrosin-like 1); PXMP2 (peroxisomal membrane protein 2, 22 kDa); PGAM5 (phosphoglycerate mutase family member 5), and within 184 kb downstream, POLE (polymerase (DNA directed), epsilon) and LOC100130238 (hypothetical LOC100130238) a miscRNA.
LOC728489. rs3829109 is in low LD with a well-established locus for inflammatory bowel disease. Two recent publications reported CARD9 SNP rs10781499 (r2 = 0.29) to be associated with ulcerative colitis65, and CARD9-SNAPC4 SNP rs4077515 (r2 = 0.27) to be associated with Crohn’s disease and ulcerative colitis66,67.Several genes are located in the region, but few with high plausibility for a role in glycemia.
AMT encodes the mitochondrial aminomethyltransferase which is a critical component of the glycine cleavage system. Depending upon the AMT transcript, rs11715915 is located in 3′UTR or within coding regions, where it causes a synonymous substitution. This SNP is also located downstream of TCTA (T-cell leukemia translocation altered), which has no known metabolic function, and upstream of RHOA (ras homolog family member A). RHOA is a signaling molecule involved actin cytoskeleton stability and reorganization68 that binds and activates Rho kinase (ROCK), a regulator of insulin transcription69 and action70 that is differentially regulated in T2D71 and hypothesized to play a role in glucose homeostasis70.
GLS2 encodes liver-expressed glutaminase 2, which is required for hydrolysis of glutamine. rs2657879 causes a benign (Polyphen) amino acid change (Leu581Pro) in the GLS2 protein. The GLS2 protein is highly expressed (human protein atlas) by both liver and pancreas, and it has been shown in liver tumors that alterations in the balance of GLS2:GLS1 (the kidney-specific homolog) activity are important for regulating glutamate metabolism72. The other gene in this region SPRYD4 (SPRY domain containing 4) has no known function in metabolism.
RREB1 (ras responsive element binding protein 1) encodes a zinc finger transcription factor, with rs17762454 lying in an intron in the gene. The protein product of RREB1 binds to RAS-responsive elements (RREs) of gene promoters, including the calcitonin gene promoter. The role of RREB1 in energy metabolism is not known. An uncorrelated SNP at this locus (rs675209) was associated with serum urate levels (P = 1.0 × 10−9) in a GWAS of serum urate, gout and cardiovascular disease risk factors73. Another gene at this locus, SSR1 (signal sequence receptor, alpha), encodes a glycosylated endoplasmic reticulum membrane receptor associated with protein translocation across the ER membrane. Reactome pathway analysis places this gene in a module with key roles in the synthesis and function of insulin, insulin-like growth factors and ghrelin, making this gene a plausible biological candidate at this locus (REACTOME: REACT_15380). A third gene at this locus, CAGE1, encodes cancer antigen 1. CAGE1 has no known role in metabolism.
Fasting Insulin
TET2 encodes the tet oncogene family member 2, isoform b, which catalyzes the conversion of methylcytosine to 5-hydroxymethylcytosine. The enzyme is involved in myelopoiesis, and defects in this gene have been associated with several myeloproliferative disorders (NCBI RefSeq). Perhaps more relevant to glycemic regulation is PPA2, which encodes the inorganic pyrophosphatase 2 isoform 1 precursor. Its protein product is localized to mitochondria; it has high homology to members of the inorganic pyrophosphatase family, including the signature sequence essential for its catalytic activity (NCBI RefSeq). Pyrophosphatases catalyze the hydrolysis of pyrophosphate to inorganic phosphate.
HIP1 encodes the huntingtin interacting protein 1, a membrane-associated protein that colocalizes with huntingtin. It is ubiquitously expressed with the highest level in brain. Loss of normal huntingtin-HIP1 interaction in Huntington’s disease may contribute to a defect in membrane-cytoskeletal integrity in the brain. Of interest to insulin action, HIP1 is involved in clathrin-mediated endocytosis and trafficking. Mice transgenic for the mutated form of huntingtin develop diabetes74,75; however, though Hip1/Hip1r double-knockout mice have severe vertebral defects, suffer from dwarfism and die in early adulthood, they do not show any fasting glucose abnormalities76. The lead SNP (rs1167800) is only 104 bp away from a missense SNP (rs1167801), encoding a Gln to His amino acid change; however, LD between them is low (r2 = 0.196).
FAM13A (family with sequence similarity 13, member A) encodes a protein with unknown function. Previous GWAS for the study of lung function measures77 and chronic obstructive pulmonary disease78 described variants in FAM13A that affect these traits. SPP1, encoding osteopontin, a secreted matrix glycoprotein and pro-inflammatory cytokine involved in cell-mediated immunity, is within 1 Mb. Mice exposed to high fat diet show increased circulating osteopontin and over-expression of Spp1 in the macrophages recruited into adipose tissue improved insulin sensitivity79, while SPP1 was highly expressed in obese twins relative to their non-obese siblings80. Recent work linked osteopontin to β-cell function through the GIP pathway81. In carriers of the GIPR variant associated with impaired glucose and GIP-stimulated insulin secretion, osteopontin levels were lower compared to non carriers. In addition, both GIP and osteopontin prevented cytokine-induced apoptosis and osteopontin-stimulated cell proliferation of functional β-cell mass.
PEPD (peptidase D) encodes a member of the peptidase family. The protein forms a homodimer that hydrolyzes dipeptides or tripeptides with C-terminal proline or hydroxyproline residues. The enzyme serves an important role in the recycling of proline, and may be rate limiting for collagen production. CEBPA (CCAAT/enhancer binding protein (C/EBP) alpha) is ~100 kb downstream of the lead SNP and encodes a transcription factor expressed in adipose tissue that regulates a number of genes involved in lipid and glucose metabolism genes. A SNP in low LD with our lead SNP was previously associated with triglyceride levels82. Cells from Cebpa (-/-) mice show a complete absence of insulin-stimulated glucose transport, secondary to reduced gene expression and tyrosine phosphorylation of the insulin receptor and IRS1 (ref. 83). CEBPA also modulates expression of leptin by binding to the promoter of the gene84, and our lead SNP showed modest association with BMI in previous GIANT meta-analyses (P = 0.005).
YSK4 (Sps1/Ste20-related kinase homolog) contains rs1530559 in an intron. This gene has no known function in human energy metabolism. Three other genes at this locus also have no known role in energy metabolism, including RAB3GAP1 (RAB3 GTPase activating protein subunit 1 (catalytic), encoding the catalytic subunit of a Rab GTPase activating protein and mutated in Warburg micro syndrome; CCNT2 (cyclin T2), belonging to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle; and ACMSD (aminocarboxymuconate semialdehyde decarboxylase), involved in the de novo synthesis pathway of NAD from tryptophan. ACMSD has been implicated in the pathogenesis of several neurodegenerative disorders.
2h Glucose
ERAP2 (endoplasmic reticulum aminopeptidase 2) encodes an aminopeptidase that hydrolyzes N-terminal amino acids of proteins or peptide substrates. The lead SNP is strongly associated with ERAP2 expression in liver (P = 1.1 × 10−55) and in lymphoblastoid cell lines in individuals from the CEU (P = 8 × 10−21) and YRI samples (P = 2 × 10−15). Also near to this lead SNP is LNPEP (leucyl/cystinyl aminopeptidase), which is widely expressed and well characterised in muscle and fat cells. In response to insulin, LNPEP translocates to the cell surface and co-localizes with GLUT4 (ref. 85). Although the role it plays in insulin action is unknown, this translocation is impaired in individuals with T2D85. PCSK1 is also within 500 kb of the lead SNP, although on the other side of a recombination hotspot (Supplementary Fig. 4d).
Figure 1.
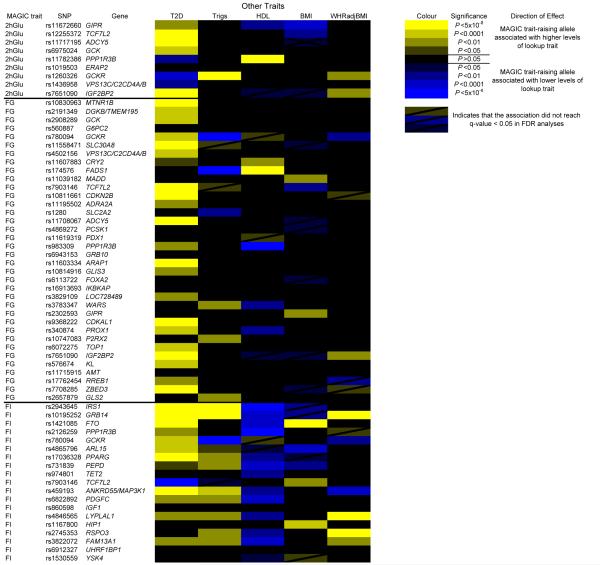
Associations between glycemic loci and T2D, HDL-cholesterol and triglycerides, BMI and WHR. Loci associated with the above traits (P < 0.05) are highlighted. Those with positively correlated effect directions are colored yellow, and those with negative correlations are colored blue. Those which did not reach a q-value < 0.05 in FDR analyses are also marked.
We also identified new associations with fasting glucose in regions previously associated with other metabolic traits or disease outcomes, including T2D6,16 (ARAP1, CDKN2B, GRB10, CDKAL1, IGF2BP2 and ZBED3, which was identified in BMI-adjusted models) and 2hGlu2 (GIPR), as well as confirming the recently identified signals for fasting glucose17-19 at FOXA2, PPP1R3B, PCSK1 and PDX1. FOXA2 is a forkhead transcription factor that regulates PDX1 expression, while PDX1 encodes a transcription factor critical for pancreatic development20. PDX1 mutations have been linked to MODY4 (ref. 21), pancreatic agenesis22 and permanent neonatal diabetes23, although we observed no significant association with T2D in DIAGRAM Metabochip analyses24 (Fig. 1).
Given the overlap between genetic loci for fasting glucose and other metabolic traits, we performed a systematic look-up of all glycemic loci and their associations with other metabolic traits using data available through other consortia25-27. In DIAGRAM Metabochip analyses24, 22 (>60%) of the now 36 genome-wide significant fasting glucose loci showed association (P < 0.05; FDR q < 0.05) with T2D (Fig. 1). In all cases, the glucose-raising allele was associated with increased risk of T2D, yet the fasting glucose effect size and T2D odds ratio were weakly correlated (Fig. 2a).
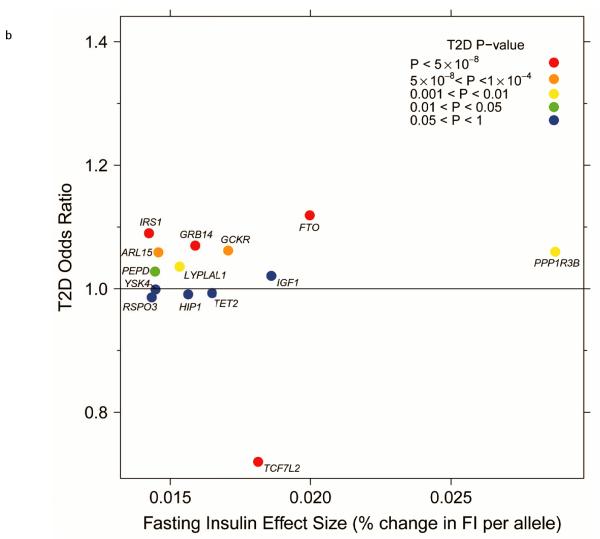
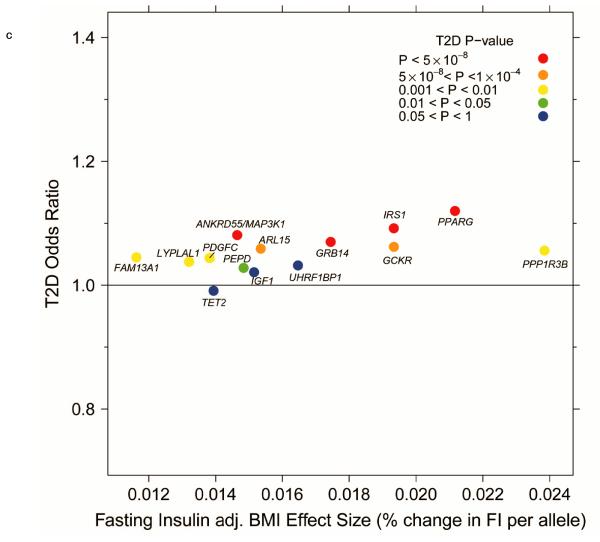
Figure 2.
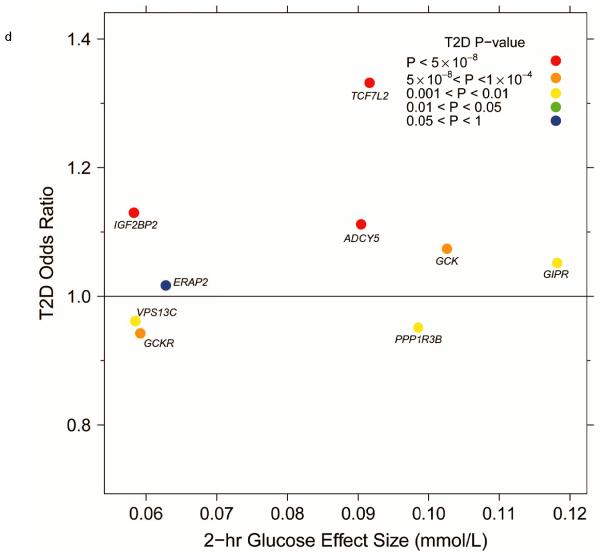
Per-allele beta-coefficients for glucose and insulin concentrations vs. odds ratios for T2D. (a) Fasting glucose vs. T2D. (b) Fasting insulin vs. T2D. (c) Fasting insulin adjusted for BMI vs. T2D. (d) 2-h glucose vs. T2D.
Gene-based analyses confirmed many of the loci identified in individual SNP analyses (Supplementary Table 3a) and identified another nine genomic regions (containing 14 genes) with significant association signals (P < 5 × 10−6), including some with biological candidacy, such as the HKDC1 gene, encoding a putative hexokinase28.
Fasting insulin
In 108,557 individuals, we identified 17 additional loci with genome-wide significant associations to fasting insulin and confirmed known associations1. These newly identified loci include variants in or near HIP1, TET2, YSK4, PEPD and FAM13A1 (Table 1, Box 1 and Supplementary Figs. 3 and 4), as well as SNPs near loci previously associated with other metabolic traits, including T2D6 (TCF7L2, PPARG), BMI29 (FTO), waist-hip ratio26 (WHR) (LYPLAL1, RSPO3, GRB14), triglycerides27 (ANKRD55-MAP3K1) and adiponectin30 (ARL15). We also confirmed the recent associations with fasting insulin at GRB14, PPP1R3B, LYPLAL1, IRS1, UHRF1BP1 and PDGFC19. The ANKRD55-MAP3K1 association is of interest as MAP3K1 regulates expression of IRS1 (ref. 31) as well as activation of NF-κB32,33 and the JNK pathway34, both centrally implicated in insulin resistance35,36. Furthermore, data from DIAGRAM Metabochip analyses show that the insulin-raising allele at this SNP is strongly associated with increased risk of T2D24.
In contrast to fasting glucose (Supplementary Fig. 5a), in fasting insulin analyses adjusted for BMI, we observed a systematic decrease in the standard errors of the SNP effect estimates (Supplementary Fig. 5b), perhaps because BMI explains more of the variance in fasting insulin (R2 = 32.6%) than in fasting glucose (R2 = 8.6%) or 2hGlu (R2 = 11.0%) (data from the Fenland study). Therefore, BMI adjustment removes more variance in fasting insulin, thereby rendering genetic associations more readily detectable. This is supported by the identification of five additional loci in BMI-adjusted models by this approach (Table 1 and Supplementary Figs. 3 and 4). As expected, BMI-adjustment abolished fasting insulin associations at FTO (P = 0.71; Supplementary Table 2b), suggesting that the association with fasting insulin is mediated entirely through association with BMI.
In total, 13 of the 19 loci associated with fasting insulin also showed associations with T2D (P < 0.05; FDR q < 0.05) (Fig. 1), with the insulin-raising allele associated with higher risk of T2D, except for TCF7L2 (Fig. 2b,c), where the allele associated with lower fasting insulin is associated with higher fasting glucose (Table 1). Notably, the loci associated with fasting insulin showed a pattern of association with lipid traits consistent with insulin resistance, and not observed for either fasting glucose or 2hGlu loci (Fig. 1). Thirteen (~68%) of the 19 loci were associated with HDL-cholesterol (q < 0.05): all insulin-raising alleles associated with lower HDL levels, nine of which were also associated with higher triglycerides (q < 0.05) (Fig. 1). Further, the insulin-raising alleles of four SNPs were associated with higher WHR (adjusted for BMI) (q < 0.05) (Fig. 1), another trait linked to insulin resistance, while five SNPs were also associated with BMI, although with inconsistent direction (q < 0.05) (Fig. 1).
In gene-based analyses, we focused on BMI-adjusted results to account for the variance in fasting insulin explained by BMI. Beyond those loci containing genome-wide significant SNPs, we identified 7 distinct regions (containing 22 genes) after Bonferroni-correction (P < 5 × 10−6). Among these genes, we identified many for which prior biological evidence suggests their role in pathways involved in insulin secretion or action (Supplementary Table 3b). While the lead SNP in PPARD was not genome-wide significant (P = 3.9 × 10−6), the PPARD gene, a regulator of adipose, hepatic and skeletal muscle metabolism37, reached the gene-based significance threshold (P < 1 × 10−6). PPARD agonists have also been shown to induce insulin sensitising effects in a mouse model38. In addition, we identified PTEN to be associated (Supplementary Table 3b), a gene previously suggested to affect glucose metabolism through regulation of insulin signalling39, and in which a muscle-specific deletion protected mice from insulin resistance and diabetes resulting from high fat feeding40.
2-h glucose
In 42,854 individuals, we identified four additional loci to be associated with 2hGlu (Table 1 and Supplementary Figs. 3 and 4), including a signal near ERAP2 and three signals near loci previously associated with fasting glucose1 (GCK), HDL-cholesterol27 (PPP1R3B) and T2D6 (IGF2BP2), as well as confirming the five previous associations2. To determine whether these associations reflected differences in the response to a glucose challenge, or were partly driven by effects on fasting glucose, we also performed analyses adjusted for fasting glucose. No additional loci were identified as genome-wide significant after adjustment for fasting glucose, although the GCK association with 2hGlu was severely attenuated (β = 0.04 (SE = 0.016) mmol/L/allele, P = 0.005 vs. β = 0.1 (0.016) mmol/L/allele, P = 5.3 × 10−11 in the model unadjusted for fasting glucose), suggesting that the association with 2hGlu is driven, at least in part, by a primary association with fasting glucose (Supplementary Table 2d). The association of SNPs near GCK with both fasting glucose and 2hGlu suggests a generalized raising of the glucose set point, consistent with inactivating mutations of GCK that cause MODY41. As for fasting glucose, when 2hGlu models were adjusted for BMI, no systematic differences were observed, although again the IGF2BP2 SNP rs7651090 reached genome-wide significance (Table 1).
Eight of the 9 SNPs associated with 2hGlu at genome-wide levels of significance were also associated with T2D (q < 0.05) (Fig. 1), although the 2hGlu-raising alleles at PPP1R3B, GCKR and VPS13C-C2CD4A/B were associated with lower risk of T2D (Fig. 2d), consistent with their association with lower fasting glucose levels (Table 1 and Supplementary Table 2e).
In addition to SNPs that were genome-wide significant in individual SNP analyses, we identified three regions (containing six genes) showing association with 2hGlu in gene-based analyses. These included the HKDC1 gene, as well as an association signal at CRHR1 (P = 2 × 10−6) (Supplementary Table 3c), mostly driven by the lead SNP in this gene (rs17762954), which approached genome-wide significance (P = 7.4 × 10−7). CRHR1, together with GIPR, belongs to the family of class B GPCRs and is highly expressed in pancreatic β-cells, where stimulation of the receptor potentiates insulin secretion in response to glucose42.
Fine-mapping of established loci
Analyses at higher SNP density around previously established loci did not generally yield stronger associations or more plausible functional variants (Supplementary Table 4). For fasting glucose, markedly more significant SNPs or larger effect size than the previous lead SNP were observed for four of the 16 loci: PROX1, GCK, ADRA2A and VPS13C-C2CD4A/B (Supplementary Table 4). Regional plots for these loci are shown in Supplementary Figure 6. While the new lead SNP near ADRA2A was not markedly more significant than the previous lead SNP, the effect size is almost double that of the previous lead SNP (Supplementary Table 4). However, this and other new lead SNPs were without more plausible functionality. The new lead SNP at VPS13C-C2CD4A/B, previously associated with proinsulin43, is far more significant and of larger effect size than the previous lead SNP (β = 0.0273 (SE = 0.0035) mmol/L/allele, P = 4.8 × 10−15 vs. β = 0.0057 (0.0036) mmol/L/allele, P = 0.111; r2 = 0.27). For fasting insulin, another SNP downstream of IGF1 was found to be more significant and with a larger effect size, although with no known functionality (Supplementary Table 4 and Supplementary Fig. 6). For 2hGlu, again, another SNP at VPS13C-C2CD4A/B was more significant than the previous lead SNP (Supplementary Table 4 and Supplementary Fig. 6) and was previously associated with diabetes in Chinese individuals44.
Pathway analysis
Next, we explored whether glycemic loci were enriched for connectivity between genes representing particular pathways or processes. To do this, we used GRAIL software45 and investigated both an excess of connectivity between the established loci (genome-wide significant) and then between established loci and those loci that did not reach genome-wide significance but that showed a lower level of association (P < 0.0005) (Online Methods). We aimed to establish whether there were any biologically relevant genes among this longer list of suggestively-associated loci. This more liberal threshold yielded 218, 155 and 100 regions for fasting glucose, fasting insulin and 2hGlu, respectively. To further assess whether the established loci represented common biological pathways, we used MAGENTA to undertake gene-set enrichment analyses (Online Methods).
We found that genes near the 36 loci associated with fasting glucose had a high degree of connectivity (refer to Online Methods for definition of how genes were selected), with eight genes demonstrating highly significant similarity to genes in other loci at a Pgrail < 0.01 level, connected by keywords such as “glucose”, “insulin”, “pancreatic” and “diabetes” (Supplementary Table 5a and Supplementary Fig. 7), and more than expected by chance (Ppermutation = 0.003). We observed less connectivity among the genome-wide significant fasting insulin and 2hGlu loci, with no genes reaching Pgrail < 0.01 for fasting insulin (Supplementary Table 5b) and only one out of nine genes for 2hGlu (Ppermutation = 0.07) (Supplementary Table 5c).
Among the list of 218 suggestively-associated fasting glucose loci (P < 0.0005), we observed 13 genes to be connected to those in the genome-wide significant loci at Pgrail < 0.01, more than expected by chance (Ppermutation = 0.003) (Supplementary Table 6a). These included genes such as GLP1R (P = 3.3 × 10−7) (a glucagon receptor that mediates the GLP-1 incretin effect and stimulates insulin release), IRS2 (P = 6.9 × 10−5; central to development and maintenance of β-cell mass and function46,47) and INS (P = 2.5 × 10−6; the insulin gene encoding proinsulin). The presence of these and other genes support our conjecture that many of the SNPs approaching genome-wide significance are likely to represent true associations. Of the 155 suggestively-associated loci for fasting insulin (adjusted for BMI), we observed seven to be connected to the genome-wide significant loci at Pgrail < 0.01; more than expected by chance (Ppermutation = 0.002), and these included INSR (Pgrail = 1.5 × 10−4; encoding insulin receptor precursor), CD36 (Pgrail = 0.001; previously implicated in insulin resistance48), GCG (Pgrail = 0.008; glucagon gene) and HNF1A (Pgrail = 0.005; mutations in which are associated with MODY3)49 (Supplementary Table 6b). Of the 100 suggestively-associated loci for 2hGlu (P < 0.0005), we found three to reach Pgrail < 0.01 (Ppermutation = 0.014) and the gene highlighted as most biologically connected to the genome-wide significant loci was again HNF1A (Pgrail = 3.4 × 10−4) (Supplementary Table 6c).
Using MAGENTA, we identified four pathways enriched for fasting glucose associations: GOTERM pathways lens development in camera-type eye (P = 0.004), PANTHER processes gut mesoderm development (P = 0.009), other steroid metabolism (P = 0.02) and KEGG MODY pathway (P = 0.03), although these were no longer significant after removing lead genes (P > 0.05), all of which were known fasting glucose loci: PROX1 for eye and gut, and G6PC2 and GCK for steroid and MODY pathways, respectively.
Directional consistency of associations between discovery and follow-up studies
Given the wealth of biologically plausible genes in loci near genome-wide significance (Supplementary Tables 6a-c) and the deviation of the observed distribution from the expected in QQ plots even after removing all established loci (Supplementary Fig. 8a-d), we hypothesized that additional loci not reaching genome-wide significance were likely to represent true associations with small effects. To establish the presence of further true associations that did not reach genome-wide significance, we compared SNP associations in discovery studies (those included in the original meta-analyses for 42,078 (fasting glucose), 34,230 (fasting insulin) and 15,252 (2hGlu) individuals1,2) with those in the “follow-up” studies (consisting of 85,710 (fasting glucose), 69,240 (fasting insulin) and 27,602 (2hGlu) individuals). We identified all SNPs which had a nominally significant association (P < 0.05) in the follow-up studies alone and, for these SNPs, performed a binomial test of whether more SNPs than expected by chance (50%) had a consistent direction of effect with that observed in the discovery analyses. We were also able to compare among SNPs nominated for follow-up by different consortia (Supplementary Fig. 9a-d).
For each trait, evaluation of the 66,000 Metabochip follow-up SNPs demonstrated a significant excess of SNPs showing directionally consistent associations (P < 0.05) compared to that expected by chance (fasting glucose: Pbinomial = 5.01 × 10−12; fasting insulin: Pbinomial = 7.58 × 10−13; fasting insulin (adjusted for BMI): Pbinomial = 9.76 × 10−9; 2hGlu: Pbinomial = 2.37 × 10−6; Supplementary Table 7 and Supplementary Fig. 9a-d). FDR analyses suggested that a number of these nominal associations in the follow-up studies are true positives for fasting glucose and fasting insulin in particular (fasting glucose: 23%; fasting insulin: 24%; Supplementary Table 7). Interestingly, when we evaluated consistency of association with fasting insulin (between discovery and follow-up) among SNPs submitted to the Metabochip by other consortia, SNPs submitted by GIANT (anthropometric traits) (Pbinomial = 1.52 × 10−8) and GLGC (lipid traits) (Pbinomial = 1.15 × 10−6) and for BMI and triglycerides in particular also demonstrated a marked excess of directional consistency (Supplementary Table 7 and Supplementary Fig. 9b). When we performed the same test for fasting insulin adjusted for BMI, the observed enrichment among SNPs submitted by GIANT and GLGC was attenuated (Supplementary Table 7 and Supplementary Fig. 9c), although SNPs nominated to follow up on triglyceride associations remained the most significant (P = 3.18 × 10−7, Supplementary Table 7 and Supplementary Fig. 9c). Of the 3,353 SNPs submitted for follow-up of triglyceride associations, 158 SNPs showed nominal significance (P < 0.05) in follow-up studies and consistent direction of association with fasting insulin (adjusted for BMI) in both discovery and follow-up (Supplementary Table 7). In 139 (88%) of these SNPs, the insulin-raising alleles were associated with higher levels of triglycerides, consistent with the positive correlations between fasting insulin and triglyceride associations observed among the genome-wide significant fasting insulin loci (Fig. 1).
DISCUSSION
In the current meta-analysis of ~66,000 Metabochip follow-up SNPs in up to 133,010 individuals, we identified a large number of loci to be associated with glycemic traits, explaining 4.8%, 1.2% and 1.7% of the variance in fasting glucose, fasting insulin and 2hGlu, respectively. Of the 53 glycemic loci, 33 are also associated with increased T2D risk (q < 0.05), extending the overlap between glycemic and T2D loci. Given the current DIAGRAM effective sample size of 106,953 individuals, we can exclude an effect on T2D of 1.04 with 80% power for alleles more frequent than 5%, effectively confirming that the overlap is incomplete and that many loci associated with glycemic traits have no discernible effect on T2D (Figs. 1 and 2).
Previously, we had detected only two loci associated with fasting insulin and had hypothesized that this might be due to a different genetic architecture of this trait compared to fasting glucose, with potentially smaller effect sizes, lower frequency alleles or greater environmental influence on fasting insulin1. In the current meta-analysis including up to 108,557 individuals (compared to 62,264 individuals previously), we expanded the number of loci associated with this trait to 19. Of note was the effect of BMI-adjustment on our ability to detect additional loci (five non-overlapping with unadjusted results)19. We also noted that some of the loci influencing fasting insulin uncovered after BMI-adjustment are likely to have been negatively confounded in previous efforts: at some loci, the insulin-raising allele was nominally associated with lower BMI (potentially via insulin resistance attenuating the anabolic effects of insulin). Given the positive correlation between BMI and fasting insulin, it is likely that this association previously masked their effect on fasting insulin. Fasting insulin loci showed directionally consistent association with lipid levels (HDL and triglycerides); that is, the insulin-raising allele was associated with lower HDL and higher triglyceride levels, a hallmark combination in insulin resistant individuals. We also observed some overlap between fasting insulin loci and those associated with abdominal obesity (Fig. 1). Jointly, these data suggest links of these fasting insulin loci to insulin resistance-related phenotypes. Indeed, some of the fasting insulin loci identified, such as IRS1 and PPARG, are classically known to exert effects on insulin action or sensitivity50,51.
There are now 36 established fasting glucose loci, many of which contain compelling biological candidate genes with plausible causality, including those encoding transcription factors with known roles in pancreas development (e.g. PDX1, FOXA2, PROX1, GLIS3) and genes involved in β-cell function and insulin secretion pathways (SLC2A2, GCK, PCSK1). For 2hGlu, only nine loci have been established to date, which likely reflects the smaller sample size available and consequently reduced power.
Comparing the consistency of the direction of associations for glycemic traits between “discovery” and “follow-up” studies suggests that we are observing more directionally consistent associations than expected by chance among Metabochip follow-up SNPs (Supplementary Fig. 9a-d). This, combined with the excess of biologically plausible genes among the borderline loci (Supplementary Table 6a-c), suggests that beyond the genome-wide significant loci there is a more extensive list of loci still likely to contain true associations. Indeed, some of these loci are implicated by gene-based analyses, which identify genes with compelling biological credentials. For fasting insulin, these analyses revealed additional loci with previously suggested links to insulin resistance (PPARD and PTEN). These results lend further support to the proposal that a long tail of common variants of small effect size are likely to account for a substantial proportion of the variance of complex traits7,8.
Of note is the number of glycemic loci associated with other metabolic traits (q < 0.05; 34 of 53) and also at genome-wide levels of significance (P < 5 × 10−8) (14 of 53) (Fig. 1), potentially implicating pleiotropic effects. Further support for this notion comes from the analysis of loci nominated for the Metabochip by other consortia and their associations with glycemic traits (Supplementary Fig. 9a-d). Indeed, some of the loci associated with glycemic traits at genome-wide significance levels were not originally nominated to the Metabochip for follow-up by MAGIC (Table 1). Metabochip data available across all contributing consortia will facilitate systematic exploration of these correlated phenotypes with more sophisticated statistical methods for joint analysis52-54, yielding greater insight into the underlying pathways and genetic networks they represent. As data from human genetic networks accrue, we will be better placed to test whether there is support for the notion of “hub” genes; that is, genes highly connected with others in the network and proposed by experiments in C. elegans to act as buffers for genetic variation and that could act as modifier genes for many different disorders55.
In summary, we present a large number of genome-wide significant loci influencing glycemic traits, many with a compelling biological basis for their association, as well as a number of loci not previously implicated in glycemic regulation, and for which fine-mapping and functional follow-up will expand and improve our understanding. Use of the Metabochip for deep follow-up has identified additional loci to be involved in glycemic regulation that, due to insufficient sample size and power, did not reach genome-wide significance. Consideration of such loci in future studies will better exploit data from GWAS and complimentary approaches and further improve our biological understanding of glycemic control and the etiology of diabetes.
ONLINE METHODS
Study design
The Illumina CardioMetabochip (Metabochip) is a custom Illumina iSELECT array of 196,725 SNPs. It has been designed to support efficient large-scale follow-up of putative associations for glycemic (including fasting glucose, fasting insulin and post-challenge glucose concentrations (2hGlu) and other metabolic and cardiovascular traits (Supplementary Fig. 1)9, and to enable the fine-mapping of established loci. Overall, there were 65,435 SNPs genotyped on the Metabochip for follow-up of previous associations including a total of 23 cardio-metabolic traits. Traits contributing SNPs to the Metabochip were prioritized into “primary” (including fasting glucose) and “secondary” (including fasting insulin and 2hGlu) contributing ~5K and ~1K SNPs, respectively, from the most significantly associated variants for each phenotype in the discovery meta-analyses from each contributing consortium. This included 5,055 SNPs for follow-up of fasting glucose, 1,046 for fasting insulin and 1,038 for follow up of 2hGlu associations. In the present analysis, we focused our analysis on this set of “follow-up” SNPs available on the Metabochip to establish variants among these SNP associated with glycemic traits. While we also included newly available studies genotyped on genome-wide platforms, we limited our primary analyses to only these ~66,000 SNPs.
Studies
In the present effort, collaborating studies within the Meta-Analysis of Glucose and Insulin related traits Consortium (MAGIC) provided results for the 66,000 “follow-up” SNPs genotyped on Metabochip on a maximum total of 133,010 (fasting glucose), 108,557 (fasting insulin) and 42,854 (2hGlu) individuals. As well as those newly genotyped on the Metabochip platform, in our overall meta-analysis we were able to include further studies which had genotyped or imputed the same SNPs on other platforms. The largest proportion of our entire sample was directly genotyped on the Metabochip and comprised 53,622 (fasting glucose), 42,384 (fasting insulin) and 27,602 (2hGlu) individuals from 26/21/12 studies, respectively. We were also able to recruit 11,690 (fasting glucose) and 8,813 (fasting insulin) individuals from up to four additional GWA studies (Prevend, Ascot (FG-only), Prosper and TRAILS) (Supplementary Table 1) not included in the original meta-analysis1. From another MAGIC study of sex-specific associations with glycemic traits (I. Prokopenko on behalf of the MAGIC authors, personal communication), we were able to recruit another 15/13 independent studies comprising up to 25,618 and 23,130 individuals for fasting glucose and fasting insulin, respectively. The above studies were combined in a single fixed-effects meta-analysis with those studies included in the original GWAS1,2: 20 (fasting glucose), 19 (fasting insulin) and 9 (2hGlu) studies and 42,080 (fasting glucose), 34,230 (fasting insulin) and 15,252 (2hGlu) individuals, as described previously1,2. The study and individual counts from the original GWAS excluded the family-based SardiNIA study where, initially, a large number of the individuals had imputed genotype data only. The entire sample was directly genotyped on Metabochip, so those data were included in place of the original GWAS. Some studies had genotyping data available from both Metabochip and genome-wide arrays but from entirely independent samples within the studies (Supplementary Table 1). Full study characteristics of all Metabochip studies are shown in Supplementary Table 1, while data from discovery genome-wide studies and those from the sex-specific analyses are reported elsewhere1,2 (I. Prokopenko on behalf of the MAGIC authors, personal communication). All participants of the main analysis were of European descent and mostly adults, although data from a total of 7,872 and 7,164 adolescents were also included in the fasting glucose and fasting insulin meta-analyses, respectively (NFBC86, Leipzig-childhood_IFB, TRAILS and ALSPAC studies). All studies were approved by local research ethic committees and all participants gave informed consent. Results from the CLHNS study of Filipino women (n = 1,682 and 1,635 for fasting glucose and fasting insulin, respectively) genotyped on Metabochip were also available and were included in supplementary analyses to compare effect directions with European-descent studies alone.
Phenotypes
Analyses were undertaken for fasting glucose and fasting insulin measured in mmol/l and pmol/l, respectively. 2hGlu was measured in mmol/l. Similar to the previous MAGIC discovery analysis1,2, individuals were excluded from the analysis if they had a physician diagnosis of diabetes, were on diabetes treatment (oral or insulin), or had a fasting plasma glucose equal to or greater than 7 mmol/l. Individual studies applied further sample exclusions, including pregnancy, non-fasting individuals and type 1 diabetes, as detailed in Supplementary Table 1. Individuals from case-control studies (Supplementary Table 1) were excluded if they had hospitalization or blood transfusion in the 2-3 months before phenotyping took place. 2hGlu measures were done 120 min after a glucose challenge during an oral glucose tolerance test (OGTT). Measures of fasting glucose and 2Glu made in whole blood were corrected to plasma level using the correction factor of 1.13 (ref. 87). Fasting insulin was measured in serum. Detailed descriptions of study-specific glycemic measurements are given in the Supplementary Table 1.
Trait transformations and adjustment
Analyses were performed for untransformed levels of fasting glucose, natural logarithm transformed fasting insulin and untransformed 2hGlu using a linear regression model. All analyses were adjusted for age (if applicable), study site (if applicable) and geographical covariates (if applicable) to evaluate the association using an additive genetic model at each genetic SNP variant.
BMI-adjusted analysis
In the Fenland study (Supplementary Table 1), we investigated the correlation between BMI and natural logarithm transformed fasting insulin, fasting glucose and 2hGlu to establish the variation in each trait explained by BMI. Meta-analyses for each trait were also adjusted for body mass index (BMI). Metabochip and new GWA studies performed study-level analyses adjusted for BMI. Most studies in the original GWAS (except deCode, GEMs, KORAF4, TwinsUK studies) as well as from the studies analyzed in a sex-specific manner were included in BMI-adjusted meta-analysis. The original discovery 2hGlu meta-analysis adjusted for BMI2 was also included in these analyses. We also performed an analysis for 2hGlu adjusted for fasting glucose to investigate if additional variants would be identified with an effect on 2hGlu independent of fasting glucose and also to establish whether identified 2hGlu associations were driven by fasting glucose.
Genotyping and quality control
The Metabochip or other commercial genome-wide arrays were used by individual studies for genotyping. Details are presented in Supplementary Table 1 or are reported elsewhere1,2. The quality control criteria for both Metabochip and genome-wide arrays for filtering of poorly genotyped individuals or low quality SNPs prior to imputation included: (i) call rate < 0.95; (ii) sex-discrepancies; (iii) ethnic outliers; (iv) heterozygosity (Supplementary Table 1); (v) SNP minor allele frequency < 0.01; (vi) SNP Hardy-Weinberg equilibrium P < 10−4; (vii) SNP effect estimate standard error (SE) ≥10; (viii) SNPs minor allele count (MAC) < 10 (calculated as total number of observed alleles at each SNP multiplied by MAF).
Studies with genome-wide arrays undertook imputation using the HapMap CEU reference panel using MACH and IMPUTE software (Supplementary Table 1). Parameters used in imputation and filters applied to imputed genotypes are described in Supplementary Table 1 or reported previously1,2. From a total of ~2.5M genome-wide directly genotyped or imputed autosomal SNPs, study-specific results for the ~66,000 Metabochip follow-up SNPs were considered for the present meta-analyses. SNPs with a meta-analysis result for more than a total 10,000 individuals were included in the analysis.
Statistical analysis
Analyses of previous discovery studies are reported elsewhere1,2, while those studies genotyped on the Metabochip are described in Supplementary Table 1. SNP effect estimates and their standard errors (for additive genetic model) were combined by inverse-variance weighted fixed effects meta-analysis using METAL88 and GWAMA89. Two parallel meta-analyses for each trait by different analysts were compared for consistency. Individual cohort results were corrected for residual inflation of the test statistics using lambda of genomic control (GC) estimates. The GC values were estimated for each study using either test statistics from all SNPs for the GWA studies, while for those studies genotyped on the Metabochip, GC lambda estimates were derived from test statistics for 5,041 SNPs selected for follow-up of QT-interval associations, as we perceived these to have the lowest likelihood of common architecture of associations with glycemic traits. Individual study-level lambda GC estimates are shown in Supplementary Table 1. Overall QQ plots for the QT follow-up SNPs are shown in Supplementary Figure 10.
Trait-associated signal selection strategy
Meta-analysis results for each trait were considered as genome-wide significant if they achieved P ≤ 5 × 10−8 threshold and were not in LD (r2 < 0.05) or within 500 kb of an established signal. The most significantly associated SNP (lowest P-value) in each region (500 kb) was selected as the lead SNP. Associated loci are referred to by the name of the nearest gene, unless a more biologically plausible gene was nearby, or a nearby gene was previously associated with another trait. In such cases, we maintain consistency with the previous naming, but list the nearest genes in Supplementary Tables 2a-d. To establish the variance in each trait explained by these SNPs, in the Framingham Heart Study, we included all SNPs in a model adjusted for age, sex, BMI and cohort.
Fine-mapping of known glycemic trait loci
To undertake preliminary fine-mapping analyses, we investigated the patterns of association at 17 known fasting glucose and fasting insulin loci1 and 5 known 2hGlu loci2 using meta-analysis results from 13,644, 1,309 and 1,249 SNPs genotyped on the Metabochip in 53,622, 42,384 and 27,602 individuals for fasting glucose, fasting insulin and 2hGlu, respectively. Only studies genotyped directly on the Metabochip were used for fine-mapping purposes in order to have equal sample size and availability of all SNPs. Regional plots for each locus were created using the previous lead SNP1 or a suitable proxy (r2 > 0.8) as the index SNP if that marker was not present on Metabochip. The plots were generated on the LocusZoom web-based plotting software90 using LD information from the 1000 Genomes Project (hg19/Nov2010EUR data). Prior to generating the plots, all SNP names and positions from the Metabochip-only meta-analysis files were aligned to build37 using the Lift Genome annotation tool on the UCSC website (http://genome.ucsc.edu/cgi-bin/hgLiftOver) in order to be compatible with the 1000 Genomes SNP naming format (chr:position) and allow more thorough assessment of the pairwise LD patterns around the established SNPs.
Associations of glycemic trait variants with related traits
For those SNPs which we identified to be genome-wide significant, we also investigated their association with other metabolic and disease traits. We exchanged reciprocal data for such SNPs with the latest DIAGRAM Metabochip analyses24, and checked associations of these SNPs in publicly available data from previous studies of lipid traits from the GLGC27 (triglycerides, HDL- and LDL-cholesterol; http://www.sph.umich.edu/csg/abecasis/public/lipids2010/) as well as BMI and waist-hip ratio (WHR) from the GIANT consortium25, 26 (http://www.broadinstitute.org/collaboration/giant/index.php/Main_Page). From these data, we were able to establish the presence of any association and the direction of effect for these other traits aligned to our trait-raising alleles. We highlighted associations with other traits at P < 0.05, and also performed FDR analyses. We performed FDR analyses for each trait separately (removing duplicate loci that were associated with more than one glycemic trait) and identified those where q < 0.05.
eQTL analyses
Liver gene expression data from the Advanced Study of Aortic Pathology (ASAP) study has been described previously91. In brief, liver biopsies were collected from patients at the Karolinska University Hospital, Stockholm, Sweden undergoing aortic valve surgery alone or combined with surgery for aortic aneurysm, starting from February 13, 2007. All subjects gave their informed consent and the study was approved by the ethics committee of Karolinska Institute, Stockholm, Sweden. After hybridization of extracted RNA to Affymetrix ST 1.0 Exon arrays, data was RMA normalized and log-transformed. DNA was extracted from whole blood and genotyping was carried out using the Illumina 610w-Quad bead array platform. Imputation was carried out on SNPs with a call rate exceeding 95%, using the MACH algorithm. Imputation quality scores of RSQ < 0.3 were excluded from analysis. An additive genetic model was used to test for association between SNPs and gene expression.
VEGAS
To identify genes with multiple associated SNPs, we performed gene-based analysis using VEGAS, described in detail previously10. Briefly, on all available samples and among the ~66,000 follow-up SNPs, VEGAS pooled the information for all SNPs within each gene (± 50 kb) to identify genes with higher evidence of association than expected by chance, while adjusting for gene size and the linkage disequilibrium structure of the SNPs, by simulation (maximum number of simulations used was 106). We identified genomic regions (separated by >1 Mb) showing evidence of association and described the genes contained within those regions. While we often identified multiple genes within an associated region, it is probable that some of these are significant via LD. Bonferroni correction was used to adjust for multiple testing, based on the number of independent tests (number of genes tested) (~9,300) and P-values < 5.0 × 10−6 were considered significant. While the number of genes represented was constrained by those SNPs submitted to the Metabochip, our analyses asked the question: of the genes represented on the Metabochip, all with a slightly raised prior likelihood of association, which genes show the most evidence for association with glycemic traits?
GRAIL
We used the GRAIL45 to evaluate whether genome-wide loci associated with glycemic traits were enriched for connectivity between genes representing particular pathways or molecular processes. As described in detail previously45, to define the genes near each SNP, GRAIL finds the furthest neighboring SNPs in the 3′ and 5′ direction in LD (Hapmap CEU: r2 > 0.5) and proceeds outwards in each direction to the nearest recombination hotspot92. All genes that overlap that interval are considered implicated by the SNP. If there are no genes in that region, the interval is extended by 250 kb in either direction. The method performs a text-based analysis looking at abstracts in PubMed prior to December 2006 (to avoid confounding from GWAS results arising after that date). We performed two analyses for each trait: first, we took all genome-wide signals for each trait as seed and query loci to investigate biological connectivity among those loci (fasting glucose = 35, fasting insulin = 16, 2hGlu = 9). For fasting insulin, we did not include FTO as the association with fasting insulin was entirely mediated by BMI. Secondly, we also investigated connectivity between these established signals (as seed regions) and those which did not reach genome-wide significance but were suggestively associated with each trait (P < 0.0005) (as query regions) as described previously93. For fasting insulin, we used BMI-adjusted results to define the query regions. Query regions were defined by taking all SNPs more significant than P < 0.0005, removing those associated at genome-wide levels of significance and pruning SNPs of r2 > 0.05 in each region using PLINK94. As GRAIL tests connectivity of regions, we also removed any duplicates where a region was represented by more than one SNP. For those SNPs not found by the software, we submitted the region as a 500 kb window centered at the location of the SNP. This approach identified 218, 155 and 100 query regions (representing 715, 639 and 298 genes) for fasting glucose, fasting insulin (adjusted for BMI),= and 2hGlu, respectively. The number of loci reaching Pgrail < 0.01 was identified from these analyses, and to establish the level of enrichment, we randomly sampled 1,000 random sets of matched numbers of SNPs and calculated the proportion with as many or more reaching Pgrail < 0.01 to derive a permutation based P-value (Ppermutation).
Pathway analyses
Pathway analysis was carried out for fasting glucose, fasting insulin and 2hGlu (uniform/FG-BMI adjusted) using data from previous discovery GWAS studies only1 to avoid bias towards pathways represented on the Metabochip (build36, n > 10,000 and MAF ≥ 1% cutoff used). The software used for this analysis was MAGENTA 2.4 (July 2011, http://www.broadinstitute.org/mpg/magenta/). SNPs from the meta-analysis file were assigned to a gene if they mapped within 110 kb upstream and 40 kb downstream of transcript boundaries. The smallest P-value for the set of SNPs assigned to the gene was adjusted for confounders, such as gene length, marker density and LD in a linear regression, creating a gene association score. If a top SNP was assigned to multiple genes, only the gene with the lowest score was kept to avoid positional clustering. The HLA region was removed due to high LD and gene density. Pathway terms from multiple databases (GO, PANTHER, Ingenuity, KEGG) was attached to each gene. The genes were ranked on their association score, and a GSEA test was performed testing all pathway terms using a 5% and 75% cutoff. Initially, 10,000 gene set permutations were performed for GSEA P-value estimation. This number was then increased with GSEA P < 1 × 10−4, and up to 1,000,000 permutations were performed. Results were sorted on FDR (5% cutoff), and FDR < 0.05 was considered to be significant.
Analyses of directional consistency of associations between discovery and follow-up studies
We investigated whether the Metabochip follow-up SNPs were likely to contain further true associations in addition to those SNPs which reached genome-wide significance. To do so, we meta-analyzed those studies involved in the original discovery analyses1,2 comprising 42,078 individuals for fasting glucose, 34,230 for fasting insulin and 15,252 for 2hGlu, and also then separately meta-analyzed all studies newly available to follow up, comprising 85,710 individuals for fasting glucose, 69,240 for fasting insulin and 27,602 for 2hGlu. For each trait (fasting glucose, fasting insulin, FI-BMIadj and 2hGlu), we then identified all SNPs which had a nominally significant association (P < 0.05) in the follow-up studies alone and, for these SNPs, performed a two-sided binomial test of whether more SNPs than expected by chance (50%) had a consistent direction of effect with that observed in the discovery analyses. Before performing these analyses, SNPs were filtered by LD (r2 < 0.01) to identify independent variants, and all SNPs (and those in LD, r2 ≥ 0.01) associated with glycemic traits (fasting glucose, fasting insulin, 2hGlu, HbA1c and proinsulin) at genome-wide levels of significance (including those SNPs identified in the present study) were excluded. These analyses were initially performed for all 66,000 SNPs, but we were then also able to compare across SNPs submitted to the Metabochip by different consortia and for SNPs submitted to follow up on particular traits amongst these consortia. The results of each of these tests were plotted overall, within SNPs from each consortia, and within SNPs submitted for follow-up of each trait in Supplementary Figure 9. The numbers of SNPs meeting these criteria are shown are Supplementary Table 7. We supplemented these results with FDR analyses and noted the q-value at P = 0.05 in the follow-up studies to identify the likelihood of true positives among these nominally significant SNPs (Supplementary Table 7).
Supplementary Material
ACKNOWLEDGMENTS
AGES: The AGES-Reykjavik study was supported by a contract from the National Institutes of Health (N01-AG-1-2100), National Institute on Aging Intramural Research Program, Hjartavernd (the Icelandic Heart Association) and the Althingi (the Icelandic Parliament).
ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which include interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council (Grant ref: 74882) the Wellcome Trust (Grant ref: 076467, 092731) and the University of Bristol provide core support for ALSPAC.
AMC-PAS: AMC-PAS is grateful to M.D. Trip and S. Sivapalaratnam for their input in collecting the data.
Amish: The Amish studies are supported by grants and contracts from the NIH, including R01 AG18728, R01 HL088119, U01 GM074518, U01 HL072515-06, U01 HL84756, R01 DK54261, the University of Maryland General Clinical Research Center, grant M01 RR 16500, the Mid-Atlantic Nutrition Obesity Research Center grant P30 DK72488, the Baltimore Diabetes Research and Training Center grant P60DK79637 and by the T32 training grant AG000219 (Dr. Montasser). In addition, this project was supported by National Research Initiative Competitive Grant no. 2007-35205-17883 from the USDA National Institute of Food and Agriculture. We gratefully thank our Amish community and research volunteers for their long-standing partnership in research, and acknowledge the dedication of our Amish liaisons, field workers and the Amish Research Clinic staff, without which these studies would not have been possible.
ARIC: The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. We thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by grant number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. We are grateful for resources provided by the University of Minnesota Supercomputing Institute.
ASAP: The ASAP study was funded by a donation from Fredrik Lundberg.
ASCOT: This work was supported by Pfizer (New York, NY, USA) for the ASCOT study and the collection of the ASCOT DNA repository; by Servier Research Group (Paris, France); and by Leo Laboratories (Copenhagen, Denmark). We thank all ASCOT trial participants, physicians, nurses and practices in the participating countries for their important contribution to the study. In particular, we thank Clare Muckian and David Toomey for their help in DNA extraction, storage and handling. The genotyping was funded by a Wellcome Trust Strategic Award (no. 083948).
BLSA: The BLSA was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. A portion of that support was through an R&D contract with MedStar Research Institute.
Busselton Health Study (BSN): The Busselton Health Study (BHS) acknowledges the generous support for the 1994/5 follow-up study from Healthway, Western Australia and the numerous Busselton community volunteers who assisted with data collection and the study participants from the Shire of Busselton. The BHS is supported by The Great Wine Estates of the Margaret River region of Western Australia.
CHS: This CHS research was supported by NHLBI contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133 and NHLBI grants HL080295, HL075366, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098 and AG-027058 from the NIA (see also http://www.chs-nhlbi.org/pi.htm). DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Bruce Psaty serves on a DSMB for a clinical trial of a device funded by the manufacturer (Zoll-Lifecor).
CLHNS: We thank the Office of Population Studies Foundation research and data collection teams and the study participants who generously provided their time for this study. This work was supported by National Institutes of Health grants DK078150, TW05596, HL085144, RR20649, ES10126 and DK56350.
CoLaus: The CoLaus study was supported by research grants from the Swiss National Science Foundation (grant no: 33CSCO-122661) from GlaxoSmithKline and the Faculty of Biology and Medicine of Lausanne, Switzerland. The authors also express their gratitude to the participants in the Lausanne CoLaus study and to the investigators who have contributed to the recruitment, in particular Yolande Barreau, Anne-Lise Bastian, Binasa Ramic, Martine Moranville, Martine Baumer, Marcy Sagette, Jeanne Ecoffey and Sylvie Mermoud, for data collection. Peter Vollenweider and Gérard Waeber received an unrestricted grant from GlaxoSmithKline to build the CoLaus study
CROATIA-Korcula: The work is supported by European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947); grant ~216-1080315-0302 (to IR) from the Croatian Ministry of Science, Education and Sport. Studies carried out in the Croatian island of Korcula were supported by Medical Research Council UK. The authors collectively thank a large number of individuals for their help in organizing, planning and carrying out the field work related to the project: Stipan Jankovic and staff at the University of Split Medical School; Branka Salzer from the biochemistry lab “Salzer”, Croatia; local general practitioners and nurses; and the employees of several other Croatian institutions who participated in the field work, including but not limited to the University of Rijeka, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia. SNP Genotyping of the Korcula samples was carried out by Helmholtz Zentrum München, GmbH, Neuherberg, Germany.
CROATIA-Split: The work is supported by grant ~216-1080315-0302 (to I.R.) from the Croatian Ministry of Science, Education and Sport. Studies carried out in the Croatian City of Split were supported by Medical Research Council UK. The authors collectively thank a large number of individuals for their help in organizing, planning and carrying out the field work related to the project: Stipan Jankovic and staff at the University of Split Medical School; Branka Salzer from the biochemistry lab “Salzer”, Croatia; local general practitioners and nurses; and the employees of several other Croatian institutions who participated in the field work. including but not limited to the University of Rijeka, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia. SNP Genotyping of the Split samples was carried out by AROS Applied Biotechnology AS, Aarhus N, Denmark.
CROATIA-Vis: The work is supported by European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947); grant ~216-1080315-0302 (to IR) from the Croatian Ministry of Science, Education and Sport. Studies carried out in the Croatian island of Vis were supported by Medical Research Council UK. The authors collectively thank a large number of individuals for their help in organizing, planning and carrying out the field work related to the project: Pavao Rudan and staff of the Institute for Anthropological Research in Zagreb, Croatia; Stipan Jankovic and staff at the University of Split Medical School; Ariana Vorko-Jovic and staff and medical students of the Andrija Stampar School of Public Health of the Faculty of Medicine, University of Zagreb, Croatia; Branka Salzer from the biochemistry lab “Salzer”, Croatia; local general practitioners and nurses; and the employees of several other Croatian institutions who participated in the field work. including but not limited to the University of Rijeka, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia. SNP Genotyping of the Vis samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh, Scotland.
DESIR: We thank all the participants of the D.E.S.I.R study, Elodie Eury and Stéphane Lobbens for technical support for the genotyping, Olivier Lantieri and Michel Marre from the D.E.S.I.R study. The genotyping was supported by the “Conseil Régional Nord-Pas-de-Calais Fonds européen de développement économique et regional” CPER axe Cartdiodiabéte 2010-2011 grant to N.B.-N.
deCODE: We thank the individuals who participated in the study and whose contribution made this work possible. The research performed at deCODE genetics was part funded through the European Community’s Seventh Framework Programme (FP7/2007-2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413.
DIAGEN: The presented study was supported by the Commission of the European Communities, Directorate C - Public Health and Risk Assessment, Health & Consumer Protection, Grant Agreement number - 2004310 and by the Dresden University of Technology Funding Grant, Med Drive. We are grateful to all of the patients who cooperated in this study and to their referring physicians and diabetologists in Saxony.
DPS: The DPS has been financially supported by grants from the Academy of Finland (117844 and 40758, 211497, and 118590), the EVO funding of the Kuopio University Hospital from Ministry of Health and Social Affairs (5254), Finnish Funding Agency for Technology and Innovation (40058/07), Nordic Centre of Excellence on Systems biology in controlled dietary interventions and cohort studies, SYSDIET (070014), the Finnish Diabetes Research Foundation, Yrjö Jahnsson Foundation (56358), Sigrid Juselius Foundation, Juho Vainio Foundation and TEKES grants 70103/06 and 40058/07.
DR’s EXTRA: The DR’s EXTRA Study was supported by grants from the Ministry of Education and Culture of Finland (627;2004-2011), Academy of Finland (102318; 123885), Kuopio University Hospital, Finnish Diabetes Association, Finnish Heart Association, Päivikki and Sakari Sohlberg Foundation, European Commission FP6 Integrated Project (EXGENESIS), LSHM-CT-2004-005272, City of Kuopio and Social Insurance Institution of Finland (4/26/2010).
EAS (Metabochip): EAS was supported by the British Heart Foundation. Genotyping was supported by a grant from the Chief Scientist Office, Scotland and performed at the Wellcome Trust Clinical Research Facility in Edinburgh.
EGCUT: EGCUT received financing by FP7 grants (201413, 245536), grant from Estonian Government SF0180142s08, from theEU through the European Regional Development Fund, in the frame of Centre of Excellence in Genomics and University of Tartu grant SP1GVARENG.
Ely: The Ely Study was funded by the MRC and Diabetes UK. We are grateful to all the volunteers, and to the staff of St. Mary’s Street Surgery, Ely and the study team. Genotyping in the Ely and Fenland studies was supported in part by an MRC-GSK pilot programme grant (ID 85374).
ERF: This study is financially supported by the Netherlands Organization for Scientific Research (NWO), the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947), the International Stichting Alzheimer Onderzoek (ISAO), the Hersenstichning Nederland (HSN), and the Centre for Medical Systems Biology (CMSB 1&2) in the framework of the Netherlands Genomics Initiative (NGI). We thank the participants from the Genetic Research in Isolated Populations, Erasmus Rucphen Family and their GPs, who made this work possible.
FamHS: The Family Heart Study (FamHS) was supported by NIH grants RO1-HL-087700 and RO1-HL-088215 (Michael A. Province, PI) from HNLBI, and RO1-DK-8925601 and RO1-DK-075681 (Ingrid B. Borecki, PI) from NIDDK.
Fenland: The Fenland Study is funded by the Wellcome Trust and the Medical Research Council. We are grateful to all the volunteers for their time and help, and to the General Practitioners and practice staff for help with recruitment. We thank the Fenland Study Investigators, Fenland Study Co-ordination team and the Epidemiology Field, Data and Laboratory teams. Biochemical assays were performed by the National Institute for Health Research, Cambridge Biomedical Research Centre, Core Biochemistry Assay Laboratory, and the Cambridge University Hospitals NHS Foundation Trust, Department of Clinical Biochemistry.
FIN-D2D 2007: The FIN-D2D study has been financially supported by the hospital districts of Pirkanmaa, South Ostrobothnia, and Central Finland, the Finnish National Public Health Institute (current National Institute for Health and Welfare), the Finnish Diabetes Association, the Ministry of Social Affairs and Health in Finland, the Academy of Finland (grant number 129293),Commission of the European Communities, Directorate C-Public Health (grant agreement no. 2004310) and Finland’s Slottery Machine Association.
FINRISK/DILGOM: DILGOM study received support from Etelä-Pohjanmaa Hospital District, Pohjois-Pohjanmaa Hospital District, Keski-Suomi Hospital District, Pirkanmaa Hospital District and Pohjois-Savo Hospital District. The DILGOM survey was funded by the Academy of Finland (grant number 118065). V.S. was supported by the Academy of Finland (grant numbers 139635 and 129494),
Framingham Heart Study: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01 HC 25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02 HL 6 4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The study is also supported by National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK) R01 DK078616 to Drs. Meigs, Dupuis and Florez, and NIDDK K24 DK080140 to Dr. Meigs.
FUSION: Support for FUSION was provided by NIH grants R01-DK062370 (to M.B.), R01-DK072193 (to K.L.M.) and intramural project number 1Z01-HG000024 (to F.S.C.). Genome-wide genotyping was conducted by the Johns Hopkins University Genetic Resources Core Facility SNP Center at the Center for Inherited Disease Research (CIDR), with support from CIDR NIH contract no. N01-HG-65403.
GEMS: The GEMS study was sponsored in part by GlaxoSmithKline. We thank the investigators Scott Grundy, Phil Barter, Ruth McPherson, Robert Mahley, Tom Bersot and Antero Kesaniemi for the collection of the samples.
GENOA: Genetic Epidemiology Network of Arteriopathy (GENOA) study is supported by the National Institutes of Health, grant numbers HL087660 and HL100245 from National Heart, Lung, Blood Institute. We thank Eric Boerwinkle (Human Genetics Center and Institute of Molecular Medicine and Division of Epidemiology, University of Texas Health Science Center, Houston, Texas, USA) and Julie Cunningham (Department of Health Sciences Research, Mayo Clinic College of Medicine, Rochester, Minnesota, USA) for their help with genotyping.
GenomEUtwin: We acknowledge support from the European Commission under ‘Quality of Life and Management of the Living Resources’ of the Fifth Framework Program (GenomEUtwin QLG2-CT-2002-01254). The study is also supported by “ENGAGE – European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant agreement number 201413”.
GLACIER: The GLACIER Study is nested within the Northern Sweden Health and Disease Study and phenotyping was conducted as part of the Västerbotten Intervention Project. We thank the participants and the investigators from these studies for their valuable contributions, with specific thanks to Lars Weinehall, Åsa Agren, Kerstin Enquist and Thore Johansson. The GLACIER Study and part of P.W.F.’s salary were funded by grants from the Swedish Research Council, Swedish Heart-Lung Foundation, Novo Nordisk, Umeå Medical Research Foundation and the Swedish Diabetes Association (to P.W.F.). Genotyping for this specific project was funded by the Wellcome Trust Sanger Institute. Inês Barroso acknowledges funding from the Wellcome Trust grant 098051, United Kingdom NIHR Cambridge Biomedical Research Centre and the MRC Centre for Obesity and Related Metabolic Diseases. We thank Emma Gray, Douglas Simpkin, Sarah Hunt and staff of the WTSI Sample Logistics, Genotyping and Variation Informatics Facilities.
GoDARTS: The study was funded by the Wellcome Trust, Tenovus Tayside and the Medical Research Council UK.
Health ABC: This study was supported by NIA contracts N01AG62101, N01AG62103 and N01AG62106, and in part by the Intramural Research Program of the NIH, National Institute on Aging. The GWAS was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C.
Health2000: The Health 2000 Study is funded by the National Institute for Health and Welfare (THL), the Finnish Centre for Pensions (ETK), The Social Insurance Institution of Finland (KELA), The Local Government Pensions Institution (KEVA) and other organizations listed on the website of the survey (http://www.terveys2000.fi). GWAS genotyping was supported by the Wellcome Trust Sanger Institute.
InChianti: The InCHIANTI study baseline (1998-2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336).
KORA F4: The MONICA/KORA Augsburg studies were financed by the Helmholtz Zentrum München-Research Center for Environment and Health, Neuherberg, Germany and supported by grants from the German Federal Ministry of Education and Research (BMBF) the Federal Ministry of Health (Berlin, Germany), the Ministry of Innovation, Science, Research and Technology of the state North Rhine-Westphalia (Düsseldorf, Germany), the German National Genome Research Network (NGFN) and the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. We thank all members of field staffs who were involved in the planning and conduct of the MONICA/KORA Augsburg studies.
LEIPZIG_ADULT_IFB: This work was supported by grants from Integrated Research and Treatment Centre (IFB) Adiposity Diseases (K7-36 to M.S. and A.K.) and from the Clinical Research Group “Atherobesity” KFO 152 (projects BL 833/1-1 to M.B., and Stu192/6-1 to MS). We thank all those who participated in the studies.
LEIPZIG_CHILHOOD_IFB: This work was supported by grants from Integrated Research and Treatment Centre (IFB) Adiposity Diseases (K7-36 to A.K. and M.S.) and from the Clinical Research Group “Atherobesity” KFO 152 (projects KO3512/1-2 to A.K.). We are grateful to all the patients and families for contributing to the study. We highly appreciate the support of the Obesity Team and Auxo Team of the Leipzig University Children’s Hospital for management of the patients and to the Pediatric Research Center Lab Team for support with DNA banking.
LURIC: LURIC received funding through the 6th Framework Program (integrated project Bloodomics, grant LSHM-CT-2004-503485) and 7th of Framework Program (integrated project AtheroRemo, Grant Agreement number 201668) of the European Union. The authors extend appreciation to the participants of the LURIC study without their collaboration this article would not have been written. We thank the LURIC study team either temporarily or permanently involved in patient recruitment, sample and data handling, and the laboratory staff at the Ludwigshafen General Hospital and the Universities of Freiburg and Ulm, Germany.
METSIM: The METSIM study was funded by the Academy of Finland (grants no. 77299 and 124243).
MICROS: For the MICROS study, we thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the research project. In South Tyrol, the study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano, and the South Tyrolean Sparkasse Foundation.
NFBC66: NFBC1986(1966) received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, the Medical Research Council, UK (G0500539, G0600705, PrevMetSyn/SALVE) and the Wellcome Trust (project grant GR069224), UK. The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. We thank Paula Rantakallio (launch of NFBC1966 and 1986) and Outi Tornwall and Minttu Jussila (DNA biobanking). The authors would like to acknowledge the contribution of the late Academian of Science Leena Peltonen.
NFBC86: The research of Vasiliki Lagou is funded in part through the European Community’s Seventh Framework Programme (FP7/2007-2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413. The research of Inga Prokopenko is funded in part through the European Community’s Seventh Framework Programme (FP7/2007-2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413.
NTRNESDA: Funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW, 904-61-090, 985-10-002, 904-61-193, 480-04-004, 400-05-717, Addiction-31160008, Middelgroot-911-09-032); Spinozapremie (SPI 56-464-14192); CMSB: Center for Medical Systems Biology (NWO Genomics); NBIC/BioAssist/RK/2008.024); BBMRI–NL: Biobanking and Biomolecular Resources Research Infrastructure (184.021.007); the VU University: Institute for Health and Care Research (EMGO+ ) and Neuroscience Campus Amsterdam (NCA); the European Science Foundation (ESF): Genomewide analyses of European twin and population cohorts (EU/QLRT-2001-01254); European Community’s Seventh Framework Program (FP7/2007-2013): ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC) Genetics of Mental Illness (230374); Rutgers University Cell and DNA Repository cooperative agreement (NIMH U24 MH068457-06); Collaborative study of the genetics of DZ twinning (NIH R01D0042157-01A); the Genetic Association Information Network, a public–private partnership between the NIH and Pfizer Inc., Affymetrix Inc. and Abbott Laboratories.
ORCADES: ORCADES was supported by the Scottish Executive Health Department and the Royal Society and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the invaluable contributions of Lorraine Anderson, the research nurses in Orkney, and the administrative team in Edinburgh.
PIVUS: Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). We thank Tomas Axelsson, Ann-Christine Wiman and Caisa Pöntinen for their excellent assistance with genotyping. The SNP Technology Platform is supported by Uppsala University, Uppsala University Hospital and the Swedish Research Council for Infrastructures. E.I. is supported by grants from the Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Foundation for Strategic Research, and the Royal Swedish Academy of Science.
PREVEND: PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the National Institutes of Health (grant LM010098), The Netherlands organisation for health research and development (NWO VENI grant 916.761.70) and the Dutch Inter University Cardiology Institute Netherlands (ICIN).
PROCARDIS: The PROCARDIS study was supported by the European Community Sixth Framework Program (LSHM-CT-2007-037273), AstraZeneca, the British Heart Foundation, the Oxford BHF Centre of Research Excellence, the Wellcome Trust (075491/Z/04), the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Heart-Lung Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Program of Karolinska Institutet and Stockholm County Council, the Foundation for Strategic Research and the Stockholm County Council (560283).
PROSPER: The PROSPER/PHASE study has received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° HEALTH-F2-2009-223004. PROSPER/PHASE is supported by grants from the Interuniversity Cardiology Institute of the Netherlands (ICIN) and the Durrer Center for Cardiogenetic Research both Institutes of the Netherlands Royal Academy of Arts and Sciences (KNAW), the Center for Medical Systems Biology (CMSB), a center of excellence approved by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO) and the Netherlands Consortium for Healthy Ageing (NCHA). The research leading to the PROSPER study was sponsored by Bristol Myers Squibb (New York, USA).
Rotterdam Study: The generation and management of GWAS genotype data for the Rotterdam Study are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
SardiNIA: The authors thank all the volunteers and the major of the four towns involved. This work was supported in part by the Intramural Research Program of the National Institute on Aging (NIA), National Institutes of Health (NIH), and by contract NO1-AG-1-2109, from the NIA, to the SardiNIA (“ProgeNIA”) team.
SCARFSHEEP: European Commission (LSHM-CT-2007-037273), the Swedish Heart-Lung Foundation, the Swedish Research Council (2669, 8691), the Knut and Alice Wallenberg Foundation, the Foundation for Strategic Research, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Programme of Karolinska Institutet and the Stockholm County Council and the Stockholm County Council (560183).
SORBS: This work was supported by grants from Integrated Research and Treatment Centre (IFB) Adiposity Diseases (K7-36 to M.S. and A.K.), from the Clinical Research Group “Atherobesity” KFO 152 (projects BL 833/1-1 to MB, and Stu192/6-1 to M.S.). We thank all those who participated in the studies. Reedik Mägi acknowledges financial support from the European Commission under a Marie Curie Intra-European Fellowship. Peter Kovacs acknowledges financial support from the Boehringer Ingelheim Foundation.
SUVIMAX : This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Institut National de la Recherche Agronomique, the Université Paris 13, the Centre National de Génotypage and the Commissariat à L’Energie Atomique
Swedish Twin Registry: This work was supported by grants from the US National Institutes of Health (AG028555, AG08724, AG04563, AG10175, AG08861), the Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Foundation for Strategic Research, the Royal Swedish Academy of Science and ENGAGE (within the European Union Seventh Framework Programme, HEALTH-F4-2007-201413). Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (http://www.genotyping.se). We thank Tomas Axelsson, Ann-Christine Wiman and Caisa Pöntinen for their excellent assistance with genotyping. The SNP Technology Platform is supported by Uppsala University, Uppsala University Hospital and the Swedish Research Council for Infrastructures.
THISEAS: Recruitment for THISEAS was partially funded by a research grant (PENED 2003) from the Greek General Secretary of Research and Technology. We thank all the dieticians and clinicians for their contribution to the project.
TRAILS: TRAILS (TRacking Adolescents’ Individual Lives Survey) is a collaborative project involving various departments of the University Medical Center and University of Groningen, the Erasmus University Medical Center Rotterdam, the University of Utrecht, the Radboud Medical Center Nijmegen, and the Parnassia Bavo group, all in The Netherlands. TRAILS has been financially supported by grants from the Netherlands Organization for Scientific Research NWO (Medical Research Council program grant GB-MW 940-38-011; ZonMW Brainpower grant 100-001-004; ZonMw Risk Behavior and Dependence grants 60-60600-98-018 and 60-60600-97-118; ZonMw Culture and Health grant 261-98-710; Social Sciences Council medium-sized investment grants GB-MaGW 480-01-006 and GB-MaGW 480-07-001; Social Sciences Council project grants GB-MaGW 457-03-018, GB-MaGW 452-04-314, and GB-MaGW 452-06-004; NWO large-sized investment grant 175.010.2003.005; NWO Longitudinal Survey and Panel Funding 481-08-013); the Sophia Foundation for Medical Research (projects 301 and 393), the Dutch Ministry of Justice (WODC), the European Science Foundation (EuroSTRESS project FP-006) and the participating universities. We are grateful to all adolescents, their parents and teachers who participated in this research and to everyone who worked on this project and made it possible. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation.
TwinsUK: The study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007-2013), ENGAGE project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London. T.D.S. is an NIHR senior Investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234). The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young).
ULSAM: Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). We thank Tomas Axelsson, Ann-Christine Wiman and Caisa Pöntinen for their excellent assistance with genotyping. The SNP Technology Platform is supported by Uppsala University, Uppsala University Hospital and the Swedish Research Council for Infrastructures. E.I. is supported by grants from the Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Foundation for Strategic Research, and the Royal Swedish Academy of Science.
Whitehall II: The WHII study has been supported by grants from the Medical Research Council; Economic and Social Research Council; BHF; Health and Safety Executive; Department of Health; National Heart Lung and Blood Institute (HL36310), US, NIH: National Institute on Aging (AG13196), US, NIH; Agency for Health Care Policy Research (HS06516); and the John D and Catherine T MacArthur Foundation Research Networks on Successful Midlife Development and Socioeconomic Status and Health. Genotyping in WHII was supported by BHF grant PG/07/133/24260 and by a MRC-GSK pilot programme grant (ID 85374).
Soumya Raychaudhuri is supported by the National Institutes of Health (K08AR055688, S.R.).
Footnotes
COMPETING FINANCIAL INTERESTS Ines Barroso and spouse own stock in GlaxoSmithkline and Incyte Ltd.
EdSumm (same for AOP and issue): Jose Florez, Claudia Langenberg, Erik Ingelsson, Inga Prokopenko, Inês Barroso and colleagues perform large-scale association analyses using the Metabochip to gain further insights into the genetic architecture of glucose regulation. They identify 38 new loci influencing one or more glycemic traits and show that many of these loci also modify risk of type 2 diabetes.
REFERENCES
- 1.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker A, et al. Association of genetic loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60:1805–1812. doi: 10.2337/db10-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingelsson E, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voight BF, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. doi: 10.1371/journal.pgen.1002793. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuro-o M, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi M, Kato S, Akiyoshi J, Atfi A, Razzaque MS. Dietary and genetic evidence for enhancing glucose metabolism and reducing obesity by inhibiting klotho functions. FASEB J. 2011;25:2031–2039. doi: 10.1096/fj.10-167056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utsugi T, et al. Decreased insulin production and increased insulin sensitivity in the klotho mutant mouse, a novel animal model for human aging. Metabolism. 2000;49:1118–1123. doi: 10.1053/meta.2000.8606. [DOI] [PubMed] [Google Scholar]
- 14.Rhee EJ, et al. Relationship between polymorphisms G395A in promoter and C1818T in exon 4 of the KLOTHO gene with glucose metabolism and cardiovascular risk factors in Korean women. J. Endocrinol. Invest. 2006;29:613–618. doi: 10.1007/BF03344160. [DOI] [PubMed] [Google Scholar]
- 15.Paroni G, et al. Klotho locus, metabolic traits, and serum hemoglobin in hospitalized older patients: a genetic association analysis. Age. doi: 10.1007/s11357-011-9273-x. published online 22 June 2011 (doi: 10.1007/s11357-011-9273-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rampersaud E, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 17.Tabassum R, Chavali S, Dwivedi OP, Tandon N, Bharadwaj D. Genetic variants of FOXA2: risk of type 2 diabetes and effect on metabolic traits in North Indians. J. Hum. Genet. 2008;53:957–965. doi: 10.1007/s10038-008-0335-6. [DOI] [PubMed] [Google Scholar]
- 18.Xing C, Cohen JC, Boerwinkle E. A weighted false discovery rate control procedure reveals alleles at FOXA2 that influence fasting glucose levels. Am. J. Hum. Genet. 2010;86:440–446. doi: 10.1016/j.ajhg.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 21.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 22.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 23.Nicolino M, et al. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes. 2010;59:733–740. doi: 10.2337/db09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AP, et al. Large-scale association analysis of the Metabochip provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. doi: 10.1038/ng.2383. in the press. [CE: ref for McCarthy] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin DM, Tan H. Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2008;3:96–107. doi: 10.1016/j.cbd.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards JB, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5:e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yujiri T, et al. MEK kinase 1 interacts with focal adhesion kinase and regulates insulin receptor substrate-1 expression. J. Biol. Chem. 2003;278:3846–3851. doi: 10.1074/jbc.M206087200. [DOI] [PubMed] [Google Scholar]
- 32.Meyer CF, Wang X, Chang C, Templeton D, Tan TH. Interaction between c-Rel and the mitogen-activated protein kinase kinase kinase 1 signaling cascade in mediating kappaB enhancer activation. J. Biol. Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 33.Lee FS, Hagler J, Chen ZJ, Maniatis T. Activation of the IkappaB alpha kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 34.Yujiri T, Sather S, Fanger GR, Johnson GL. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 35.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 36.Cai D, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat. Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J. Clin. Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler M, et al. Specific inhibition of PTEN expression reverses hyperglycemia in diabetic mice. Diabetes. 2002;51:1028–1034. doi: 10.2337/diabetes.51.4.1028. [DOI] [PubMed] [Google Scholar]
- 40.Wijesekara N, et al. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol. Cell. Biol. 2005;25:1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 42.Schmid J, et al. Modulation of pancreatic islets-stress axis by hypothalamic releasing hormones and 11beta-hydroxysteroid dehydrogenase. Proc. Natl. Acad. Sci. USA. 2011;108:13722–13727. doi: 10.1073/pnas.1110965108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui B, et al. A genome-wide association study confirms previously reported loci for type 2 diabetes in Han Chinese. PLoS ONE. 2011;6:e22353. doi: 10.1371/journal.pone.0022353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 47.Withers DJ, et al. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nat. Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 48.Aitman TJ, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 49.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 50.White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 51.Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 52.Curtis RE, Yin J, Kinnaird P, Xing EP. Finding genome-transcriptome-phenome association with structured association mapping and visualization in genamap. Pac. Symp. Biocomput. 2012:327–338. [PubMed] [Google Scholar]
- 53.Kim S, Xing EP. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 2009;5:e1000587. doi: 10.1371/journal.pgen.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, Sohn KA, Xing EP. A multivariate regression approach to association analysis of a quantitative trait network. Bioinformatics. 2009;25:i204–i212. doi: 10.1093/bioinformatics/btp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 56.Anderson SL, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiromura M, et al. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J. Biol. Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 58.Shimoyama Y, Nishio K, Hamajima N, Niwa T. KLOTHO gene polymorphisms G-395A and C1818T are associated with lipid and glucose metabolism, bone mineral density and systolic blood pressure in Japanese healthy subjects. Clin. Chim. Acta. 2009;406:134–138. doi: 10.1016/j.cca.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Oguro R, et al. Association of carotid atherosclerosis with genetic polymorphisms of the klotho gene in patients with hypertension. Geriatr. Gerontol. Int. 2010;10:311–318. doi: 10.1111/j.1447-0594.2010.00612.x. [DOI] [PubMed] [Google Scholar]
- 60.Freathy RM, et al. The functional “KL-VS” variant of KLOTHO is not associated with type 2 diabetes in 5028 UK Caucasians. BMC. Med. Genet. 2006;7:51. doi: 10.1186/1471-2350-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mullin BH, et al. Klotho gene polymorphisms are associated with osteocalcin levels but not bone density of aged postmenopausal women. Calcif. Tissue Int. 2005;77:145–151. doi: 10.1007/s00223-004-0291-x. [DOI] [PubMed] [Google Scholar]
- 62.Zarrabeitia MT, et al. Klotho gene polymorphism and male bone mass. Calcif. Tissue Int. 2007;80:10–14. doi: 10.1007/s00223-006-0233-x. [DOI] [PubMed] [Google Scholar]
- 63.Reue K, Xu P, Wang XP, Slavin BG. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 2000;41:1067–1076. [PubMed] [Google Scholar]
- 64.Aulchenko YS, et al. LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes. 2007;56:3020–3026. doi: 10.2337/db07-0338. [DOI] [PubMed] [Google Scholar]
- 65.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang AT, Campbell WB, Nithipatikom K. ROCK1 feedback regulation of the upstream small GTPase RhoA. Cell Signal. 2012;24:1375–1380. doi: 10.1016/j.cellsig.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakamura Y, et al. Marked increase of insulin gene transcription by suppression of the Rho/Rho-kinase pathway. Biochem. Biophys. Res. Commun. 2006;350:68–73. doi: 10.1016/j.bbrc.2006.08.192. [DOI] [PubMed] [Google Scholar]
- 70.Furukawa N, et al. Role of Rho-kinase in regulation of insulin action and glucose homeostasis. Cell Metab. 2005;2:119–129. doi: 10.1016/j.cmet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 71.Chun KH, et al. In vivo activation of ROCK1 by insulin is impaired in skeletal muscle of humans with type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 2011;300:E536–E542. doi: 10.1152/ajpendo.00538.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Q, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ. Cardiovasc. Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurlbert MS, et al. Mice transgenic for an expanded CAG repeat in the Huntington’s disease gene develop diabetes. Diabetes. 1999;48:649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 75.Bjorkqvist M, et al. The R6/2 transgenic mouse model of Huntington’s disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum. Mol. Genet. 2005;14:565–574. doi: 10.1093/hmg/ddi053. [DOI] [PubMed] [Google Scholar]
- 76.Bradley SV, et al. Degenerative phenotypes caused by the combined deficiency of murine HIP1 and HIP1r are rescued by human HIP1. Hum. Mol. Genet. 2007;16:1279–1292. doi: 10.1093/hmg/ddm076. [DOI] [PubMed] [Google Scholar]
- 77.Hancock DB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cho MH, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nomiyama T, et al. Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J. Clin. Invest. 2007;117:2877–2888. doi: 10.1172/JCI31986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pietilainen KH, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lyssenko V, et al. Pleiotropic effects of GIP on islet function involve osteopontin. Diabetes. 2011;60:2424–2433. doi: 10.2337/db10-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olofsson LE, et al. CCAAT/enhancer binding protein alpha (C/EBPalpha) in adipose tissue regulates genes in lipid and glucose metabolism and a genetic variation in C/EBPalpha is associated with serum levels of triglycerides. J. Clin. Endocrinol. Metab. 2008;93:4880–4886. doi: 10.1210/jc.2008-0574. [DOI] [PubMed] [Google Scholar]
- 83.Wu Z, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 84.Hollenberg AN, et al. Functional antagonism between CCAAT/Enhancer binding protein-alpha and peroxisome proliferator-activated receptor-gamma on the leptin promoter. J. Biol. Chem. 1997;272:5283–5290. doi: 10.1074/jbc.272.8.5283. [DOI] [PubMed] [Google Scholar]
- 85.Keller SR. Role of the insulin-regulated aminopeptidase IRAP in insulin action and diabetes. Biol. Pharm. Bull. 2004;27:761–764. doi: 10.1248/bpb.27.761. [DOI] [PubMed] [Google Scholar]
- 86.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 87.D’Orazio P, et al. Approved IFCC recommendation on reporting results for blood glucose (abbreviated) Clin. Chem. 2005;51:1573–1576. doi: 10.1373/clinchem.2005.051979. [DOI] [PubMed] [Google Scholar]
- 88.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Folkersen L, et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:365–373. doi: 10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- 92.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 93.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.