At one time, the structure of DNA was blissfully simple. It was elegant, regular, and universal—quite unlike the structures of proteins, which were complicated, highly varied, and full of peculiarities consistent with their multifarious functions. DNA was a plectonemic double helix of 10 nucleotide pairs per turn (a nice round number) containing planar base pairs stacked neatly perpendicular to the helix axis and, of course, capable of accommodating any conceivable sequence of nucleotides within its regular structure so that it could encapsulate any kind of biologically meaningful sequence information within its consummate regularity (1). This simple and unifying concept was so compelling, and the imaginative representations printed in textbooks or devised for the media were so visually attractive, that the “ideal” B-form helix dominated the thinking of molecular biologists in the early years and has probably done more than any other single icon to attract brilliant students to venture into biology.
But it could not last. Even at the outset it was known that fibers of DNA would give the relatively fuzzy B-form x-ray diffraction pattern at 92% relative humidity, supposed to be the biologically relevant condition, but would switch reversibly to a more crystalline state (the A-form) at 75% relative humidity, characterized by sharper reflections and altered helical parameters like 11 tilted base pairs per turn (2, 3). It was also known that binding of aminoacridines such as proflavine to DNA would modify the B-form pattern and destroy its helical regularity, although not the perpendicular stacking of the base pairs, and thus the intercalation hypothesis was born (4).
About this time, as a Ph.D. student in Cambridge working on ethidium bromide among other things, it seemed to me that ethidium ought to be a better, or at least cleaner, intercalator than proflavine. I enlisted the help of Watson Fuller, then working in Maurice Wilkins' lab at King's College London, to try x-ray diffraction on ethidium–DNA complexes (5). We thought that ethidium ought to impede the B ↔ A transition (which it does, actually), but our first experiments were a spectacular failure. Fibers precipitated from an ethidium–DNA complex underwent the B → A transition on lowering the relative humidity from 92 to 75% as if nothing had happened, and vice versa, but the control fibers—DNA alone that had never seen the drug—did not. Watson reported this disappointing result, with a characteristically jokey final comment, in a letter dated 5 March 1964:
… . “As you see the [ethidium] complex behaved as normal Na DNA and the diffraction patterns are indistinguishable from those of normal Na DNA… [several notional explanations]… One thing—the DNA control did not undergo the A → B transition (I tried a few times). This does happen from time to time and is probably not important. However, it may be that the ethidium makes the A to B transition easier in some peculiar way: How about a Paper entitled Ethidium Bromide—a molecular grease?”
I mention this tale because it could well be the first time, albeit erroneously, that anyone advanced the idea that drugs might be able to induce or facilitate long-range structural transitions in DNA.
Another current of thinking that can be traced back to early days is the notion that the biological function of DNA, and in particular differential gene expression, might actually require the molecule to adopt different helical states. How else could regulators quickly locate and identify such elements as start/stop signals in the huge length of DNA present in a cell nucleus? Searching for sequence-dependent structural variations led to the discovery of an impressive variety of odd things that DNA can do under defined conditions, often related to bending, although for some time at least the base pairing and right-handed helical screw sense remained inviolate. Not for long. One of the biggest upsets to orthodoxy was the discovery of Z-DNA, the left-handed double-helical conformation with a dinucleotide repeat reported in 1979–1980 (6, 7). And the last bastion crumbled just a few years later when it was found that Watson–Crick base pairs could be induced to flip into the long-outlawed Hoogsteen pairing configuration (8) by binding of bis-intercalating quinoxaline antibiotics (9, 10). Add to that the recent excitement generated by G-quadruplex structures adopted by guanine-rich sequences present in the telomeres of mammalian chromosomes (11), and you could be forgiven for thinking that nothing is sacred nowadays. However, it would be foolish to dismiss even outrageous noncanonical structures as mere aberrations or artifacts; Nature has a remarkable way of using rare phenomena to perform biological roles.
In many of these discoveries, the binding of drugs played a significant part, as I have indicated. Interestingly, the x-ray crystallographic evidence for a left-handed helical form of poly (dG⋅dC) at high salt concentration was foreshadowed by the remarkable finding of Pohl, Jovin, and colleagues (12) that ethidium serves as a powerful cooperative allosteric effector modulating the structure of this artificial DNA by virtue of its huge preference for binding to the right-handed double-helical conformation preferred at low salt concentration as opposed to the “L-form” preferred in the presence of high salt. A “molecular grease” indeed? No, because the drug drives the left-handed Z-form back to B-form (or, more accurately, an intercalated right-handed form) because its binding to the polynucleotide duplex perturbs the equilibrium between the two forms at high salt in favor of the latter. A “grease” would act like an enzyme and merely hasten the attainment of thermodynamic equilibrium.
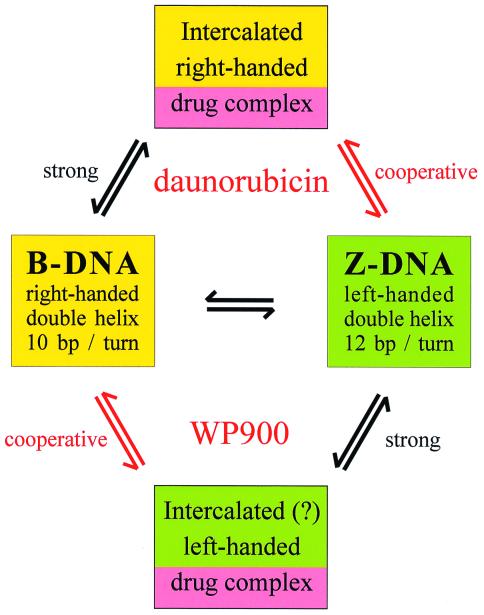
Against this background, the report of Qu, Chaires, and colleagues (13) in this issue of PNAS is of classic significance. Starting from the observation that the widely used antitumor antibiotic (+)-daunorubicin (daunomycin) appears well tailored to bind intercalatively to right-handed B-form DNA, the authors decided to synthesize its optical antipode (−)-daunorubicin in the expectation that it might bind selectively to left-handed double-helical DNA. The synthesis of the enantiomer, code-named WP900, was no easy undertaking; it required some 37 steps. Qu et al. first used an ingenious method to determine whether natural daunorubicin and WP900 might bind to double helices of opposite handedness by mixing solutions of the ligands in equal proportions and then seeing which isomer would predominate after dialysis against a sample of poly(dG⋅dC) in either the right-handed (low salt) or left-handed (high salt) form. At low salt, it was WP900 that predominated in the dialysate, whereas at high salt there was more daunorubicin left behind, so to speak—clear qualitative evidence that in simple competition the isomers preferred to bind to different forms of the polynucleotide. Binding measurements conducted by fluorescence titration indicated a 21-fold preference for daunorubicin to bind to the right-handed DNA and a more modest 5-fold preference for WP900 to bind to the left-handed form. But the crucial test, to see whether each ligand would actually drive the polynucleotide to adopt a specific conformational form, is the real strength of this paper and betokens the rigorous understanding and application of thermodynamic principles that are characteristic of the Chaires laboratory. Qu et al. placed poly (dG⋅dC) in moderately high salt such that the polymer existed as a mixture of the two helical forms, poised close to the thermodynamic equilibrium, and looked to see what happened to the CD spectrum on addition of either ligand. The spectra showed clearly that WP900 did selectively drive the nucleic acid to adopt a left-handed helical form, providing unambiguous evidence of structural selectivity opposite to that of (+)-daunorubicin. A best estimate of its preference for left-handed DNA over right-handed DNA under these conditions was again a factor of 5, compared with something like 44 for daunorubicin in the opposite direction, and it is this difference in affinity that drives the allosteric conversion of the polynucleotide to a left-handed form (Fig. 1).
Figure 1.
Structural transitions of poly(dG⋅dC) in 2.1–3.0 M NaCl. The structures of B-DNA, Z-DNA, and various intercalated oligonucleotide–daunorubicin complexes have been established experimentally by x-ray diffraction. Qu et al. (13) have performed computer simulations by using molecular dynamics to build an energetically and stereochemically feasible model of a left-handed duplex containing intercalated WP900. Simulated B-form DNA–WP900 and Z-form DNA–daunorubicin complexes were not stable; they resulted in disruption of the duplexes and loss of intercalation.
This is not the first time that a stereoselective ligand capable of binding to Z-DNA has been described, but it does seem to be the first unambiguous demonstration of true selectivity for left-handed DNA over right-handed DNA backed up with thermodynamically credible numbers. In absolute terms, the binding constants are not enormous, but they are sufficient to nudge the polymer in one direction or the other in what appears to be a cooperative fashion. Much effort has been devoted in earlier work to seeking a similar goal by using chiral metal chelates, which can certainly bind with differential affinity to helices of opposite handedness (14, 15), but the evidence for genuine selectivity favoring left-handed DNA has been considered equivocal (16), and there have been no reports of such compounds triggering conversion of B-DNA to Z-DNA. Of course, it must be remembered that left-handed Z-DNA is by no means the optical antipode of right-handed B-DNA (or indeed any other right-handed double-helical form), so the artificial synthesis of an exact enantiomer of a known B-DNA-binding compound is not guaranteed to produce a left-handed helix-selective ligand. In that sense, the authors may have been lucky. One wonders what would have happened had they made an enantiomer of actinomycin D (17), which appears to be exquisitely tailored to fit within the minor groove of an intercalatively distorted B-DNA helix. Quite possibly nothing at all. And by the same token, it may well be possible in future to design ligands de novo that will specifically recognize the curious dinucleotide repeat motif of Z-DNA and thereby manifest substantial selectivity for a left-handed double helix.
The potential uses of a left-handed helix-selective probe in biology are obvious. To be able to use such compounds as tools to investigate the significance and function of left-handed DNA in vivo, and even to modulate biological properties by shifting the balance in favor of left-handed helices, would be of great interest to cell biologists. And what about therapeutic applications and the prospect of a new class of useful drugs? Qu et al. leave us with the tantalizing observation that WP900 is cytotoxic in vitro, albeit less potent than daunorubicin against a standard human carcinoma cell line, but retains cytotoxic properties against a multidrug-resistant variant. They note that its biological activities must presumably include action via mechanisms other than topoisomerase poisoning normally attributed to anthracycline antibiotics (18). If these findings are confirmed, they could herald a bright future for WP900 and an exciting new approach to cancer therapy based on sound molecular principles (19).
Acknowledgments
I thank Jared Lawrence for his help in preparing Fig. 1 and the Cancer Research Campaign for supporting research in my laboratory.
Footnotes
See companion article on page 12032.
References
- 1.Watson J D, Crick F H C. Nature (London) 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Calladine C R, Drew. H R. Understanding DNA. 2nd Ed. London: Academic; 1997. [Google Scholar]
- 3.Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. New York: Oxford Univ. Press; 1999. [Google Scholar]
- 4.Lerman L S. J Mol Biol. 1961;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- 5.Fuller W, Waring M J. Ber Bunsenges Phys Chem. 1964;68:805–808. [Google Scholar]
- 6.Wang A H-J, Quigley G J, Kolpak F J, Crawford J L, van Boom J H, van der Marel G, Rich A. Nature (London) 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 7.Drew H, Takano T, Tanaka S, Itakura K, Dickerson R E. Nature (London) 1980;286:567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- 8.Hoogsteen K. Acta Crystallogr. 1959;12:822–823. [Google Scholar]
- 9.Wang A H-J, Ughetto G, Quigley G J, Hakoshima T, van der Marel G A, van Boom J H, Rich A. Science. 1984;225:1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- 10.Quigley G J, Ughetto G, van der Marel G A, van Boom J H, Wang A H-J, Rich A. Science. 1986;232:1255–1258. doi: 10.1126/science.3704650. [DOI] [PubMed] [Google Scholar]
- 11.Patel D J, Bouaziz S, Kettani A, Wang Y. Oxford Handbook of Nucleic Acid Structure. New York: Oxford Univ. Press; 1999. pp. 389–453. [Google Scholar]
- 12.Pohl F M, Jovin T M, Baehr W, Holbrook J J. Proc Natl Acad Sci USA. 1972;69:3805–3809. doi: 10.1073/pnas.69.12.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu X, Trent J O, Fokt I, Priebe W, Chaires J B. Proc Natl Acad Sci USA. 2000;97:12032–12037. doi: 10.1073/pnas.200221397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton J K, Basile L A, Danishefsky A, Alexandrescu A. Proc Natl Acad Sci USA. 1984;81:1961–1965. doi: 10.1073/pnas.81.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton J K. Science. 1986;233:727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 16.Nordén B, Lincoln P, Åkerman B, Tuite E. Metal Ions in Biological Systems. 1996;33:177–252. [PubMed] [Google Scholar]
- 17.Gale E F, Cundliffe E, Richmond M H, Reynolds P E, Waring M J. The Molecular Basis of Antibiotic Action. 2nd Ed. London: Wiley; 1981. pp. 314–333. [Google Scholar]
- 18.Ralph R K, Judd W, Pommier Y, Kohn K W. In: Molecular Aspects of Anticancer Drug–DNA Interactions. Neidle S, Waring M J, editors. Vol. 2. London: Macmillan; 1994. pp. 1–95. [Google Scholar]
- 19.Waring M J, Ponder B A J. The Search for New Anticancer Drugs. Dordrecht, The Netherlands: Kluwer; 1992. [Google Scholar]