SUMMARY
Treatment of tuberculosis, like other infectious diseases, is increasingly hindered by the emergence of drug resistance. Drug discovery efforts would be facilitated by facile screening tools that incorporate the complexities of human disease. Mycobacterium marinum-infected zebrafish larvae recapitulate key aspects of tuberculosis pathogenesis and drug treatment. Here we develop this model for rapid in vivo drug screening using fluorescence-based methods for serial quantitative assessment of drug efficacy and toxicity. We provide proof-of-concept that both traditional bacterial-targeting antitubercular drugs as well as newly identified host-targeting drugs would be discovered using this model. We demonstrate the model’s utility for identification of synergistic combinations of antibacterial drugs and demonstrate synergy between bacterial and host targeting compounds. Thus the platform can be used to identify new antibacterial agents and entirely new classes of drugs that thwart infection by targeting host pathways. The methods developed here should be widely applicable to small molecule screens for other infectious and noninfectious diseases.
INTRODUCTION
The zebrafish has emerged as a favorite organism for wide-ranging studies of development and disease owing to its genetic tractability and facile husbandry (Mione and Trede, 2010; Trede et al., 2004). Moreover, zebrafish larvae are an attractive tool for drug screens as their small size (~ 2 mm) and optical transparency allow for husbandry in multiwell plates and evaluation by fluorescence detection techniques, respectively (Cao et al., 2009; Milan et al., 2009; Ou et al., 2012; Peterson and Fishman, 2004; Stern et al., 2005; Tan and Zon, 2011; Zon and Peterson, 2005). Such screens have been used to discover drugs that modulate developmental and disease pathways shared with humans (Cusick et al., 2012; Milan et al., 2003; Peterson and Fishman, 2004; Rudner et al., 2011).
Zebrafish possess a complex immune system akin to that of humans (Traver et al., 2003; Trede et al., 2004), and have been instrumental in identifying pathways that mediate disease pathogenesis (Etchin et al., 2011; Martin et al., 2011; Mione and Trede, 2010). The zebrafish larva infected with fluorescently labeled Mycobacterium marinum (Mm) has proved an excellent model for the in vivo dissection of tuberculosis (TB) pathogenesis. Use of this model enabled the identification and mechanistic dissection of host and bacterial determinants, thus yielding new insights into TB (Davis and Ramakrishnan, 2009; Tobin and Ramakrishnan, 2008; Tobin et al., 2010; Volkman et al., 2010). In particular, discoveries about the roles of macrophages, granulomas and inflammation have suggested completely new approaches to treating this ancient disease (Clay et al., 2007; Davis and Ramakrishnan, 2009; Tobin et al., 2012; Tobin et al., 2010; Volkman et al., 2010).
Existing TB therapies are increasingly limited by the emergence of drug resistant disease, including Mycobacterium tuberculosis (Mtb) strains that are resistant to all known antitubercular drugs (Udwadia et al., 2012; Zignol et al., 2012). There has been a resurgence of interest in antimicrobial drug discovery, and modern genomic and chemical synthetic methods are now being used to search for new classes of drugs. While a few compounds have emerged for select organisms, these approaches have been largely unsuccessful in identifying new antibiotics (Payne et al., 2007; Pethe et al., 2010). While the causes for this failure are many, two main problems are 1) compounds identified by in vitro, target-based screens may not inhibit bacterial growth, and 2) compounds identified in “whole cell” screens using bacteria in axenic culture may not be active against bacteria in vivo. Thus, a major bottleneck in drug discovery is the lack of a facile, yet relevant, model for drug screening. In vivo screening is particularly important because TB is a complex disease in which the bacteria sense and exploit host defenses (Volkman et al., 2004). The mouse model of TB has been widely used for antitubercular drug testing and has predicted the behavior of many drugs in humans (Dorman et al., 2009; Mitchison and Chang, 2009), including the presence of tolerance. However, its use is expensive, time-consuming and laborious: the discovery of the antitubercular activity of isoniazid (INH) involved the use of 50,000 mice to screen 5000 compounds and only assessed efficacy—toxicity studies then required additional animals (Tweedy and Lesney, 2000). More recently, high content screening (HCS) platforms have been applied to intracellular pathogens such as Salmonella (Kuijl et al., 2007) and Mtb (Christophe et al., 2009), in which compound screening is performed on infected macrophages in multiwell format, allowing the assessment of drug activity against intracellular bacteria. Given the high attrition of lead compounds due to host toxicity and lack of efficacy in vivo, a screening platform that replicates the complexities of in vivo treatment, yet is amenable to high throughput methodologies, is highly desirable.
Recently, we found that first line antitubercular drugs were active against Mm infection in zebrafish larvae, recapitulating important clinical features of human disease including early bactericidal activity (EBA), the localized responsiveness of individual granulomas, drug synergies, and drug tolerance (Adams et al., 2011). Here, we present the Mm-zebrafish larval model as a platform for in vivo antitubercular drug discovery. We describe new high throughput methodologies for larval husbandry and imaging, as well as a novel fluorimetry platform that allows rapid, serial quantitation of both drug efficacy and toxicity. We demonstrate the utility of the platform for compound screening of both bacterial- and host-targeting drugs as well as for synergistic drug combinations. The high throughput methods we have developed should have broad utility for compound screening in the zebrafish.
RESULTS
High throughput husbandry and cryo-anesthesia of zebrafish larvae
A versatile compound-screening platform would ideally allow maintenance and serial imaging of larvae in situ, require only a single application of compound and permit observation over the course of several days. In the Mm-larval model, infected larvae are maintained in petri dishes with regular water changes, and bacterial burdens are enumerated by imaging and quantification by Fluorescence Pixel Count (FPC)(Adams et al., 2011). Because larval motility interferes with imaging, they must be transferred into tricaine-containing medium, then to glass slides for imaging, and finally returned to tricaine-free maintenance conditions (Adams et al., 2011; Cosma et al., 2006a; Tobin et al., 2010; Volkman et al., 2010). To remove these barriers to high-throughput implementation we first established that 3 days post fertilization (dpf) larvae could be maintained in optical-bottom 96-well plates, without water changes or feeding, for 10 days with no mortality (Figure S1A, and data not shown). Next, we found that cryo-anesthesia of larvae was an effective and reversible immobilization technique. Incubation of the 96-well plates on ice for 10 minutes prevented larval movement for an additional 10 minutes at room temperature, without affecting larval viability (Figure S1A) nor altering the course of Mm infection as judged by overall bacterial burdens and the distribution of infection (Figure S1B and S1C). Finally, we automated fluorescence microscopy, image collection and analysis (see Experimental Procedures, Documents S1 and S2), allowing an entire plate to be imaged in only 4 minutes. In summary, 96-well husbandry and automated imaging of cryo-anesthetized larvae allows for rapid, serial, and high-throughput data collection.
Automated plate fluorimetry (APF) for rapid dual-assay drug screening
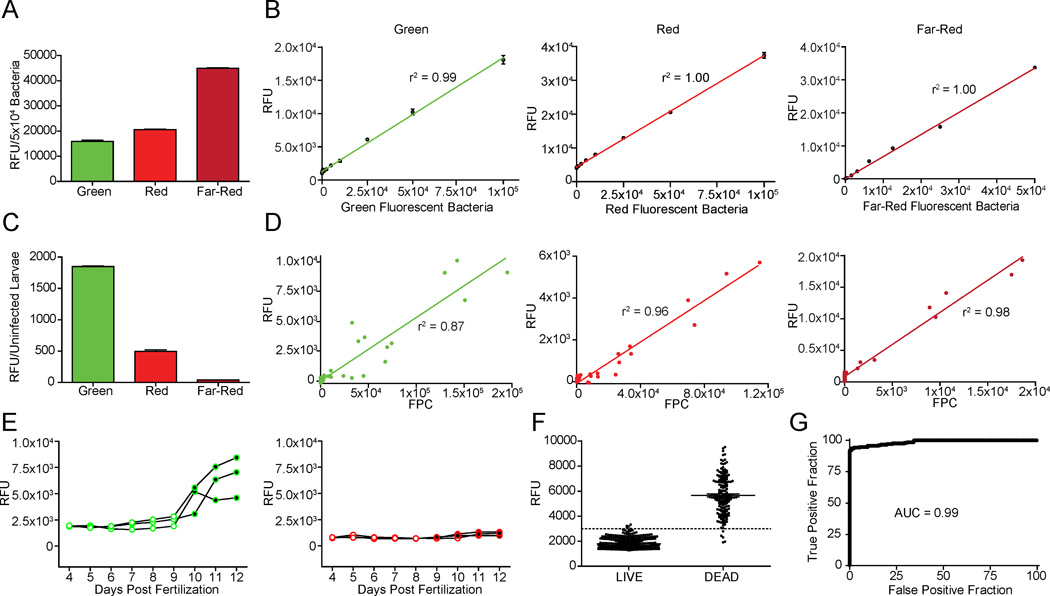
Because of the relatively low cost and wider availability of microplate fluorimeters as compared to automated fluorescence microscopes and specialized image analysis software, we additionally developed fluorimetric assays for in situ fluorescence monitoring of larvae in 96-well plates. Preliminary analyses of uninfected larvae revealed more background fluorescence at shorter wavelengths, consistent with the longstanding observation of less biological autofluorescence at longer wavelengths in a variety of biological tissues. Therefore, we assessed Mm strains constitutively expressing the widely used fluors GFPmut3 (“green”) and dsRed2 (“red”) as well as the newly available tdKatushka2 (Shcherbo et al., 2009), which fluoresces in the far-red range (Table S1). Of the three strains tested, tdKatushka2 provided the brightest signal, approximately three-fold that of GFPmut3, and twice that of dsRed2 (Figure 1A). Additionally, fluorimetry was a reliable indicator of relative bacterial number, showing a linear relationship between fluorescence intensity and the number of bacterial cells in suspension (Figure 1B). We next assessed the background noise resulting from autofluorescence of uninfected larvae in all three channels. As expected, the degree of autofluorescence was inversely correlated with wavelength (Figure 1C). Finally, we examined larvae with varying Mm infection burdens by APF and compared the results with our established method, FPC (Figure 1D). The two methods correlated well for all three fluors, with the best correlation being seen with tdKatushka2, consistent with its superior signal to noise ratio. Importantly, this assessment did not require anesthesia of the larvae, further increasing its throughput.
Figure 1. Automated Plate Fluorimetry.
(A,B) Fluorescence of bacteria expressing GFPmut3 (“green”), DsRed2 (“red”) or tdKatushka2 (“far-red”) was measured by APF. (A) Comparison of mean fluorescence per 5×104 bacteria, n=3. (B) Linear relationship between number of bacteria and RFU measurements. (C) Mean autofluorescence of live uninfected 7-dpf larvae was measured at wavelengths used for detection of the three fluors (n=10). (D) Larvae were infected with varying doses of fluorescent Mm and bacterial burdens were quantified by FPC and APF. (E) Autofluorescence of individual uninfected larvae was measured at wavelengths used for detection of green and red-fluorescent Mm from 4 to 12 dpf. Larvae were also assessed daily for viability by cardiac assessment; alive (○), dead (●). (F,G) Forty larvae with varying levels of infection were housed in 96-well plates beginning at 2-dpf and were monitored daily for green autofluorescence via APF and survival state via cardiac assessment for 13 days, for a total of 520 APF measurements. (F) Graph shows larval autofluorescence, stratified by viability. Line represents 3000 RFU, or ~50% of the mean fluorescence of dead larvae. (G) ROC curve showing accuracy of green autofluorescence as predictor of viability. See also Figures S1, S2 and Table S1.
Tracking larval survival during a screen can provide information regarding both compound efficacy and toxicity. While characterizing larval autofluorescence, we observed that green, but not red, autofluorescence was much higher in dead larvae, as compared to live ones (Figure 1E). To examine the timing and predictive value of green autofluorescence for the automated assessment of survival, larvae with varying levels of far-red Mm infection were monitored daily for 13 days. Viability was scored by the standard method of assessing the presence of a heart beat by stereomicroscopy (“cardiac assessment”) and by green autofluorescence by APF (Figure 1F). The mean fluorescence of dead larvae was 3 times higher than that of live larvae, and Receiver-Operator Characteristic (ROC) analysis showed that autofluorescence is highly predictive of viability (AUC = 0.99) (Figure 1G). Using a cutoff of 50% of the mean fluorescence of dead larvae, only four false positives and five false negatives occurred out of a total of 520 APF measurements, for an error rate of 1.7%. Moreover, even these false negatives represented measurements taken in the window between cardiac arrest and the ensuing increase in autofluorescence that results from tissue decay: when these 5 dead larvae were re-examined 24 hours later, their autofluorescence had increased, moving above the cutoff (Figure S2A). We confirmed that this method can also be used in the presence of compounds, like RIF, that absorb light and thus partially reduce autofluorescence measurements in the green channel (Figure S2B and S2C). In sum, green autofluorescence provides a versatile, rapid, and automated readout to distinguish live from dead larvae.
Because of its superior signal to noise ratio and compatibility with the autofluorescence-based live/dead assay, we chose tdKatushka2-expressing Mm for further experiments. However, we note that the other fluors can also be used reliably, for example, in assays evaluating drug effects on green or red fluorescent markers in transgenic fish.
APF reliably detects antitubercular drug efficacy
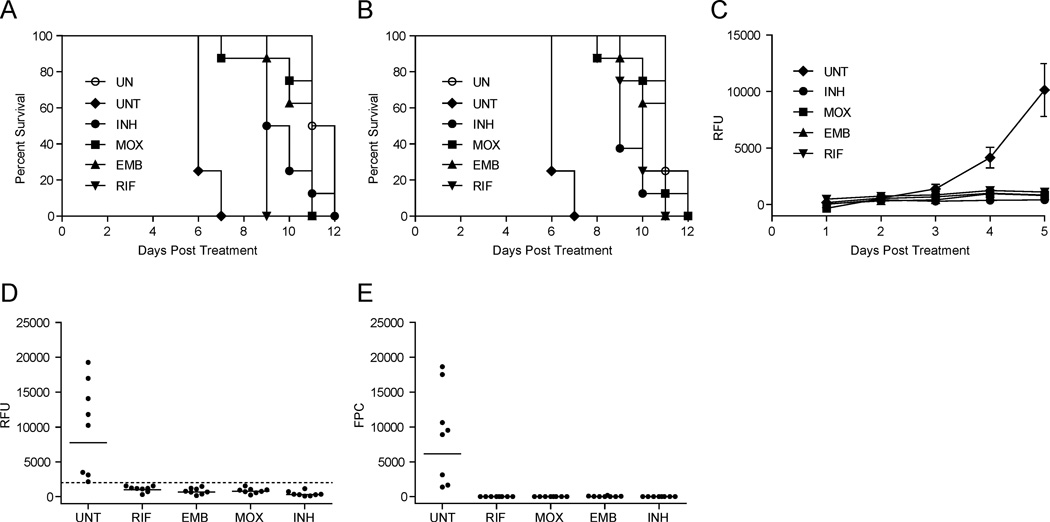
We previously showed that first line antitubercular drugs are active against larval Mm infection, and recapitulate the efficacy and tolerance profiles observed during human treatment (Adams et al., 2011). To validate the 96-well APF format for antitubercular compound screening, we first examined whether the activities of known drugs could be detected using 400 µM drug per well. This represents the equivalent of 100 nmol, an amount that is feasible for one-time use in a screen. Larvae one day post-infection (dpi) with far-red fluorescent Mm were maintained in 96-well plates, in the presence and absence of the drugs isoniazid (INH), rifampicin (RIF), ethambutol (EMB) and moxifloxacin (MOX), and were monitored daily for viability and bacterial burdens. Larval viability was scored by both cardiac assessment and green-channel APF. Survival of treated larvae was significantly different from the untreated controls and the median times to death agreed closely by the two methods (Figure 2A and 2B, and Table S2). Simultaneous daily assessment of the larvae by far-red APF revealed that drug treatment also prevented the bacterial expansion that occurs in untreated animals (Figure 2C), and was as sensitive as FPC in detecting the differences between treated and untreated larvae (Figure 2D and 2E). ROC analysis of the pooled APF data from treated versus untreated larvae shows that the platform possesses strong predictive value for the detection of antibiotic efficacy (AUC=1, data not shown). For example, using 20% of the mean untreated fluorescence as a cutoff for drug efficacy would yield 32/32 true positives and zero false positives. A screen in which 50% of the mean fluorescence of untreated larvae is used as the threshold would yield 32/32 true positives and 3/8 false positives.
Figure 2. Automated in vivo assessment of antimycobacterial drug efficacy by APF.
Larvae 1 dpi with 300 far-red fluorescent Mm were placed in wells of 96-well plates (one larva per well) containing no antibiotic (UNT), EMB, INH, MOX or RIF (all at 400 µM). Uninfected (UN) larvae were also included, and were not exposed to antibiotic; N=8. (A, B) Larval survival during the 12 day treatment period was determined using APF of green-autofluorescence (A) or cardiac assessment of each larva by stereomicroscopy (B). Each drug treated group survived significantly longer than the UNT group as determined by individual log-rank tests (P values ranged from <0.0001 to 0.0002; see also Table S2). (C) Bacterial burdens were measured daily by far-red APF. Mean RFU with SEM are shown. (D, E) Fluorescence of individual untreated and treated larvae (from panel C) at 5 dpt as measured by APF (D) and FPC (E). Bars represent geometric means, and dotted line represents 20% of the mean untreated RFU. See also Figures S3, S4 and Table S2.
Finally, we considered the possibility that tuberculous granulomas, which form at 3 dpi in this model, could potentially impact drug efficacy by altering bacterial physiology or limiting drug access t o sites of infection. Therefore, we asked whether APF could be used for drug screening after granulomas had formed, and found that it could. AFP revealed that treatment initiated 3 dpi was similarly effective at improving larval survival (Figure S3A, and S3B) and reducing bacterial burdens (Figure S3C and S3D), as it was when larvae were treated 1 dpi. ROC analysis confirms that the platform remained highly predictive for drug efficacy, even in the context of granulomatous infection (AUC=0.87, P=0.016, Figure S3E).
In summary, the simultaneous assessment of host survival and bacterial burdens allows for the identification of traditional antimicrobial drugs as well as those that could potentially modify host outcome without changing bacterial burdens. Finally, a parallel screen for host toxicity is inherently built into the platform, providing information on an additional critical parameter in drug development.
APF can identify synergistic anti-bacterial drug combinations
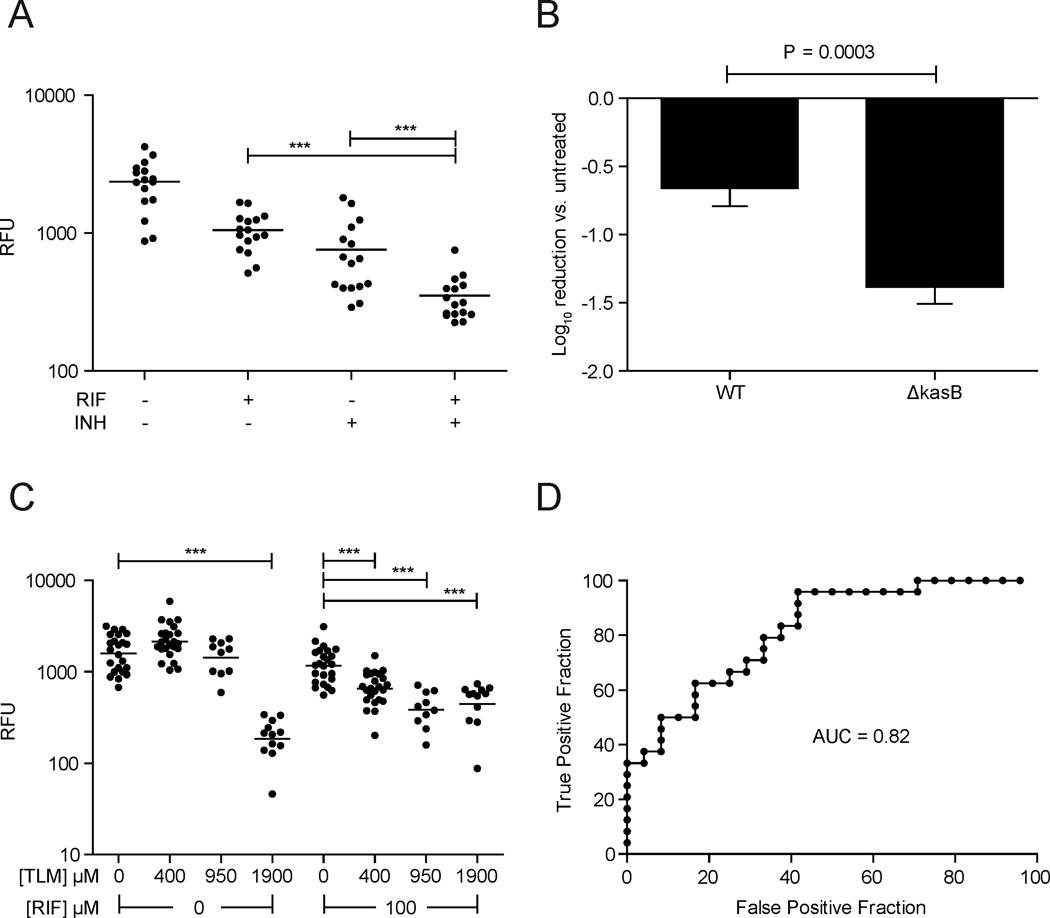
In the context of antimicrobial therapy, synergy is generally defined as a response achieved by a drug combination, that is superior to that seen with the individual antibiotics therein (Pratt and Fekety, 1986). For human TB therapy, drug synergisms have been central in shortening time to sterilization and treatment length, as exemplified by the addition of RIF to INH-containing regimens (Donald and McIlleron, 2009). This synergistic effect was apparent early; within days of treatment a further reduction in sputum bacterial counts was observed with the drug combination compared to INH alone (Jindani et al., 2003). In the larval model, the addition of RIF to INH similarly revealed a further reduction in bacterial burdens by FPC, suggesting the utility of the zebrafish to detect drug synergies (Adams et al., 2011). We confirmed that APF could also detect the INH-RIF synergy (Figure 3A).
Figure 3. Synergistic drug interactions in the zebrafish larval infection model.
(A) Larvae 1dpi with 300 far-red fluorescent Mm were treated with RIF (50 µM), INH (100 µM), or both. Bacterial burdens were measured by APF at 5 dpt. Statistical analysis performed via ANOVA on log10 transformed data, with Bonferroni’s post-hoc testing for individual comparisons. (B) Larvae were infected with 100–200 WT or 500–1000 ΔkasB Mm expressing GFPmut3 and then treated with 290 µM RIF for 4 days prior to quantification of bacterial burden by FPC. The reduction of bacterial burden is expressed as log10 FPC counts – mean log10 FPC of the untreated controls. Results analyzed by a Student’s unpaired t-test. (C) Larvae 1dpi with 300 far-red fluorescent Mm and then treated with RIF (100 µM), TLM (400, 950, or 1900 µM), or both. Bacterial burdens were measured by APF at 5 dpt. Bars represent geometric mean RFU. Log10 transformed RFU were compared using ANOVA with a Dunnett’s post-test. TLM treated groups were compared to their respective no-TLM controls (either with or without RIF). (D) ROC analysis showing the predictive value of the model to detect the activity of 400 µM TLM in the presence of RIF (calculated from 100 µM- RIF only group vs. 100 µM RIF/400 µM TLM group).
We next predicted the existence of a possible synergy based on a phenotype associated with a known mycobacterial mutation. KasB is an acyl carrier protein involved in mycolic acid biosynthesis and kasB mutations in both Mtb and Mm render the bacteria hypersusceptible to RIF in culture, presumably by increasing cell wall permeability (Bhatt et al., 2007; Gao et al., 2003). The Mm ΔkasB mutant was also hypersusceptible to RIF in the context of zebrafish infection (Figure 3B). Next we examined whether thiolactomycin (TLM), an antibiotic which targets KasB (Kremer et al., 2000), would synergize with RIF in vivo. We tested a range of TLM concentrations with and without RIF to determine both the minimal effective concentration of TLM and the concentration at which it would synergize with RIF (Figure 3C). We found that 1900 µM TLM was required when used alone, whereas in combination with a subtherapeutic dose of RIF (100 µM), synergy was observed with as little as 400 µM TLM, our suggested screening concentration. ROC analysis yielded an AUC of 0.82 (P=0.0001; Figure 3D). These results suggest the feasibility of using APF to identify new compounds that synergize with existing antitubercular agents.
APF can identify host-targeting compounds and synergies between host and bacterial targeting compounds
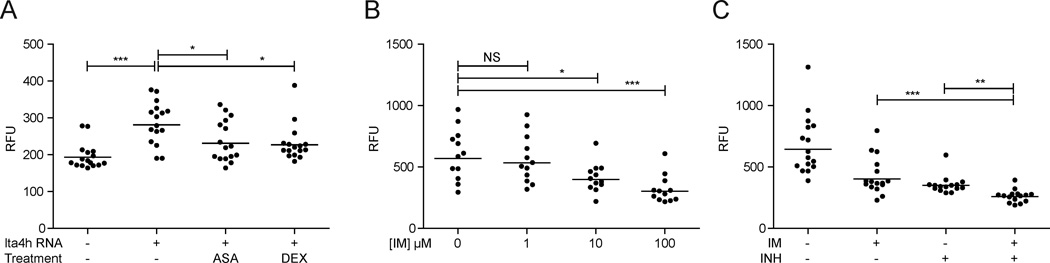
Next we asked if APF could detect the indirect antitubercular activity of host-targeting compounds. We have recently shown that an increased susceptibility to mycobacterial infection is associated with a hyperinflammatory state stemming from overexpression of the leukotriene A4 hydrolase (lta4H) gene (Tobin et al., 2010). The hyperinflammatory state permits increased bacterial growth, which can be reduced by treatment with anti-inflammatory compounds such as dexamethasone (DEX) and acetylsalicylic acid (ASA) (Tobin et al., 2012). The APF assay detected the efficacy of both compounds to reduce infection burdens in the context of the hyperinflammatory state (Figure 4A). Another recent study showed that the ABL tyrosine kinase family inhibitor imatinib (Gleevec) reduces Mm and Mtb burdens in mice by targeting host pathways (Napier et al., 2011). APF identified imatinib efficacy at concentrations as low 10 µM (Figure 4B). Furthermore, we were able to demonstrate that imatinib synergizes with traditional antibacterial drugs: combination treatment with 100µM each imatinib and INH was more efficacious than either drug alone (Figure 4C). Together these results highlight the ability of the APF platform to detect antitubercular activity of host-targeting compounds, and synergies with traditional antimicrobials; interactions that would not be detected in axenic culture models.
Figure 4. Host-targeting antimicrobials and synergies.
(A) WT and LTA4H overexpressing larvae were infected with 300 far-red fluorescent Mm and then immediately treated for 2 days with 0.75µM DEX or 1µM ASA. (B, C) Larvae were infected with 300 far-red fluorescent Mm and then treated for 3 days with various concentrations of imatinib (B), or 100 µM imatinib, 100µM INH or a combination of both. For all panels, bars represent geometric mean RFU. For statistical analyses, data were log10 transformed and compared by one-way ANOVA with appropriate post-hoc testing to accommodate multiple comparisons (A and C, Bonferroni post-tests for shown comparisons and B, Dunnett’s post-test).
DISCUSSION
We have developed and validated a comprehensive platform for rapid, in vivo antitubercular drug discovery that combines husbandry and transient, reversible anesthesia in 96-well plates, with automated in situ assays for drug efficacy. These assays monitor drug efficacy by two parameters—quantification of bacterial burdens and host survival—within a compressed time frame of 5 days. These are the key parameters by which antibiotic efficacy is assessed in mammalian models, usually over an extended time frame of weeks to months. In addition, the automated microscopy platform provides more in-depth information regarding the distribution and quality of infection, for example whether or not granulomas form (Volkman et al., 2004). The fluorimetry platform is faster and potentially more versatile, as it also provides information on acute drug toxicity in the context of the whole animal. However, the platform is not appropriate for the study of long-term drug effects, either sterilization activity or chronic toxicity. Compounds emerging from this platform would still need to be tested in adult animal models.
Both of the platforms we describe would have quickly identified anti-tubercular agents in use today, and are also sensitive enough to detect drug synergies, even at subtherapeutic concentrations of the individual agents. For example, our identification of a previously unrecognized drug synergism, between TLM and RIF, was demonstrated in less than a week. The quest for additional drugs that synergize with the current regimen continues with the goal of further shortening treatment of drug-susceptible TB. At the same time, it is important to identify new synergistic regimens for drug-resistant TB. This larval platform can serve both needs.
Utility for discovery of in vivo specific and host-targeting compounds
Mycobacteria, like other pathogens, express host-induced determinants important for in vivo survival and virulence (McKinney, 2000; Tobin and Ramakrishnan, 2008; Valdivia et al., 2000). Indeed, the failure of in vitro target-based and whole cell screens to identify compounds that are active in animal infection models (Payne et al., 2007; Pethe et al., 2010) has highlighted the importance of developing screening platforms that better replicate conditions in vivo. The development of high content screening (HCS), which utilizes bacteria grown in cultured cells, may partly address the shortcomings of older methods (Brodin and Christophe, 2011; Christophe et al., 2009). HCS methodologies, which also incorporate toxicity screening, certainly provide higher throughput than the larval platform described here. However, the larval platform has some distinct advantages for primary screening as well. While some mycobacterial determinants are activated upon infection of macrophages in culture, others are only induced upon granuloma formation in vivo (Davis et al., 2002; Ramakrishnan et al., 2000). Therefore, additional druggable targets may be revealed only in the context of in vivo infection
Furthermore, there is a growing appreciation that pathogenic organisms induce and exploit host determinants for their own benefit, and that such factors can be targeted pharmacologically to thwart infection (Agarwal et al., 2009; Chen et al., 2008; Kuijl et al., 2007; Kumar et al., 2010; Schwegmann and Brombacher, 2008; Tobin et al., 2012; Tobin et al., 2010; Volkman et al., 2010). For example, mycobacteria require host kinases for intracellular growth, inhibitors of which curb mycobacterial growth in cultured macrophages, mice (Napier et al., 2011), and as we now show, in zebrafish larvae. Likewise, mycobacteria are sensitive to the fine balance of inflammatory responses, modification of which can impact bacterial survival (Tobin et al., 2012; Tobin et al., 2010). These observations suggest the advantage of unbiased screens in the context of in vivo infections. The utility of this approach is highlighted by recent work in the zebrafish larva showing that bacterial expansion is mediated in part by interactions of a bacterial virulence determinant with the host matrix metalloproteinase MMP9 (Volkman et al., 2010). As MMP9 is induced not in the infected macrophages, but in the surrounding epithelium, inhibitors of this pathogen-beneficial pathway can only be identified in the context of whole animals. The discovery of host- targeting “anti-microbial” compounds has the potential to improve treatment both of drug susceptible TB and drug-resistant disease. Thus, our demonstration that the platform can identify host-targeting compounds is an important validation of its unique potential.
The larval platform should also prove valuable in secondary screening of lead compounds identified by a variety of other methods. Use of the larvae to identify effective drugs and their metabolic derivatives in the context of an active and intact physiological system would reduce the number of leads subjected to expensive in vivo toxicity and efficacy testing, and would also serve as a facile tool for lead optimization.
Practical advantages
The platforms we describe further allow rapid and extensive compound characterization in vivo using limited samples. For example, evaluating the effect of INH treatment using five larvae over the course of 10 days requires 69 µg of INH at the 400 µM concentration used. In contrast, 25 mg would be required for a similar assessment of compound efficacy in mice, assuming a 25 mg kg−1day−1 dosing regimen (Stover et al., 2000). Derivative compounds synthesized in small amounts for structure-activity relationships can also be rapidly characterized for differences in their biological activity.
Another advantage of the larval model may be that pharmacokinetic restrictions such as compound bioavailability are less relevant, allowing for a primary assessment of pure biological efficacy. For example, the efficacy of streptomycin can be detected simply by soaking larvae in the drug (Adams et al., 2011), but must be administered by injection in humans and mice. The discovery of parent compounds without bioavailability constraints in the zebrafish may then allow for their optimization for human use. In this way, the larval platform represents the best of both worlds—complex enough to identify drugs that can only be identified in the context of in vivo infection, but simple enough to bypass bioavailability constraints.
Finally, the newer HCS methodologies require expensive and computationally intensive microscopy (Brodin and Christophe, 2011), whereas the larval APF format delivers the same information (efficacy and toxicity) more quickly, and using more widely available equipment. Given the failure of large pharmaceutical companies to identify new antimicrobial drugs, and ready access to drug libraries, drug discovery is increasingly shifting to academic and non-profit research groups, and smaller biotechnology companies (Moellering, 2011). Thus, the larval platform we describe may be accessible, though perhaps with lower throughput, to a wider range of academic and non-profit research groups.
Algorithm for drug discovery
A combination of fluorimetric and microscopic techniques can be used in the model to interface rapid drug screening with detailed characterization of drug effects on disease pathogenesis (Figure S4). The screen should yield host-targeting compounds as well as antibacterial compounds, the latter of which may be selectively effective in vivo, or also effective against bacteria in culture. In this context, we note that many host targeting drugs, both in the context of TB and for other diseases have been found effective at concentrations < 10 µM (Becker et al., 2012; Cusick et al., 2012; Das et al., 2010; Laggner et al., 2012; Ni et al., 2011; Tobin et al., 2012; Tobin et al., 2010; White et al., 2011). So it is possible that parallel screens with high and low compound concentrations can be used to enrich for bacterial and host targeting drugs, respectively. As we have detailed for INH, identification of a candidate drug by APF can be rapidly followed by survival studies and/or low magnification microscopy to determine EBA, dose-dependent efficacy and tolerance. Synergy screens can be performed de novo to identify combinations of compounds with increased EBA. Rapid evaluation of drug toxicity is also feasible in this model (McGrath and Li, 2008).
In summary, this work highlights the relevance, versatility and ease of the zebrafish larval platform for the discovery of traditional antitubercular drugs, and suggests its potential for the discovery of entirely new types of anti-infectives. Zebrafish are proving to be versatile models for infectious diseases and are now being used to study a variety of human pathogens where drug resistance poses great difficulties. These include the bacterial pathogens Burkholderia cenocepacia, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes and Francisella species, the fungal pathogen Candida albicans and the viral pathogen herpes simplex virus type 1 (Brannon et al., 2009; Chao et al., 2010; Clatworthy et al., 2009; Prajsnar et al., 2008; Szabady et al., 2009; Vergunst et al., 2010; Wiles et al., 2009). The methods described here should be easily extended for drug discovery efforts focused on these pathogens.
The use of these platforms is not limited to antibiotic screens; indeed, they should be applicable to any screen involving fluorescence readouts in transgenic larvae. The use of green autofluorescence to detect larval death by fluorimetry should be useful for screens of drug toxicity and toxin screens (Peterson and Macrae, 2012). Similarly, the temporary and reversible cryo-anesthesia of zebrafish larvae should be widely applicable for a variety of other drug screens involving older, motile larvae with the obvious exception of cardiac or motility-based drug screens.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Methods
M. marinum strains M (ATCC BAA-535) and 4E4 (ΔkasB (Gao et al., 2003)) expressed either GFPmut3 (plasmid pGFPHYG2, (Cosma et al., 2006b)), dsRed2 (plasmid pR2HYG, a hygromycin-resistant derivative of pMSP12::dsRed2 (Cosma et al., 2004)), or tdKatushka2 (plasmid pTEC22, a derivative of pGFPHYG2 in which the tdKatushka2 open reading frame (Axxora, San Diego, CA) was used to replace gfpmut3). All plasmids are available from AddGene. Bacteria were grown at 33°C in Middlebrook 7H9 medium (Difco) supplemented with 0.5% bovine serum albumin, 0.005% oleic acid, 0.2% glucose, 0.2% glycerol, 0.085% sodium chloride, 0.05% Tween 80 and 50 µg/mL hygromycin. Single-cell suspensions of bacteria were prepared for experimental infections as follows: 100 mL bacteria were grown to mid-log phase, harvested by centrifugation, and resuspended in 1 ml medium. The bacterial suspension was passed through a 27 gauge needle 10 times to disrupt cell clumps, centrifuged at 100×g, and the turbid supernatant collected in a clean tube. The pellet was again resuspended in 1 mL and the process repeated 3 times. The collected supernatants, which consist of single cells and partially disrupted bacterial clumps, were then passed through a sterile 5 µM filter. The resulting single-cell suspensions were stored in 10 µL aliquots at −80°C.
Zebrafish Infection, Husbandry, and Drug Treatment
Wild-type AB zebrafish were infected via caudal vein injections at 36–48 hours post-fertilization (Cosma et al., 2006a) with thawed single-cell suspensions. Embryo maintenance and injection procedure were previously described (Cosma et al., 2006a). LTA4H overexpressing larvae were generated by injection of lta4h RNA into one-cell stage embryos as previously described (Tobin et al., 2012). Larvae were housed in optical bottom 96-well plates (Nunc 265301) containing a single larva per well in 250 µL filter-sterilized fish water (Cosma et al., 2006a). Larval feeding was omitted, thus improving water quality and avoiding the need to transfer larvae to fresh 96 well plates. Relying on the yolk as their nutrient source, uninfected larvae survive up to 12 days dpf. All drug treatment experiments were performed in fish water containing 1% DMSO, except for DEX and ASA treatment experiments which were performed in 0.5% DMSO.
Microscopy
Wide-field microscopy was performed using a Nikon Eclipse Ti-E equipped with a C-HGFIE 130W mercury light source and 2x/0.10 Plan Apochromat objective. Fluorescence images were captured with a CoolSNAP HQ2 Monochrome Camera (Photometrics) using NIS-Elements (version 3.22). Fluorescence filter cubes sets included Chroma FITC (41001), TRITC (41002), and ETmCherry (49008), for detection of green, red and far-red light, respectively. Automated microscopy was performed using the Nikon Ti-S-E Motor XY Stage and custom macros programmed into AutoHotkey version 1.1 (Document S1: Supplemental Software for High-Throughput Microscopy).
Fluorescent Pixel Count
Quantification of fluorescent-Mm infection using images of individual embryos has been described (Adams et al., 2011), and is detailed in Document S2: Supplemental Software for Image Analysis.
Fluorimetry
Fluorimetry was performed using a Tecan GENios Pro microplate reader equipped with 485/20 nm and 535/25 nm, 535/25 nm and 590/20 nm, or 595/10 nm and 650/10 nm filters for green, red, and far-red fluorescence measurements. Bacterial fluorimetry readings were taken using 96-well black-bottom microplates (Costar 3915) loaded with single-cell bacterial suspensions resuspended in 100 µL PBS. For automated plate fluorimetry (APF), a 12-point 4×4 beam pattern was used (see Document S3: APF Settings) to maximize the measured surface area per well. Gain was optimized for individual experiments and fluorescence intensity reported as Relative Fluorescence Units (RFU). For experiments measuring infection burden via APF, the baseline fluorescence of uninfected larvae was measured and subtracted from the fluorescence of infected larvae.
Statistics
Statistical analyses were performed using Prism 5.01 (GraphPad). For data sets requiring log10 transformation prior to ANOVA, embryos with no detectable fluorescence above background, or with no detectable CFU were assigned a value of 0.9, with 1 being the limit of detection, prior to log10 transformation. Post-test P values are as follows: *P < 0.05; **P < 0.01; ***P < 0.001.”
Supplementary Material
Highlights.
A larval zebrafish-M. marium platform for in vivo antitubercular drug discovery
High throughput husbandry and reversible anesthesia in 96 well format
Fluorimetric in situ measurement of drug efficacy and toxicity
Detection of host-acting compounds and identification of a novel drug synergy
ACKNOWLEDGEMENTS
We thank C.E. Barry, III for the gift of TLM, L.-Y. Gao for the kasB mutant strain, J. Cameron for fish facility management and help with the cryo-anesthesia protocol, H.D. Kulasekara and B.A. Landis for technical advice on microplate reading and fluorescent proteins, K. Winglee for software design, G. Davis and D. Fong for advice on automated microscopy, P. Edelstein for discussion, J. Szumowski for manuscript review, P. Greenberg for use of microplate reader, J. P. Ray for help with figure preparation. This work was supported by a Gates Foundation TB drug accelerator grant, a University of Washington Royalty Research Fund Grant, and NIH grants RO1 AI036396, RO1 AI54503 and U54AI057141 to LR. L.R. is a recipient of the NIH Director’s Pioneer Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR. Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature. 2009;460:98–102. doi: 10.1038/nature08123. [DOI] [PubMed] [Google Scholar]
- Becker JR, Robinson TY, Sachidanandan C, Kelly AE, Coy S, Peterson RT, Macrae CA. In vivo natriuretic peptide reporter assay identifies chemical modifiers of hypertrophic cardiomyopathy signalling. Cardiovasc Res. 2012;93:463–470. doi: 10.1093/cvr/cvr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A, Fujiwara N, Bhatt K, Gurcha SS, Kremer L, Chen B, Chan J, Porcelli SA, Kobayashi K, Besra GS, et al. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc Natl Acad Sci U S A. 2007;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cellular Microbiology. 2009;11:755–768. doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Christophe T. High-content screening in infectious diseases. Curr Opin Chem Biol. 2011;15:534–539. doi: 10.1016/j.cbpa.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Cao Y, Semanchik N, Lee SH, Somlo S, Barbano PE, Coifman R, Sun Z. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21819–21824. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CC, Hsu PC, Jen CF, Chen IH, Wang CH, Chan HC, Tsai PW, Tung KC, Lan CY, Chuang YJ. Zebrafish as a model host for Candida albicans infection. Infection and Immunity. 2010;78:2512–2521. doi: 10.1128/IAI.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Divangahi M, Gan H, Shin DS, Hong S, Lee DM, Serhan CN, Behar SM, Remold HG. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. The Journal of experimental medicine. 2008;205:2791–2801. doi: 10.1084/jem.20080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, et al. High content screening identifies decaprenyl-phosphoribose 2' epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000645. e1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Lee JS, Leibman M, Kostun Z, Davidson AJ, Hung DT. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infection and Immunity. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Davis JM, Swaim LE, Volkman H, Ramakrishnan L. Current Protocols in Microbiology. John Wiley and Sons, Inc.; 2006a. Zebrafish and Frog Models of Mycobacterium marinum Infection; pp. 10B.12.11–10B.12.33. [DOI] [PubMed] [Google Scholar]
- Cosma CL, Humbert O, Ramakrishnan L. Superinfecting mycobacteria home to established tuberculous granulomas. Nat Immunol. 2004;5:828–835. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- Cosma CL, Klein K, Kim R, Beery D, Ramakrishnan L. Mycobacterium marinum Erp is a virulence determinant required for cell wall integrity and intracellular survival. Infect Immun. 2006b;74:3125–3133. doi: 10.1128/IAI.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick MF, Libbey JE, Trede NS, Eckels DD, Fujinami RS. Human T cell expansion and experimental autoimmune encephalomyelitis inhibited by Lenaldekar, a small molecule discovered in a zebrafish screen. J Neuroimmunol. 2012;244:35–44. doi: 10.1016/j.jneuroim.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Das BC, McCartin K, Liu TC, Peterson RT, Evans T. A forward chemical screen in zebrafish identifies a retinoic acid derivative with receptor specificity. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010004. e10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald PR, McIlleron H. Antituberculosis Drugs. In: Schaff HS, Zumla A, editors. Tuberculosis: A Comprehensive Clinical Reference. Saunders Elsevier; 2009. pp. 608–617. [Google Scholar]
- Dorman SE, Johnson JL, Goldberg S, Muzanye G, Padayatchi N, Bozeman L, Heilig CM, Bernardo J, Choudhri S, Grosset JH, et al. Substitution of moxifloxacin for isoniazid during intensive phase treatment of pulmonary tuberculosis. American Journal Of Respiratory And Critical Care Medicine. 2009;180:273–280. doi: 10.1164/rccm.200901-0078OC. [DOI] [PubMed] [Google Scholar]
- Etchin J, Kanki JP, Look AT. Zebrafish as a model for the study of human cancer. Methods in cell biology. 2011;105:309–337. doi: 10.1016/B978-0-12-381320-6.00013-8. [DOI] [PubMed] [Google Scholar]
- Gao LY, Laval F, Lawson EH, Groger RK, Woodruff A, Morisaki JH, Cox JS, Daffe M, Brown EJ. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol Microbiol. 2003;49:1547–1563. doi: 10.1046/j.1365-2958.2003.03667.x. [DOI] [PubMed] [Google Scholar]
- Jindani A, Dore CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. American Journal Of Respiratory And Critical Care Medicine. 2003;167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- Kremer L, Douglas JD, Baulard AR, Morehouse C, Guy MR, Alland D, Dover LG, Lakey JH, Jacobs WR, Jr, Brennan PJ, et al. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. The Journal of biological chemistry. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJ, Geluk A, et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Laggner C, Kokel D, Setola V, Tolia A, Lin H, Irwin JJ, Keiser MJ, Cheung CY, Minor DL, Jr, Roth BL, et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat Chem Biol. 2012;8:144–146. doi: 10.1038/nchembio.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Moriyama A, Zon LI. Hematopoietic stem cells, hematopoiesis and disease: lessons from the zebrafish model. Genome medicine. 2011;3:83. doi: 10.1186/gm299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P, Li CQ. Zebrafish: a predictive model for assessing drug-induced toxicity. Drug Discov Today. 2008;13:394–401. doi: 10.1016/j.drudis.2008.03.002. [DOI] [PubMed] [Google Scholar]
- McKinney JD. In vivo veritas: the search for TB drug targets goes live. Nat Med. 2000;6:1330–1333. doi: 10.1038/82142. [DOI] [PubMed] [Google Scholar]
- Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan DJ, Peterson TA, Ruskin JN, Peterson RT, MacRae CA. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107:1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- Mione MC, Trede NS. The zebrafish as a model for cancer. Dis Model Mech. 2010;3:517–523. doi: 10.1242/dmm.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison DA, Chang KC. Experimental models of tuberculosis: can we trust the mouse? American Journal Of Respiratory And Critical Care Medicine. 2009;180:201–202. doi: 10.1164/rccm.200905-0708ED. [DOI] [PubMed] [Google Scholar]
- Moellering RC., Jr Discovering new antimicrobial agents. Int J Antimicrob Agents. 2011;37:2–9. doi: 10.1016/j.ijantimicag.2010.08.018. [DOI] [PubMed] [Google Scholar]
- Napier RJ, Rafi W, Cheruvu M, Powell KR, Zaunbrecher MA, Bornmann W, Salgame P, Shinnick TM, Kalman D. Imatinib-sensitive tyrosine kinases regulate mycobacterial pathogenesis and represent therapeutic targets against tuberculosis. Cell host & microbe. 2011;10:475–485. doi: 10.1016/j.chom.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni TT, Rellinger EJ, Mukherjee A, Xie S, Stephens L, Thorne CA, Kim K, Hu J, Lee E, Marnett L, et al. Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem Biol. 2011;18:1658–1668. doi: 10.1016/j.chembiol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou H, Simon JA, Rubel EW, Raible DW. Screening for chemicals that affect hair cell death and survival in the zebrafish lateral line. Hear Res. 2012;288:58–66. doi: 10.1016/j.heares.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Fishman MC. Discovery and use of small molecules for probing biological processes in zebrafish. Methods Cell Biol. 2004;76:569–591. doi: 10.1016/s0091-679x(04)76026-4. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Macrae CA. Systematic approaches to toxicology in the zebrafish. Annu Rev Pharmacol Toxicol. 2012;52:433–453. doi: 10.1146/annurev-pharmtox-010611-134751. [DOI] [PubMed] [Google Scholar]
- Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun. 2010;1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cellular Microbiology. 2008;10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- Pratt W, Fekety R. The antimicrobial drugs. New York: Oxford University Press; 1986. [Google Scholar]
- Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- Rudner LA, Brown KH, Dobrinski KP, Bradley DF, Garcia MI, Smith AC, Downie JM, Meeker ND, Look AT, Downing JR, et al. Shared acquired genomic changes in zebrafish and human T-ALL. Oncogene. 2011;30:4289–4296. doi: 10.1038/onc.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann A, Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Sci Signal. 2008;1:re8. doi: 10.1126/scisignal.129re8. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1:366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- Szabady RL, Lokuta MA, Walters KB, Huttenlocher A, Welch RA. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000320. e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2011;105:493–516. doi: 10.1016/B978-0-12-381320-6.00021-7. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cellular Microbiology. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148:434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Vary JC, Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Tweedy BL, Lesney MS. The Pharmaceutical Century. American Chemical Society; 2000. 1950s: Prescriptions and Polio. [Google Scholar]
- Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally drug-resistant tuberculosis in India. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54:579–581. doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Cirillo DM, Lee AK, Bouley DM, Falkow S. mig-14 is a horizontally acquired, host-induced gene required for salmonella enterica lethal infection in the murine model of typhoid fever. Infection and Immunity. 2000;68:7126–7131. doi: 10.1128/iai.68.12.7126-7131.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Meijer AH, Renshaw SA, O'Callaghan D. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infection and Immunity. 2010;78:1495–1508. doi: 10.1128/IAI.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Bower JM, Redd MJ, Mulvey MA. Use of zebrafish to probe the divergent virulence potentials and toxin requirements of extraintestinal pathogenic Escherichia coli. PLoS Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000697. e1000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zignol M, vanGemert W, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007–2010. Bulletin of the world Health Organization. 2012;90:111–119. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.