Abstract
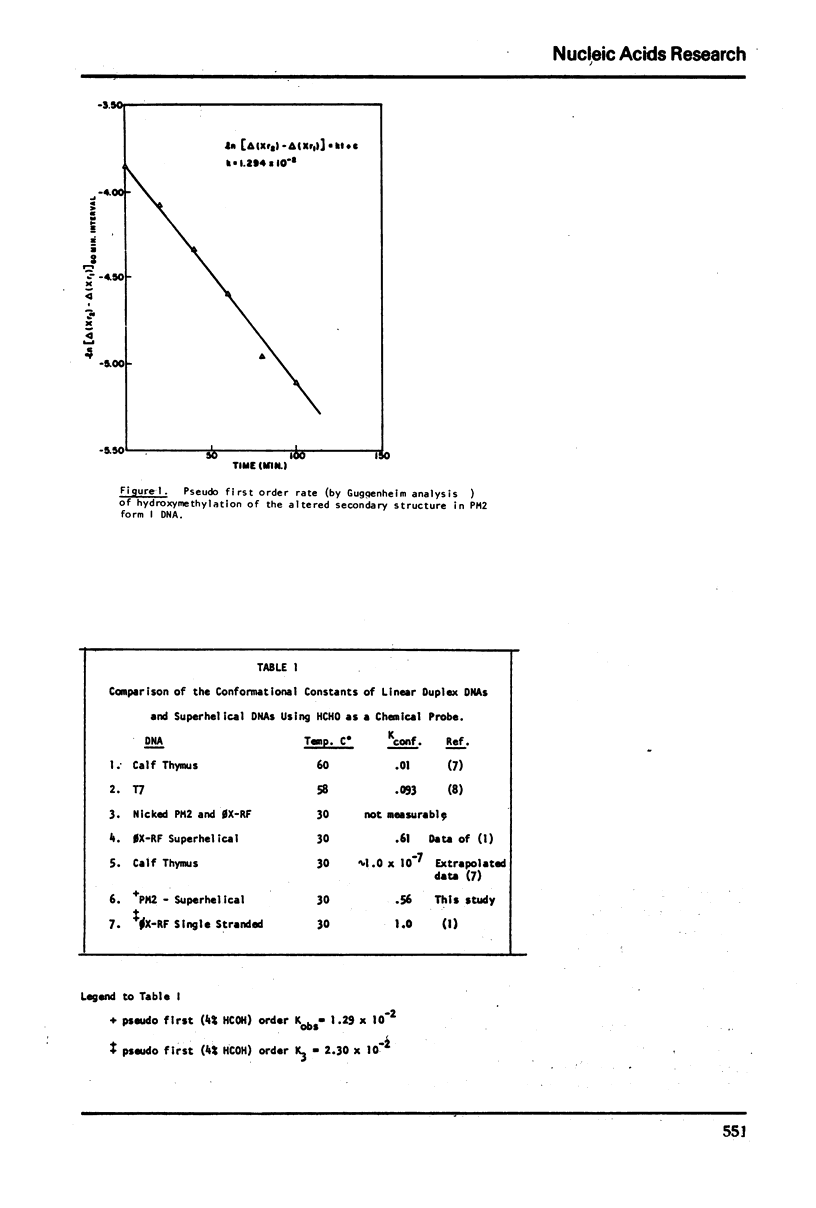
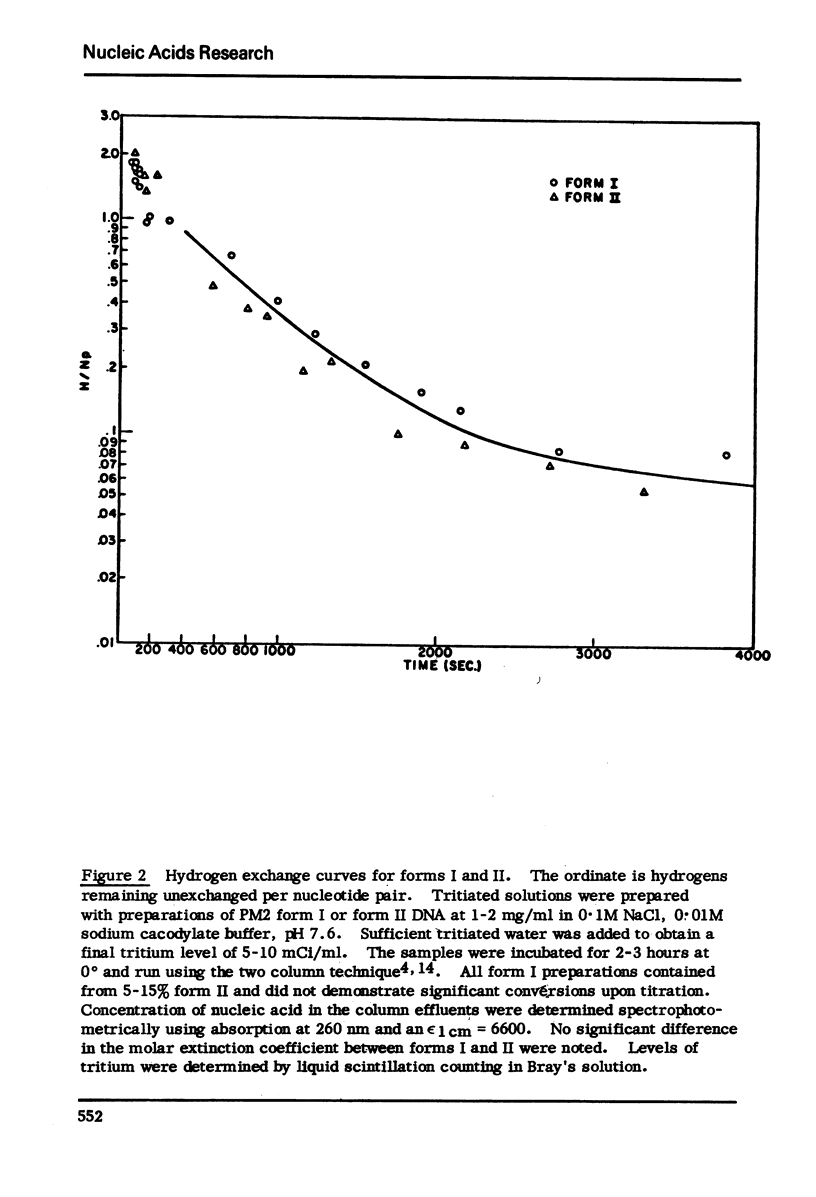
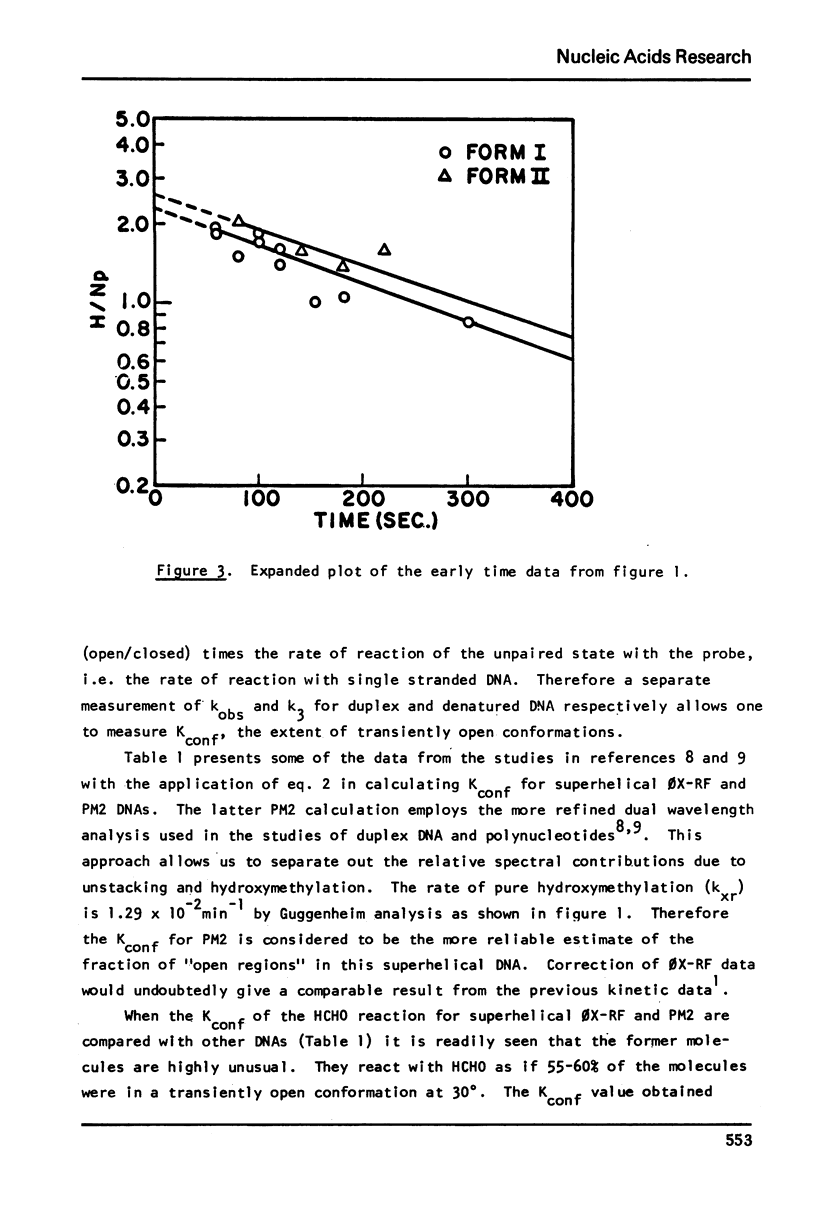
Kinetic analysis of the early, fast reaction of superhelical DNA with formaldehyde reveals that this region or regions is 56% “single strand like” in character. Hydrogen-tritium exchange studies coupled with other considerations show that this reaction is not due to a difference in conformational motility between form I and form II molecules, but is due to unpaired or weakly hydrogen bonded, localized region(s) of the form I allomorph of circular DNA.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Beerman T. A., Lebowitz J. Further analysis of the altered secondary structure of superhelical DNA. Sensitivity to methylmercuric hydroxide a chemical probe for unpaired bases. J Mol Biol. 1973 Sep 25;79(3):451–470. doi: 10.1016/0022-2836(73)90398-7. [DOI] [PubMed] [Google Scholar]
- Englander J. J., Von Hippel P. H. Slow exchange of the "outside" amino hydrogens of DNA. J Mol Biol. 1972 Jan 14;63(1):171–177. doi: 10.1016/0022-2836(72)90528-1. [DOI] [PubMed] [Google Scholar]
- Hanson C. V. A study of rapid hydrogen exchange in nucleic acids. J Mol Biol. 1971 Jun 28;58(3):847–863. [PubMed] [Google Scholar]
- Hvidt A., Nielsen S. O. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Kato A. C., Bartok K., Fraser M. J., Denhardt D. T. Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim Biophys Acta. 1973 Apr 21;308(7):68–78. doi: 10.1016/0005-2787(73)90123-8. [DOI] [PubMed] [Google Scholar]
- McConnell B., von Hippel P. H. Hydrogen exchange as a probe of the dynamic structure of DNA. I. General acid-base catalysis. J Mol Biol. 1970 Jun 14;50(2):297–316. doi: 10.1016/0022-2836(70)90194-4. [DOI] [PubMed] [Google Scholar]
- McConnell B., von Hippel P. H. Hydrogen exchange as a probe of the dynamic structure of DNA. II. Effects of base composition and destabilizing salts. J Mol Biol. 1970 Jun 14;50(2):317–332. doi: 10.1016/0022-2836(70)90195-6. [DOI] [PubMed] [Google Scholar]
- Printz M. P. Tritium-hydrogen exchange studies of polynucleotides. Double-stranded polyriboadenylic acid. Biochemistry. 1970 Jul 21;9(15):3077–3087. doi: 10.1021/bi00817a022. [DOI] [PubMed] [Google Scholar]
- Printz M. P., von Hippel P. H. On the kinetics of hydrogen exchange in deoxyribonucleic acid. pH and salt effects. Biochemistry. 1968 Sep;7(9):3194–3206. doi: 10.1021/bi00849a023. [DOI] [PubMed] [Google Scholar]
- Rosenfeld A., Stevens C. L., Printz M. P. Studies on the secondary structure of phenylalanyl transfer ribonucleic acid. Biochemistry. 1970 Dec 8;9(25):4971–4980. doi: 10.1021/bi00827a022. [DOI] [PubMed] [Google Scholar]
- Utiyama H., Doty P. Kinetic studies of denaturation and reaction with formaldehyde on polydeoxyribonucleotides. Biochemistry. 1971 Mar 30;10(7):1254–1264. doi: 10.1021/bi00783a024. [DOI] [PubMed] [Google Scholar]
- Von Hippel P. H., Wong K. Y. Dynamic aspects of native DNA structure: kinetics of the formaldehyde reaction with calf thymus DNA. J Mol Biol. 1971 Nov 14;61(3):587–613. doi: 10.1016/0022-2836(71)90066-0. [DOI] [PubMed] [Google Scholar]



