Abstract
Keratin 8 (K8) is a type II keratin that is associated with the type I keratins K18 or K19 in single layered epithelia. We generated a bacterial artificial chromosome (BAC) transgenic mouse line that expresses the tamoxifen inducible CreERT2 inserted into the endogenous murine K8 gene. The transgenic mouse line contains two copies of the BAC transgene. To determine the expression specificity and inducibility of CreERT2, the K8–CreERT2 mice were bred with a Gt(ROSA26)ACTB–tdTomato–EGFP fluorescent protein-based reporter transgenic mouse line. We demonstrated that CreERT2 and the endogenous K8 gene share the same patterns of expression and that the enzymatic activity of CreERT2 can be efficiently induced by tamoxifen in all K8-expressing tissues. This mouse line will be useful for studying gene function in development and homeostasis of simple epithelia, and investigating both tissue lineage hierarchy and the identity of the cells of origin for epithelial cancers.
Keywords: Keratin 8, CreERT2, BAC transgenesis, Simple epithelia
Keratins belong to intermediate filaments that are one of the major components of the cytoskeleton in vertebrate epithelial cells (Coulombe 1993). There are two types of keratins: the type I acidic and the type II basic keratins. They form obligate non-covalent heteropolymers that are differentially expressed in various epithelial cells (Moll et al. 1982). Keratin 8 (K8) is a type II keratin that is associated with the type I keratins K18 or K19 in single layered epithelia. For example, intermediate filaments in intestinal epithelial cells are mainly composed of both K8–K18 and K8–K19 heterpolymers, while those in the kidney, prostate and mammary gland are predominantly K8–K18 heteropolymers (Moll et al. 1982).
Keratin 8 (K8) is found in various carcinomas developed in a wide spectrum of tissues (Moll et al. 1982; Quinlan et al. 1985) such as small intestine, prostate and mammary gland, suggesting that the cell lineages that express K8 may serve as one of the cells-of-origin for those cancers. This hypothesis can be tested by activating oncogenic signaling specifically in K8 positive epithelial cells. K8 is expressed at the morula stage and in the trophectoderm of blastocysts during early development (Chisholm and Houliston 1987). Therefore, when generating such a mouse model it is essential that not only is the Cre expression under the control of the K8 regulatory sequences, but also the Cre activity should be constitutively inactivated and will only be switched on upon induction as desired. This can be achieved by expressing a tamoxifen-inducible CreERT2 (Vasioukhin et al. 1999) driven by the K8 promoter.
Keratin 8 (K8) plays an essential role in vertebrate development, since K8 null mice are embryonic lethal on the C57BL/6 background and develop hepatitis and colonic hyperplasia on the FVB/N background (Baribault et al. 1993, 1994). Additionally, it has been shown that not only the 5′ and 3′ flanking sequences, but also the intragenic sequences are essential for sufficient and specific expression of the endogenous K8 gene (Casanova et al. 1995). These facts present huge caveats to generating the K8–CreERT2 mouse model using the knock-in approach which usually inactivates the endogenous allele, or the traditional transgenic approach. Nevertheless, a 3.5 kb fragment upstream of the start codon of murine K8 gene was reported to drive the expression of CreER in a lineage specific manner in a K8–CreER transgenic mouse strain (Van Keymeulen et al. 2011). In contrast to this strategy, we chose to generate the K8–CreERT2 transgenic mouse model using a bacterial artificial chromosome (BAC) transgenic approach (Heintz 2001). The BAC transgenic approach combines the advantages of the conventional transgenic and the knock-in gene-targeting strategies, while at the same time avoiding the aforementioned caveats.
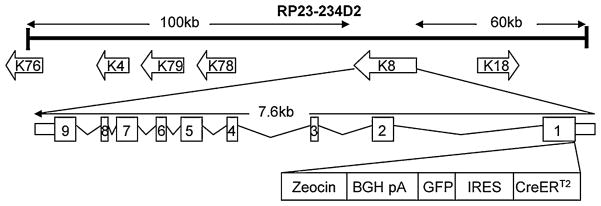
A BAC clone RP23-234D2 that contains the full-length K8 gene and 60 kb of 5′ as well as 100 kb of 3′ flanking sequences was purchased from the BACPAC resource center as the backbone for the transgenic unit. As shown in Fig. 1, we used red-ET cloning and inserted the CreERT2 coding sequence at the translational start codon of the K8 gene. Additionally, a GFP expressing cassette driven by an internal ribosomal entry site (IRES–GFP) was inserted downstream of the CreERT2 cDNA, so that GFP will be expressed simultaneously to fluorescently label the K8-expressing cells. The recombined BAC clone was verified by restriction enzyme digestion and a PCR based approach described in Methods to confirm that the transgene cassette was inserted in the expected locus. The resulting BAC construct was linearized, purified and microinjected into fertilized eggs derived from C57BL/6 mice.
Fig. 1.
Diagram of the strategy generating the K8–CreERT2 targeting BAC clone. Transgene, including CreERT2 cDNA, an IRES–GFP expression cassette, and the Zeocin resistant gene, was inserted at the AUG translational initiation codon of the K8 gene in BAC clone RP23-234D2. Zeocin serves as a positive selection marker during BAC recombineering. IRES internal ribosomal entry site. GFP green fluorescence protein
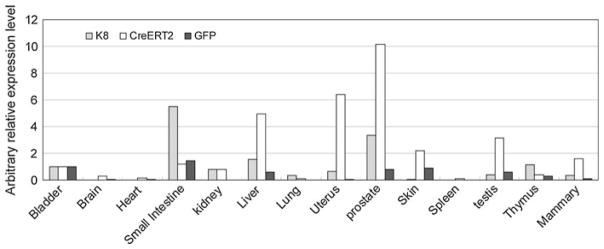
A preliminary RT–PCR based assay was utilized to screen for the mouse lines that expressed the CreERT2 transgene with a pattern similar to that of the endogenous K8 gene. Based on this assay, one transgene positive mouse line was selected for further extensive characterization. Major organs were collected from offspring of this founder and quantitative RT–PCR analyses were performed to measure the expression levels of CreERT2, GFP and the endogenous K8 gene. Figure 2 shows that the endogenous K8 expression is detected in bladder, kidney, liver, lung, mammary gland, prostate, skin, testis, thymus and uterus. Consistently, CreERT2 and GFP are expressed at a very similar pattern in this line, demonstrating that transgene expression recapitulates that of the endogenous K8 gene. Though GFP expression can be detected by PCR and Western blot analyses, green fluorescence was not detectable on frozen tissue sections by fluorescent microscopy or using dissociated prostate cells by fluorescence activated cell sorting analysis (data not shown), suggesting that the expression level of GFP is below the threshold for detection by fluorescent microscopy and flow cytometry.
Fig. 2.
A representative quantitative RT–PCR analysis of the expression of CreERT2, GFP and the endogenous K8 gene in a spectrum of tissues in 5-week-old K8–CreERT2 BAC transgenic mice. Bar graph shows the arbitrary relative expression level of each gene, normalized by the expression level in the bladder
To further confirm the expression pattern of CreERT2 at the protein level and test the inducible properties of the Cre recombinase, K8–CreERT2 mice were bred to Gt(ROSA)26ACTB–tdTomato–EGFP transgenic mice (hereafter referred to as the mTmG mice), generating the K8–CreERT2; mTmG dual transgenic mice. The mTmG mouse line is a fluorescent protein-based reporter line that possesses loxP sites on either side of a membrane-targeted tomato–Red (mT) cassette and expresses strong red fluorescence in all tissues (Muzumdar et al. 2007). Upon excision of the mT expression cassette by Cre-mediated homologous recombination, a previously silenced expression cassette for a membrane-targeted enhanced green fluorescence protein (mG) is activated. Tamoxifen (2 mg/ 40 g/day for 4 consecutive days) and the vehicle solvent (corn oil) were injected intraperitonially into 5- or 8-week old male and female mice. Four days after the last injection, various organs were collected for analysis of GFP expression. The IRES–GFP expression cassette in the transgene expresses GFP at a level that is undetectable by fluorescent microscopy, hence it did not interefere with visualization and interpretation of the expression of the mTmG reporter.
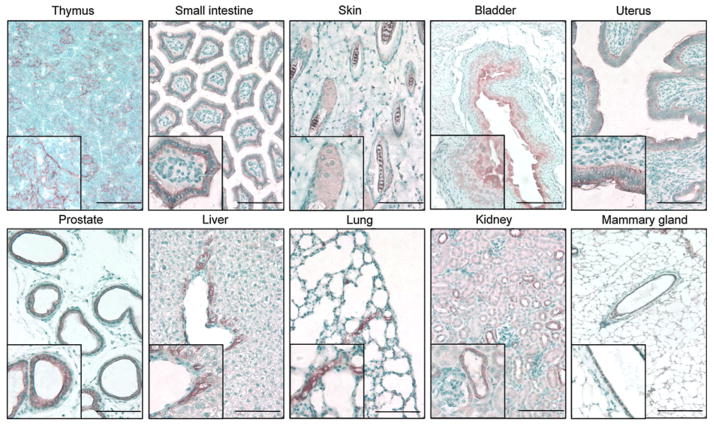
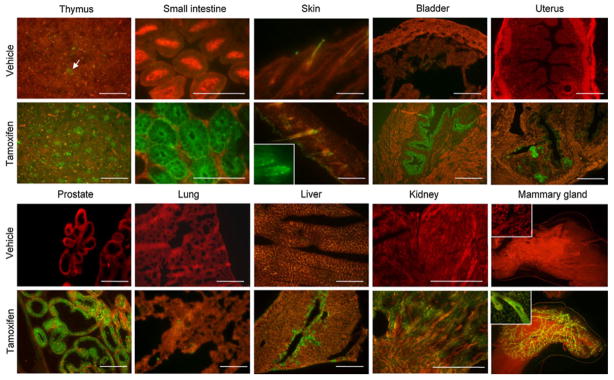
The immunohostochemical analyses of the expression of the endogenous K8 gene in the tissues where the K8 transcript is detectable by quantitative RT-PCR are shown in Fig. 3. K8 is expressed in hepatocytes and biliary ducts in the liver, epithelial cells in distal tubules in the kidney, epithelial reticular cells in the thymus, hair follicles in the skin, and epithelial cells in the small intestine, bladder, prostate, mammary gland, and uterus. Consistently, in the tamoxifen-treated group, GFP positive epithelial cells were observed in the prostate, thymus, small intestine, mammary gland, bladder, uterus, liver, kidney, lung and skin. Figure 4 shows representative fluorescent images of tissue sections from various organs in the vehicle and tamoxifen treated groups. GFP is expressed in the same pattern with that of the endogenous K8 in all tissues except the testis. Though PCR analysis showed that K8 and CreERT2 are expressed in the testis, IHC analysis showed ambiguous K8 staining and GFP fluorescence was not observed in the testis of tamoxifen treated K8–CreERT2; mTmG mice (data not shown). No GFP signal was observed in other major organs where K8 is not expressed (data not shown). In summary, the expression pattern of the reporter gene is consistent with that of the endogenous K8 gene (Casanova et al. 1995). Residual leaky Cre activity was observed in the thymus and small intestine in the vehicle treated K8–CreERT2; mTmG animals, but not in any other organs. These data demonstrated that CreERT2 expression faithfully recapitulates that of the endogenous K8 and its activity is efficiently induced by tamoxifen.
Fig. 3.
Immunohistochemical analysis of the endogenous K8 expression in various murine tissues. Bars = 100 μm
Fig. 4.
Fluorescent microscopic analyses of GFP expression in tissues from K8 to CreERT2; mTmG mice treated with tamoxifen (2 mg/40 g/mouse/day for 4 consecutive days) and the vehicle (corn oil). White arrow shows residual leaky CreERT2 activity in thymus. Bars = 100 μm
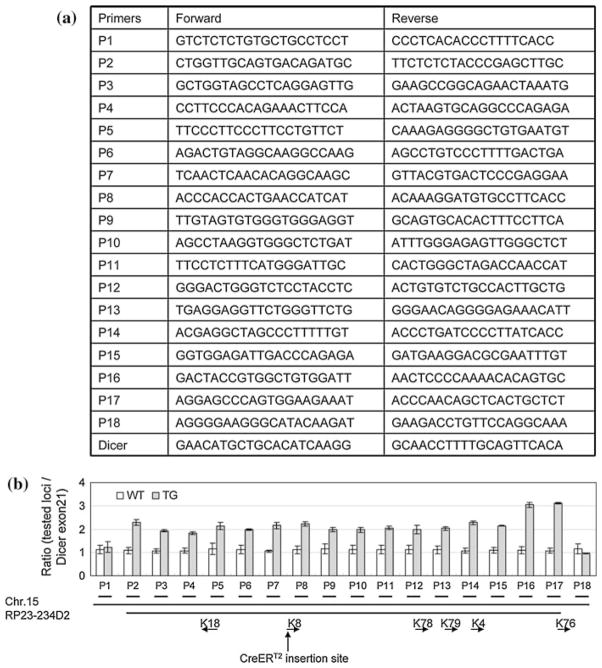
We determined the BAC copy number using a qPCR based method. As schematically illustrated in Fig. 5a, 18 pairs of primers for quantitative RT–PCR analysis were designed approximately every 10 kb spanning the 190 kb genomic locus that includes all the 170 kb sequences covered by the RP23-234D2 BAC clone. The first and the last pairs of the primers recognize sequences that are upstream and downstream of the sequences covered by the BAC clone, respectively. Therefore, the copy numbers of the sequences amplified by these primer sets should reflect that of the BAC transgene. We also designed primers that generate a 79 bp amplicon from the exon21 of mouse Dicer1 genomic locus. This sequence is unrelated with the BAC clone sequence and serves as the two-copy gene control. Figure 5b showed that the K8–CreERT2 transgenic line contains twofold of the amount of the first 150 kb genomic sequences covered by the BAC clone as compared with the wild type control littermates. Surprisingly, transgenic mice possess threefold of the amount of the last 20 kb sequence covered by the BAC clone. This may be caused during formation of trangene concatemer. Overall, our result demonstrates that the BAC clone was inserted into the genome intact, at least at a resolution of 10 kb. Since the transgenic line is maintained in heterozygosis, it should contain 2 copies of the BAC transgene. F1 progeny from different litters were tested in this assay and the same results were obtained, suggesting that the two copies of the BAC transgene do not segregate and that the line contains a single insertion site.
Fig. 5.
Quantitative genomic DNA PCR analysis demonstrated that the K8–CreERT2 line contains 2 copies of the transgene. Error bars represent means and STD from results generated from 2 wild type and 3 transgenic mice
In summary, we have generated a BAC transgenic mouse line that expresses the tamoxifen-inducible CreERT2 transgene under the regulation of K8 genetic apparatus included in the 170 kb BAC construct. The CreERT2 expression faithfully recapitulates that of the endogenous K8 gene and its activity is efficiently activated by tamoxifen. This mouse line will be useful for studying gene function in development and homeostasis of simple epithelia by systemic or topical gene activation or inactivation. Additionally, this mouse line is also of great value for investigating tissue lineage hierarchy by lineage tracing as well as the identity of the cells of origin for cancer.
Methods
BAC recombineering and generation of the K8–CreERT2 mouse line
The BAC clone RP23-234D2 containing the murine K8 gene and 60 kb 5′ as well as 100 kb 3′ flanking sequences was purchased from the BACPAC resources center (Children’s Hospital Oakland Research Institute). The primers used to generate 5′ and 3′ homology regions (HR) are as below: 5′ HR sense: 5′-ATGGCG CGCCAGGGTACTTATACCCCCAAAGGG-3′; 5′HR antisense: 5′-TTGGCGCGCCGGTGAAGTCTGGAGCGAAGGCAGTGAGC-3′; 3′ HR sense: 5′-ATGCTAGCATGTCCATCAGGGTGACTCAGAAATCC-3′; 3′HR antisense: 5′-ATGCTAGCTGCACGGGGCACCATGAGGAAACC-3′. Amplicons were cloned into a modified pUC19 plasmid using the restriction sites within the oligos. The 5′HR amplicon ends at the start codon of the K8 gene. CreERT2 cDNA followed by an IRES–GFP cassette with bovine growth factor poly-adenylation sequence was then cloned into the plasmid so that CreERT2 is immediately downstream of the start codon of the K8 gene. Finally, an expression cassette encoding zeocin was put downstream of BGH polyA site for positive selection during recombineering. The linearized plasmid was electroporated into RP23-234D2 containing bacteria and the recombination procedures were performed using the quick and easy BAC modification kit (Gene Bridges GmbH) according to the manufacturer’s instructions. Recombined BACs were verified by enzymatic digestion. In addition, a PCR based methodology was also employed to confirm the correct insertion of the CreERT2 expression cassette in the BAC clone. A forward verification primer (5′-GCTGGTAAAGGC CGGTGCCGGC-3′) upstream of the 5′ HR region was used in conjunction of a reverse primer within CreERT2 (5′-CATGTTTAGCTGGCCCAAAT-3′) for this purpose. A 900 bp amplicon was generated from correctly recombined BAC clones.
Bacterial artificial chromosome (BAC) DNA was prepared using NucleoBond BAC 100 kit (Macherey–Nagel). After digestion with the PI–SceI restriction enzyme at 37°C overnight, the linearized BAC DNA was dialyzed against a microinjection buffer (10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, and 100 mM NaCl) at 4°C for 4 h. The linearized BAC fragment was then injected at a concentration of 1 ng/ul into fertilized mouse oocytes isolated from the C57BL/6J strain. Oocytes that survived injection were transferred into both oviducts of pseudo-pregnant mothers.
Mouse genotyping
Transgenic mice were identified by PCR analysis with genomic DNA isolated from tail biopsies using the DNeasy blood and tissue kit (Qiagen). The primers used to genotype transgenic mice are as below: Cre forward: 5′-CCTGACAGTGACGGTCCAAAG-3′, Cre reverse: 5′-CATGACTCTTCAACTCAAACT-3′; mTmG primer1:5′-CTCTGCTGCCTCCTGGCTTCT-3′;mTmG primer 2:5′-CGAGGCGGATCACAAGCAATA-3′ and mTmG primer 3: 5′-TCAATGGGCGGGGGTCG TT-3′. A 700 bp amplicon should be generated from K8 to CreERT2 mice. A 330 and a 250 bp amplicon should be generated from the wild type and mTmG alleles, respectively, using the mTmG primers.
RNA isolation and quantitative RT–PCR
Total RNA was isolated from cells using the RNeasy mini isolation kit (Qiagen). Reverse transcription was performed using the iScript one-step RT kit (BioRad). Quantitative RT–PCR was performed using the Taqman Power Sybrgreen master mix (Applied Biosystems) on a StepOne plus real-time PCR system (Applied Biosystems). Primers for real-time PCR are as below: actin forward: 5′-GACGGCCAGGTCATC ACTAT-3′; actin reverse: 5′-CGGATGTCAACGTCACACTT-3′; Cre forward: 5′-TCCAGCACCCTGA AGTCTCT-3′; Cre reverse: 5′-GCCATCAGGTGGA TCAAAGT-3′; GFP forward: 5′-GACGTAAACGGC CACAAGTT-3′; GFP reverse: 5′-GAACTTCAGGG TCAGCTTGC-3′; K8 forward: 5′-AGCTGAGGCTG AAACCATGT-3′; K8 reverse: 5′-TTGATGTTGCGG TTCATCTC-3′;
Copy number
Genomic DNA was isolated from tail biopsies from 4-week old K8–CreERT2 and littermate control mice using the DNeasy blood and tissue Kit (Qiagen). Quantitative PCR was performed using the Taqman Power Sybrgreen master mix (Applied Biosystems) on a StepOne plus real-time PCR system (Applied Bio-systems). Primers for real-time PCR are listed in Fig. 5a.
Administration of tamoxifen
Male K8–CreERT2; mTmG mice were injected at 5–8 weeks postnatally. Four injections (1–4 mg/40 g) separated by 24 h each were given i.p. Tamoxifen was dissolved in corn oil (Fluka) at 10 mg/ml. For vehicle control, animals were injected with corn oil only.
Immunohistochemistry
Slides were made from formalin-fixed and paraffin-embedded (FFPE) tissues or from frozen tissues. Tissue sections were stained with mouse anti-cytokeratin 8 (clone 1E8, Covance). Biotinylated goat-anti-rabbit or rabbit-anti-mouse secondary antibodies and Strepavidin-conjugated horseradish peroxidase (Vector Lab Ltd.) was used for chromatic visualization. For GFP and tomato–red fluorescence visualization, frozen tissue sections were analyzed by fluorescent microscopy.
Acknowledgments
We thank Dr. Paul Overbeek for the CreERT2 cDNA. Li Xin is supported by NIH CA125937, DK092 202 and CA141497. NIH CA125937, DK092202 and CA141497.
Contributor Information
Li Zhang, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
Boyu Zhang, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
Sang Jun Han, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
Amy N. Shore, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA, Program in Developmental Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA
Jeffrey M. Rosen, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA, Dan L. Duncan Cancer Center, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA
Francesco J. DeMayo, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA, Dan L. Duncan Cancer Center, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA
Li Xin, Email: xin@bcm.edu, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA, Dan L. Duncan Cancer Center, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA, Department of Pathology and Immunology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
References
- Baribault H, Price J, Miyai K, Oshima RG. Mid-gestational lethality in mice lacking keratin 8. Genes Dev. 1993;7:1191–1202. doi: 10.1101/gad.7.7a.1191. [DOI] [PubMed] [Google Scholar]
- Baribault H, Penner J, Iozzo RV, Wilson-Heiner M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994;8:2964–2973. doi: 10.1101/gad.8.24.2964. [DOI] [PubMed] [Google Scholar]
- Casanova L, Bravo A, Were F, Ramirez A, Jorcano JJ, Vidal M. Tissue-specific and efficient expression of the human simple epithelial keratin 8 gene in transgenic mice. J Cell Sci. 1995;108(Pt 2):811–820. doi: 10.1242/jcs.108.2.811. [DOI] [PubMed] [Google Scholar]
- Chisholm JC, Houliston E. Cytokeratin filament assembly in the preimplantation mouse embryo. Development. 1987;101:565–582. doi: 10.1242/dev.101.3.565. [DOI] [PubMed] [Google Scholar]
- Coulombe PA. The cellular and molecular biology of keratins: beginning a new era. Curr Opin Cell Biol. 1993;5:17–29. doi: 10.1016/s0955-0674(05)80004-3. [DOI] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of BAC transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Quinlan RA, Schiller DL, Hatzfeld M, Achtstatter T, Moll R, Jorcano JL, Magin TM, Franke WW. Patterns of expression and organization of cytokeratin intermediate filaments. Ann NY Acad Sci. 1985;455:282–306. doi: 10.1111/j.1749-6632.1985.tb50418.x. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;34(3):121–136. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]