Abstract
Helicobacter pylori (H. pylori) infection with its vast prevalence is responsible for various gastric diseases including gastritis, peptic ulcers, and gastric malignancy. While effective, current treatment regimens are challenged by a fast declining eradication rate due to the increasing emergence of H. pylori strains resistant to existing antibiotics. Therefore there is an urgent need to develop novel antibacterial strategies against H. pylori. In this study, we developed a liposomal nanoformulation of linolenic acid (LipoLLA) and evaluated its bactericidal activity against resistant strains of H. pylori. Using a laboratory strain of H. pylori, we found that LipoLLA was effective in killing both spiral and coccoid forms of the bacteria via disrupting bacterial membranes. Using a metronidazole-resistant strain of H. pylori and seven clinically isolated strains, we further demonstrated that LipoLLA eradicated all strains of the bacteria regardless of their antibiotic resistance status. Furthermore, under our experimental conditions, the bacteria did not develop drug resistance when cultured with LipoLLA at various sub-bactericidal concentrations, whereas they rapidly acquired resistance to both metronidazole and free linolenic acid (LLA). Our findings suggest that LipoLLA is a promising antibacterial nanotherapeutic to treat antibiotic-resistant H. pylori infection.
Keywords: Helicobacter pylori, Bacterial infections, Free fatty acid, Linolenic acid, Liposome
Introduction
Helicobacter pylori (H. pylori) colonizes the stomach of more than half of the world’s population and are the etiologic agent of various gastric diseases including gastritis and peptic ulcer disease.1 H. pylori infection is also the single most common cause of gastric cancer.2 Triple-therapy, the combination of two antibiotics (clarithromycin plus amoxicillin or metronidazole) and a proton pump inhibitor, remains the standard first-line treatment of H. pylori infection worldwide.3 However, this therapy is associated with poor compliance of patients, side effects of the antibiotics, and high cost. Moreover, the increasing emergence of H. pylori strains resistant to some of the antibiotics have resulted in a progressive decline in recent years to unacceptable low eradication rates ranging from 60% to 75%.4–6 Particularly, resistance prevalence of metronidazole, a cornerstone of the triple-therapy formulation, has reached nearly 40% in developed countries and over 90% in developing countries.7 In certain geographic regions, H. pylori resistance to clarithromycin has sharply decreased the rate of eradication by over 40% 6. Even in regions where the conventional therapy remains efficacious, poor patient compliance due to the severe side effects associated with multiple antibiotics frequently leads to therapy failure.8 Therefore, there is an urgent need to develop new antibacterial agents that can effectively eradicate H. pylori with minimum drug resistance.
In this regard, a series of free fatty acids (FFAs) including lauric acid, myristoleic acid, linoleic acid, and linolenic acid have attracted much attention as they have shown antibacterial activities against a diverse range of bacteria including H. pylori.9 In addition to their high potency, these lipid-like molecules are ubiquitous, hence considered less harmful. Studies have also shown that FFAs induce drug resistance in H. pylori at a much lower frequency when compared to conventional antibiotics.10 While promising results have been reported, the use of FFAs in inhibiting H. pylori remains challenging. In particular, medium-chain FFAs that are effective in inhibiting H. pylori commonly have low water solubility. If orally administered, acidic pH in stomach further decreases their solubility, making these molecules ineffective. Even if a small amount of FFAs can be dissolved, they are subject to oxidation, esterification, and lipid-protein complexation, further compromising their bactericidal activity in vivo.9 Therefore, to fulfill the therapeutic potential of FFAs, it is essential to develop novel FFA formulations that can overcome these challenges.
Among various approaches, using nanotechnology to formulate nano-sized FFA-loaded liposomes is promising. During liposome formulation, the amphiphilic nature of FFAs allows these molecules to be directly incorporated into the hydrophobic membranes at high loading yield.11 The resulting liposomes can protect FFAs from degradation. We have demonstrated that FFAs such as lauric acid and oleic acid can be loaded into liposomes and form potent antibacterial agents against Propionibacterium acnes and drug-resistant Staphylococcus aureus, respectively.12, 13 Based on our previous development in FFA-loaded liposomes, in this study we further address challenges in H. pylori treatment with a primary focus on overcoming antibiotic resistance of the bacteria using liposomal formulations. Antibacterial activities induced by liposome-bacterial membrane fusion have been shown to be less likely to induce drug resistance.14 In addition, the approach of using liposomal formulations to inhibit H. pylori in vitro, if successful, will allow us to implement various established nanotechnological strategies, particularly those tailored for oral drug delivery, for future in vivo development.15, 16
Herein, we chose linolenic acid (LLA) as a model FFA to formulate liposomal LLA (LipoLLA), and evaluated its bactericidal activities against various resistant strains of H. pylori. On a laboratory strain of H. pylori, LipoLLA showed an antibacterial efficacy comparable with free LLA pre-dissolved in organic solvent in inhibiting both spiral and coccoid forms of the bacteria. Compared to amoxicillin, which killed the spiral form but not the coccoid form of the bacteria, LipoLLA and LLA were able to kill both forms. Using a metronidazole-resistant strain of H. pylori and seven clinically isolated strains, we further demonstrated that LipoLLA eradicated all strains of bacteria regardless of their resistance status to metronidazole. Finally, we tested LipoLLA in eliciting drug resistance in H. pylori. Under our experimental conditions, bacteria did not develop drug resistance when cultured with LipoLLA at various sub-bactericidal concentrations. On the contrary, bacteria acquired resistance to both metronidazole and LLA when cultured with drugs at comparable concentrations. Our findings suggest that LipoLLA holds strong potential to become an effective antimicrobial agent to treat H. pylori infection.
Experimental Section
Materials
Hydrogenated L-α-phosphatidylcholine (Egg PC), cholesterol, C6-NBD Sphingomyelin (C6NBD), and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl (DMPE-RhB) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). LLA was purchased from Ultra Scientific (North Kingstown, RI). Brain Heart Infusion (BHI), Columbia broth, and agar were purchased from Becton Dickinson (Sparks, MD). Alamar blue dye, tetracycline hydrochloride, amoxicillin, glutaraldehyde, along with other common reagents including tris(hydroxymethyl) aminomethane (Tris), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), phosphate buffered saline (PBS), dimethyl sulfoxide (DMSO), dithiothreitol (DTT), and glycerol were obtained from Sigma-Aldrich Co. LLC (St. Louis, MO).
H. pylori strains and bacterial culture
H. pylori Sydney strain 1 (SS1) and seven clinically isolated strains were used in this study. H. pylori SS1 is a mouse-adapted strain originally described by Lee et al.17 Metronidazole-resistant H. pylori SS1 (Mtzr SS1 mutant) and clinical isolates (Shi470, Lithuania75, SouthAfrica7, India7, Gambia94/24, PeCan4, and SJM180) were a kind gift from Dr. Douglas Berg (Washington University, St. Louis, MO).
H. pylori strains were routinely maintained on Columbia agar supplemented with 5% laked horse blood at 37°C under microaerobic conditions (10% CO2, 85% N2, and 5% O2) as previously described.18 For experiments, broth cultures of H. pylori were prepared by subculturing fresh colonies from agar plates into BHI containing 5% fetal bovine serum (FBS) and incubated overnight at 37°C under microaerobic conditions with moderate reciprocal shaking. Mtzr SS1 mutant was selected in medium containing 15 µg/mL of chloramphenicol and 20 µg/mL of kanamycin.19
Preparation of solutions containing LLA and LipoLLA
Stock solution containing 5 mg/mL LLA was prepared by dissolving LLA powder in DMSO. The experimental concentrations were achieved by diluting the stock solution of LLA with PBS or BHI growth medium containing 5% FBS. LipoLLA was prepared by a sonication and needle extrusion method as previously described.13 Specifically, a total of 4 mg of Egg PC, cholesterol, and LLA (5:1:4 weight ratio) were dissolved in 1 mL of chloroform. Then the chloroform was evaporated by blowing argon gas for 10 min and a thin lipid layer formed. The film was rehydrated by adding 2 mL PBS or BHI medium. The resulting lipid suspension was vortexed for 1 min, followed by sonication for 3 min in a bath sonicator (Fisher Scientific FS30D) to produce multilamellar vesicles (MLVs). Then the MLVs were sonicated using a Branson 450 sonifier with a Ti-probe at 20 W for 30 sec to produce small unilamellar vesicles (SUVs). The solution containing liposomes was obtained by extruding the SUVs through a 100 nm pore-sized polycarbonate membrane 11 times with a mini-extruder (Avanti polar lipids, Alabaster, AL). After removing the excess free LLA, possibly in the form of LLA micelles, from solution of LipoLLA using a Sephadex G75 column, the loading yield of LLA in the synthesized LipoLLA was determined using an Agilent 1100 series LC-MSD-Trap-SL high performance ion trap mass spectrometer (Agilent Technologies, Santa Clara, CA), equipped with an electrospray ionization source, following a previously published protocol.13 Finally, the solution containing LipoLLA was sterilized by filtration through a 0.22 µm filter unit (Millipore, Billerica, MA) prior to uses. The hydrodynamic size (diameter, nm) and surface zeta potential (mV) of LipoLLA were measured using the Malvern Zetasizer ZS (Malvern Instruments, UK).
Bactericidal activity against spiral form of H. pylori SS1
An overnight broth culture of H. pylori SS1 was centrifuged at 5000 × g for 10 min to obtain a bacterial pellet. The pellet was adjusted to an optical density at 600 nm (OD600) of 1.0, corresponding to approximately 1×108 colony forming units (CFU)/mL. 10 µL/well of bacterial suspension containing 1×106 CFU bacteria was added to a 96-well plate containing 190 µL BHI medium supplemented with 5% FBS along with various concentrations of LLA or LipoLLA. The plate was incubated at 37°C under microaerobic conditions on a reciprocal shaker. Several pilot experiments were performed at 2-, 5-, and 10-fold dilutions of LLA and LipoLLA at 0.5, 1, 6, 12, and 24 hr of incubation times to pinpoint desired concentration ranges and time points. After incubation for 30 min, a series of 10-fold dilutions of the bacterial suspension (1:10 to 1:105) was prepared, and 5 µL from each diluted sample was inoculated onto a Columbia agar plate supplemented with 5% laked horse blood. The plates were cultured in the incubator for 4 days before counting colonies.
Bactericidal activity against coccoid form of H. pylori SS1
The coccoid form of H. pylori SS1 was generated using a method described elsewhere.20 Briefly, tetracycline hydrochloride (15 µg/mL) was added to an overnight culture and cultivated for an additional 4 days. The culture was centrifuged at 600 × g for 5 min to pellet any remaining spiral form of the bacteria followed by the centrifugation of the supernatant at 11,000 × g for 5 min. The resulting pellet contained mostly the induced coccoid form of H. pylori SS1, as confirmed by observation under light microscope. Their viability was confirmed with Alamar blue dye assay.21 Briefly, 10µL Alamar blue dye (10% vol/vol) was added to each well and the plate was incubated at 37°C for 4 hr. The fluorescence intensity was measured by a fluorescence microplate reader (Synergy Mx, Biotek, Winooski, VT) with an excitation wavelength of 560 nm and an emission wavelength of 600 nm. A higher fluorescence intensity indicates a higher viability of the bacteria in the well. The density of coccoid H. pylori SS1 in solution was determined through an OD600 measurement.
To test the sensitivity of coccoid H. pylori SS1 to LLA and LipoLLA, 1×106 CFU coccoid H. pylori SS1 were added to 200 µL BHI containing amoxicillin (1, 10, or 20 µg/ml), LipoLLA (2 or 4 mg/mL, containing 400 or 800 µg/mL of LLA, respectively), and LLA (400 or 800 µg/ml). The samples were incubated in a 96 well plate at 37°C with reciprocal shaking for 0.5, 4, or 24 hr. After incubation, bacterial viability in the suspension was measured by Alarmar blue dye assay. As a comparison, the same test was carried out in parallel for spiral H. pylori SS1 under the same conditions. Finally, the bacterial viability of all samples was normalized to the control sample treated with PBS buffer.
LipoLLA– H. pylori SS1 fusion study
Florescence resonance energy transfer (FRET) was preformed to investigate the interaction mechanism between LipoLLA and H. pylori SS1. To prepare a FRET labeled LipoLLA, a fluorescent donor (C6NBD, 0.1 mol%) and a fluorescent acceptor (DMPE-RhB, 0.5 mol%) were simultaneously incorporated into the lipid bilayer membranes of the LipoLLA (0.5 mg/mL) by mixing the donor and the quencher with Egg PC, cholesterol, and LLA prior to the preparation of LipoLLA. Next, the resulting liposomes were diluted with PBS twofold into a solution containing 0.25 mg/mL of LipoLLA and 0.25 mL of the diluted solution was mixed with different amounts of H. pylori SS1. The total volume of the final solution (LipoLLA + H. pylori SS1) was adjusted to 1 mL, resulting bacterial concentrations of 8.0×107, 1.6×108, 2.4×108, 3.2×108, and 4.0×108 CFU/mL, respectively. After 10 min incubation at room temperature, samples were centrifuged at 13,500 rpm for 1 min to remove the excess amount of LipoLLA followed by re-suspending in 1 mL of PBS. Subsequently, emission spectra in the region of 500–700 nm were obtained by exciting the sample at 470 nm using a fluorescent spectrophotometer (BioTek Instrument, USA). The solution containing the same amount of LipoLLA without incubating with H. pylori SS1 was used as a negative control. Fluorescence emission of H. pylori SS1 itself at the corresponding concentrations was subtracted from of each sample before data plotting.
Scanning electron microscope (SEM) imaging of H. pylori SS1
The morphology of both spiral and coccoid forms of H. pylori SS1 treated with LLA and LipoLLA was examined with SEM. H. pylori SS1 was incubated with 200 µg/mL of LLA and 1 mg/mL of LipoLLA (containing 200 µg/mL LLA) at the same conditions as those for the bactericidal studies. At 30 min, the bacteria were harvested and visualized by an FEI XL30 Environmental SEM. To prepare SEM samples, untreated and treated bacteria were centrifuged to remove the supernatant, and the remaining pellet was fixed with 2% glutaraldehyde for 2 hr at room temperature. Post fixing, the sample was centrifuged to remove glutaraldehyde and resuspended in 100KL water. Then 5KL of bacterial suspension was dropped onto a polished silicon wafer and allowed to dry overnight in a biosafety cabinet. The samples were then coated with chromium before SEM imaging.
Bactericidal activity against antibiotics-resistant H. pylori strains
To test bactericidal activity of LLA and LipoLLA against antibiotic-resistant strains of H. pylori, including seven clinically isolated resistant strains (Shi470, Lithuania75, SouthAfrica7, India7, Gambia94/24, PeCan4, and SJM180) and metronidazole-resistant H. pylori SS1 (Mtzr SS1 mutant), 1×106 CFU of bacteria from an overnight culture was added to 200 µL BHI medium containing LLA and LipoLLA at various concentrations and cultured in a 96-well plate. The plate was incubated at 37°C under microaerobic condition with reciprocal shaking for 30 min. The bacterial suspension was then subjected to serial dilution and spotted onto Columbia agar plates supplemented with 5% laked horse blood. The plates were cultured in the incubator for 4 days before counting colonies.
Resistance acquisition of H. pylori SS1 upon treatments
To test the drug resistance development of H. pylori SS1 upon treatments, 1×107 CFU of bacteria in logarithm growth phase were added to 600 µL of BHI/5% FBS containing various concentrations of metronidazole, LLA, or LipoLLA in a 24 well plate. The plate was incubated at 37°C under microaerobic conditions with gentle shaking. After 24 hr, 100 µL sample from each well was collected and added to 500 µL fresh medium containing the same type of drug and drug concentration. Meanwhile, the bacteria from each treatment group were tested for sensitivity to the corresponding drug using the same protocol described in 2.4. This process was repeated for 10 days to observe possible drug resistance of H. pylori SS1.
Results and discussion
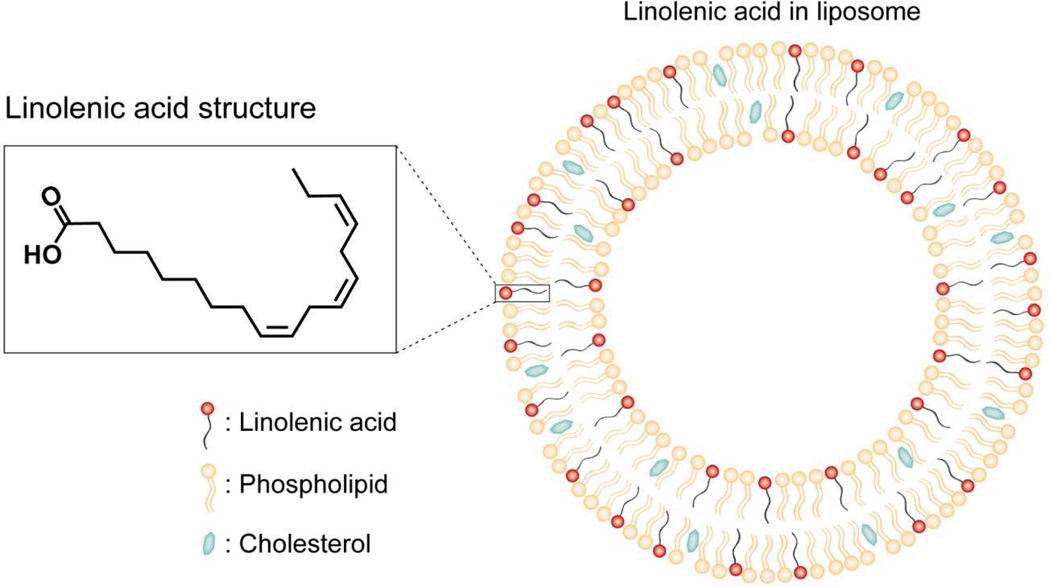
In this study, liposome was made of egg PC and cholesterol and its lipid bilayer structure was illustrated in Figure 1. Liposome offers unique physicochemical properties for delivering FFAs such as LLA for therapeutic applications. Particularly, amphiphilic LLA molecules can be readily entrapped in the hydrophobic lipid membranes of a liposome by mixing LLA with egg PC and cholesterol at desirable ratios prior to liposome preparation. As a result, the use of LipoLLA can overcome the poor solubility of LLA in aqueous solution. For antibacterial applications, the liposome formulations, as compared to other LLA formulations such as micelles or emulsions, can fuse with bacterial membranes and thus directly release the entrapped LLA molecules into bacterial membranes for efficient bactericidal activity.12, 13 The loading yield of LLA in LipoLLA formulations was determined by liquid chromatography-mass spectrometry following a procedure previously reported.13 Under our experimental conditions, 40% LLA in the initial mixture of Egg PC, cholesterol and LLA resulted in a final loading yield of approximately 20%. There was a loss of LLA during the experimental processes, including needle sonication, extrusion, and sterilization. A similar loss of FFA during liposome formulation was also observed in our previous study to encapsulate oleic acid into a liposomal formulation.13 LipoLLA formulated here has a hydrodynamic size (diameter) of 88 ± 3 nm, a polydispersity index of 0.17 ± 0.01, and a surface zeta potential of −78 ± 4 mV in deionized water, determined by dynamic light scattering measurements. It is expected that these sub-100 nm liposomes can readily fuse with bacterial membranes due to their high surface tension.
Figure 1.
A schematic drawing shows the molecular structure of LLA and the structure of LipoLLA composed of phospholipid, cholesterol and LLA.
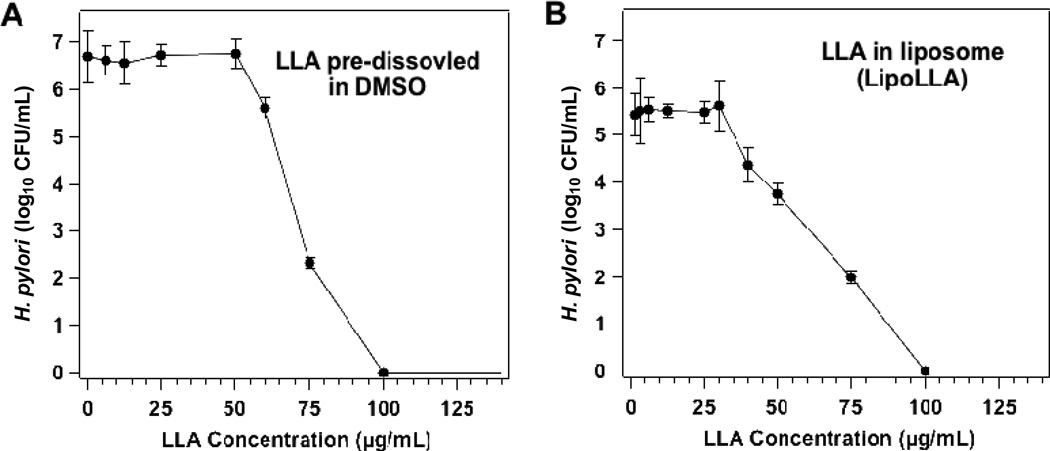
The bactericidal activity of LLA (dissolved in 5% DMSO) and LipoLLA was first tested against replicating form (also called spiral form) of H. pylori SS1 strain. This strain of bacteria was chosen because of its significance and wide applications in H. pylori research.22 When H. pylori SS1 was incubated for 30 min in broths containing various concentrations of LLA, the relationship between bacterial viability and LLA concentration was not linear (Figure 2A). Specifically, when LLA concentration was below 50 µg/mL, negligible bactericidal activity was observed. Further increasing LLA concentration above 50 µg/mL led to a sharp decrease in viable bacterial numbers. When LLA concentration reached 100 µg/mL, no viable bacteria were detected. For this study, we defined minimal bactericidal concentration (MBC) as the minimum concentration of the bactericidal agent required to kill 3 logs (99.9%) of the bacteria during a 30-min incubation. Therefore, the MBC value for LLA was determined to be 75 µg/mL (equivalent to 0.27 mM). A similar relationship between bacterial viability and LLA concentration was also observed when LipoLLA at various concentrations was added to bacterial cultures (Figure 2B). Specifically, bactericidal activity was negligible when LLA concentration was below 30 µg/mL. Above this concentration, a rapid decrease in viable bacteria number was observed. When LLA concentration was raised to 100 µg/mL, no viable bacteria were detected. From the relationship between bacterial viability and LLA concentration, the MBC value of LipoLLA was determined to be 67 µg/mL (equivalent to 0.22 mM), at which 99.9% of the bacteria were killed.
Figure 2.
In vitro bactericidal activity of (A) LLA and (B) LipoLLA at different drug concentrations against H. pylori SS1. All concentrations refer to LLA concentration, regardless of the formulation. LLA or LipoLLA was incubated with 5×106 CFU/mL H. pylori SS1 bacteria at 37°C under microaerobic conditions for 30 min followed by Columbia agar plate subculture.
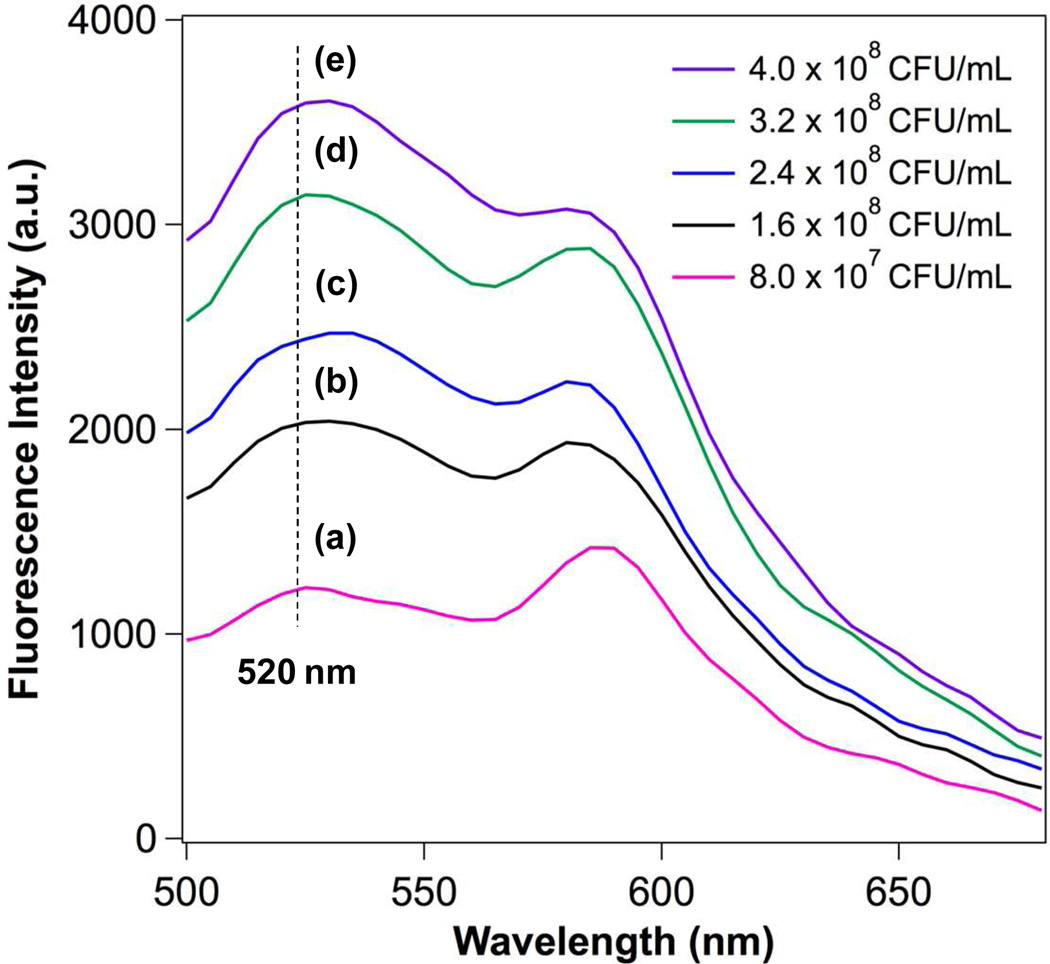
The observation of a sharp decrease in viable bacteria at the MBC values implies that the killing of H. pylori may involve a highly destructive mechanism such as lysis of the bacterial membranes. This is further supported by a killing time no longer than 30 min, relatively short when compared to 2.5 hr needed for H. pylori to complete a replication cycle.9 To better understand the interaction mechanism between LipoLLA and H. pylori bacteria at a molecular level, we labeled LipoLLA with a FRET-pair of fluorescence probes and monitored the FRET signal changes upon mixing with the bacteria at various concentrations. FRET is a sensitive technique that precisely detects the distance change between two subjects at the molecular level based on an energy transfer mechanism of two chromophores.23, 24 Herein, we incorporated a fluorescence donor (C6NBD: excitation/emission = 470 nm/520 nm) and a fluorescence acceptor (DMPE-RhB: excitation/emission = 550 nm/580 nm) into the lipid bilayer membranes of LipoLLA at a molar ratio of 1:5, at which the fluorescence emission from C6NBD was completely quenched by DMPE-RhB. We hypothesize that if the LipoLLA fuses with the bacterial membranes, the spread of the donor and acceptor within the bacterial membranes will alleviate or eliminate the FRET efficiency, resulting in a recovery of the donor florescence. To test this hypothesis, we mixed the FRET labeled LipoLLA with varying concentrations of H. pylori SS1, followed by removing the excess LipoLLA. The samples were then excited at the wavelength of 470 nm and the emission spectra at the range of 500–700 nm were obtained. As shown in Figure 3, the rise of the emission peak of C6NBD at 520 nm was detected when the concentration of H. pylori SS1 increased, indicating an increase of spatial separation between C6NBD and DMPE-RhB upon mixing LipoLLA with H. pylori bacteria. These results confirm that the interaction mechanism between LipoLLA and the bacteria was fusion as opposed to adsorption or aggregation.
Figure 3.
FRET measurements of the fusion between LipoLLA and H. pylori SS1. A fluorescent donor (C6NBD, 0.1 mol%) and a fluorescent acceptor (DMPE-RhB, 0.5 mol%) were concurrently incorporated into the lipid bilayer membranes of LipoLLA so that the acceptor completely quenched the fluorescence emission from the donor. The FRET-pair labeled LipoLLA was incubated with H. pylori at a concentration of a–e: 8.0×107, 1.6×108, 2.4×108, 3.2×108, and 4×108 CFU/mL for 10 min. After removing the excess LipoLLA, all samples were excited at 470 nm. A rise in emission intensity of C6NBD (donor) at 520 nm was observed with the increase of bacterial concentrations, indicating the occurrence of fusion between LipoLLA and H. pylori that caused the spatial separation of C6NBD and DMPE-RhB.
After having confirmed that the interaction mechanism between LipoLLA and H. pylori is fusion, we further tested the bactericidal activity of LLA and LipoLLA against the dormant form (also called coccoid form) of H. pylori SS1 and examined the effects of LLA and LipoLLA on the bacterial membrane morphology upon treatments. H. pylori bacteria adopt two major morphologies; spiral and coccoid forms. Despite the controversy regarding the vitality of non-culturable coccoid form of H. pylori, accumulating evidence suggests that H. pylori bacteria in coccoid form can play a critical role in disease transmission and subsequent treatment.25 Particularly, studies have found that H. pylori transmitted through oral–oral or oral–fecal routes may survive as coccoid forms.26 The coccoid form of the bacteria was also shown to contribute to the development of relapses following antimicrobial therapy in H. pylori infection.27 Furthermore, in our study, we induced the coccoid form of H. pylori by supplementing the culture medium with tetracycline hydrochloride, a method that has been shown to reduce CoA transferase activity and increase α-ketoglutarate oxidoreductase activity of the induced coccoid bacteria.20 Such changes in enzymatic activities involved in bacterial energy metabolism not only indicate that the bacteria remain viable during the transformation from spiral to coccoid form, but also suggest that target-specific antibiotics effective in inhibiting one form of the bacteria may not be effective in another. Hence, for successful therapy, it is essential to develop antimicrobials that can eliminate both forms of H. pylori.
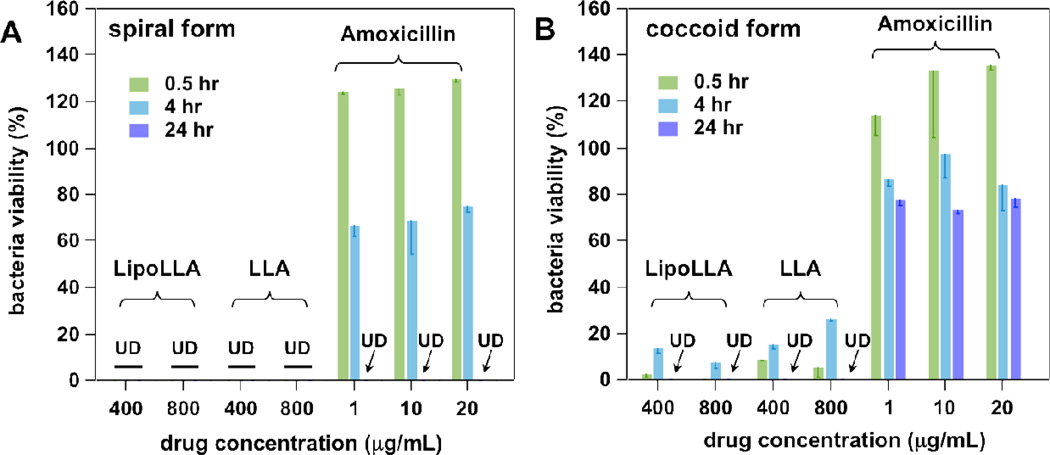
Next we examined the bactericidal activities of LLA and LipoLLA against both spiral and coccoid forms of H. pylori SS1, while using amoxicillin as a model anti-H. pylori antibiotic as a positive control (see Figure 4). We started with 400 µg/mL of LLA as this was the lowest concentration where complete killing of coccoid H. pylori was observed at 24 hr incubation time. At concentrations of 400 and 800 µg/mL (referring to LLA), LLA and LipoLLA completely killed H. pylori in spiral form at 30 min incubation time and coccoid form at 24 hr incubation time. Herein, 1 µg/mL of amoxicillin was selected as the lowest concentration, which is approximately the MIC value of amoxicillin for H. pylori in the spiral form.28, 29 As shown in Figure 3, at 0.5 hr incubation time, amoxicillin did not show any inhibition activity against either the spiral or coccoid form of H. pylori; while at 24 hr, amoxicillin killed the spiral but not the coccoid H. pylori. The lack of efficacy of amoxicillin in inhibiting the coccoid H. pylori but not the spiral form can be explained by its mechanism of action, which is to inhibit bacterial cell wall biosynthesis 30. As the coccoid form is “dormant”, the non-dividing bacteria will not be affected by amoxicillin. In contrast, the bactericidal activity of LLA results at least in part from the incorporation of LLA into bacterial membranes,10 and the liposome-bacterial membrane fusion would further enhance the antibacterial activity of LipoLLA.12 Such drug-bacterial membrane interactions are less dependent on the metabolic rate of the bacteria. Hence, when compared to amoxicillin, LLA and LipoLLA show less difference in efficacy when inhibiting spiral and coccoid forms of H. pylori. These data therefore provide evidence that LLA and LipoLLA are more efficient in killing both forms of H. pylori. in comparison to existing antibiotics.
Figure 4.
In vitro bactericidal activity of LipoLLA in comparison with LLA and Amoxicillin against (A) spiral form and (B) coccoid form of H. pylori SS1. Bacterial viability was tested by using Alarmar blue dye assay after an incubation period of 0.5, 4, or 24 hr. All concentrations of LLA and LipoLLA refer to LLA concentration, regardless of the formulation. Bacterial viability was normalized to the control sample treated with PBS buffer.
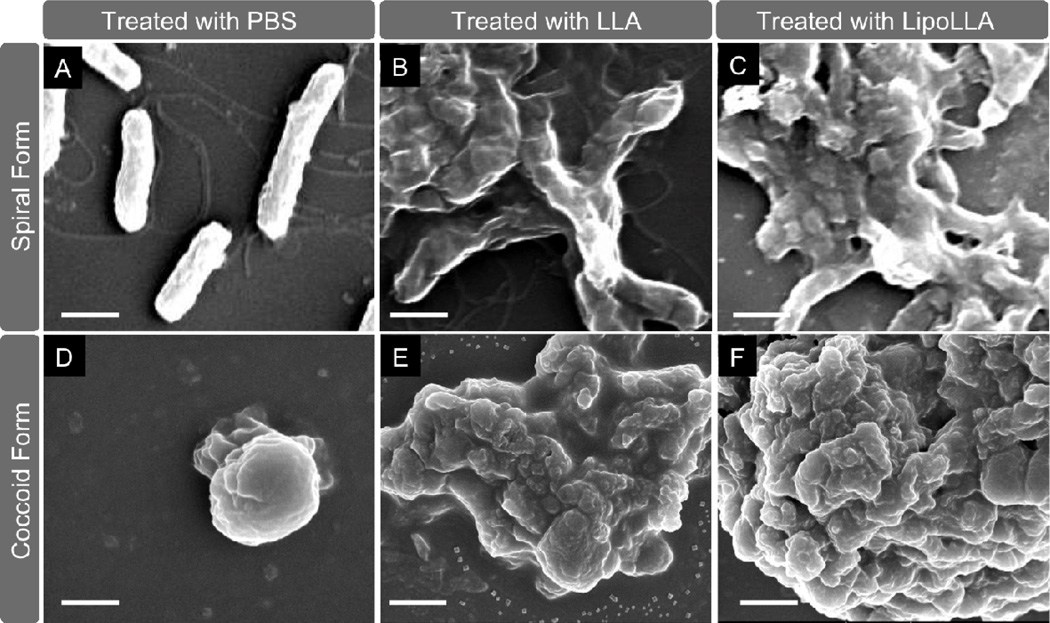
The morphology transformation of H. pylori from spiral form to coccoid form was examined using SEM, where the image showed typical spiral H. pylori cells with normal curved morphlogy and intact cell membranes (Figure 5A). A typical spiral H. pylori cell was 2 ~ 4 µm long and 0.5 ~ 0.8 µm wide with visible sheathed flagella. After the bacteria were treated with 400 µg/mL LLA for 30 min, complete killing of bacteria was confirmed by Alamar blue dye staining assay. Meanwhile, SEM image showed the bacterial morphology with a complete loss of the normal curved shape, disruption of protoplasmic cylinders, cell lysis, fragmentation of the bacterial cell membranes, and severe clustering (Figure 5B). Bacteria treated with LipoLLA under the same conditions had similar morphology as those treated with LLA (Figure 5C). Meanwhile, the SEM image of H. pylori in coccoid form showed predominantly non-colonized spherical shape with a diameter around 2 µm (Figure 5D). By using Alamar blue dye staining assay to measure viability, a concentration below 400 µg/mL of LLA (pre-dissolved in DMSO or in liposome formulation) was insufficient to completely kill coccoid form of H. pylori bacteria. In contrast, treatment with 400 µg/mL LLA for 24 hr led to complete killing of bacteria in coccoid form. SEM images showed that the morphology of dead coccoid form of H. pylori bacteria was similar to that of the dead spiral bacteria, where the bacterial membranes were completely disrupted and formed clusters (Figure 5E and F). In addition, the bacterial morphology had little discernible difference between the treatments with LLA or LipoLLA. As a control, the bacterial membrane morphology remained intact upon incubation with the same concentration of bare liposomes (without LLA).
Figure 5.
Morphology of H. pylori SS1 bacteria in their spiral form (A–C) and coccoid form (D–F) exposed to different treatments. In (A) and (D), bacteria were treated with PBS; in (B) and (E), bacteria were treated with LLA pre-dissolved in DMSO; in (C) and (F), bacteria were treated with LipoLLA. In all experiments, initial concentration of the bacteria was 5×106 CFU/mL and drug concentration was 200 µg/mL (referring to LLA). All samples were treated for 30 min before glutaralderhyde fixation. The scale bar in the image represents 1 µm.
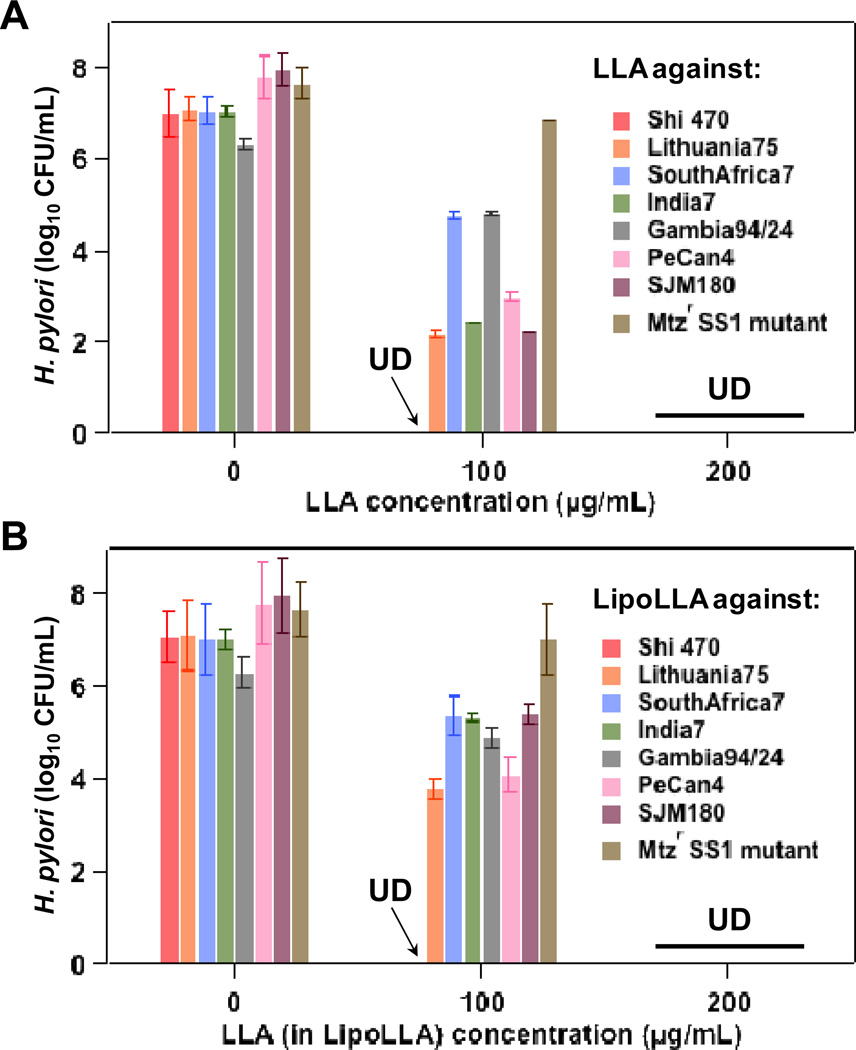
To further evaluate the bactericidal activity of LLA and LipoLLA against H. pylori, we next examined their antimicrobial efficacy against a series of clinical isolates of H. pylori, including Shi470, SouthAfrica7, India7, Gambia94/24, PeCan4, Lithuania75 and SJM180. These clinical isolates are normally resistant to existing antibiotics such as metronidazole. In addition, we also developed a metronidazole-resistant strain of H. pylori SS1 (Mtzr SS1 mutant) 19 to be included into this test. All of these antibiotic-resistant H. pylori strains were then tested for their susceptibility to LLA and LipoLLA. As shown in Figure 6, when LLA concentration was 100 µg/mL, clinical isolate Shi470 was completely killed while initial inhibition was observed on other strains. When LLA concentration was raised to 200 µg/mL, all strains, regardless of their resistance status to metronidazole, were completely killed by both LLA and LipoLLA. The non-selective bactericidal activity of LLA and LipoLLA against a broad range of H. pylori strains are of great value in developing new treatments for H. pylori infection.
Figure 6.
In vitro bactericidal activity of (A) LLA and (B) LipoLLA against various H. pylori clinical isolates and a metronidazole-resistant strain of H. pylori SS1 (Mtzr SS1 mutant). In all experiments, 5×106 CFU/mL bacteria were incubated with 0, 100, or 200 µg/mL LLA, either as free LLA pre-dissolved in DMSO or in liposome formulation (LipoLLA). Samples were incubated at 37°C under microaerobic conditions for 30 min before serial dilution followed by bacterial colony enumeration on Columbia agar plates.
In addition to the efficacy studies conducted to evaluate bactericidal activities of LLA and LipoLLA against H. pylori SS1, in both spiral and coccoid forms, and the clinical isolates of H. pylori strains, we finally investigated the potential resistance acquisition of H. pylori bacteria under the treatments of LLA and LipoLLA. In the study, we used H. pylori SS1 as an example to examine the development of drug resistance when the bacteria were cultured with various sub-bactericidal concentrations of LLA or LipoLLA over a span of 10 days. Here, metronidazole, an anti-H. pylori antibiotic in clinic use, was chosen to serve as a positive control. As shown in Table 1, H. pylori treated with 0.5 or 1 µg/mL metronidazole acquired resistance to the antibiotic on day 3, and by day 4 the bacteria in all concentration groups had developed resistance to metronidazole. In contrast, bacteria treated with LLA at concentrations of 18.8 and 28.2 µg/mL did not acquire drug resistance. However, when the LLA concentration was raised to 37.6 µg/mL (half MBC value of LLA against H. pylori SS1), drug resistance was established on day 3. In contrast to both metronidazole and LLA, bacteria treated with LipoLLA in all three different concentration groups including 16.8 µg/mL, 22.4 µg/mL, and 33.6 µg/mL (half MBC value of LipoLLA against H. pylori SS1), did not acquire drug resistance during the 10-day period of studies.
Table 1.
Resistance development of H. pylori SS1 upon incubation with different levels of metronidazole, LLA and LipoLLA over a span of 10 days. The highest concentrations selected in LLA group (37.6 µg/mL) and LipoLLA group (168 µg/mL, containing 33.6 µg/mL of LLA) were half of their MBC values, respectively. In the metronidazole group, 1 µg/mL is approximately 1/8 of its reported MIC value. In the table, ‘−’ denotes ‘non-resistant’ and ‘+’ denotes ‘resistant’.
| Treatment | LLA Concentration (µg/mL) |
Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metronidazole | 0.25 | − | − | − | + | + | + | + | + | + | + |
| 0.5 | − | − | + | + | + | + | + | + | + | + | |
| 1 | − | − | + | + | + | + | + | + | + | + | |
| LLA (pre-dissolved in DMSO) |
18.8 | − | − | − | − | − | − | − | − | − | − |
| 28.2 | − | − | − | − | − | − | − | − | − | − | |
| 37.6 | − | − | + | + | + | + | + | + | + | + | |
| LipoLLA | 16.8 | − | − | − | − | − | − | − | − | − | − |
| 22.4 | − | − | − | − | − | − | − | − | − | − | |
| 33.6 | − | − | − | − | − | − | − | − | − | − | |
The rapid resistance development of H. pylori under the treatment of metronidazole is likely attributed to the fast adaptation of bacteria to target-specific antibiotics through various mechanisms including decreased drug uptake, increased efflux, decreased drug activation, mutations in biological target, increased oxygen scavenging capabilities, and enhanced activity of DNA repair enzymes.31, 32 Similar to metronidazole, FFAs have also been hypothesized to inhibit bacteria growth through mechanisms that alternate cell survival signaling pathways. For example, it has been suggested that FFAs may enter the cells and inhibit bacterial growth by creating an acidic environment in cytoplasm that damages pH-sensitive intracellular enzyme activities or amino acid transports.33 FFAs have also been speculated to participate in chemical processes that generate toxic lipid peroxides, which in turn cause antibacterial activities.34 These mechanisms are target-specific. Therefore, treated with FFAs, H. pylori may still evolve and develop drug resistance. However, FFAs can also inhibit bacteria growth by incorporating themselves into bacterial membranes, disrupting the structural integrity of the cell membrane, and inducing abnormal cell forms and cell lysis.9, 10 Such antibacterial activities are physical, broad, and unspecific, which may explain a lower frequency of resistance development observed when bacteria are treated with FFAs. For instance, H. pylori acquired much less resistance against LLA than metronidazole as shown in Table 1.
Compared to free FFAs, liposomal formulation confines FFA molecules within the lipid bilayer. During the process of liposome-bacterial membrane fusion, FFA molecules are exclusively distributed into the bacterial membranes, limiting their interaction with cells’ intracellular machineries. As liposomal formulation of FFAs limits chemical alterations on cell survival pathways but promotes physical-structural disruption of cell membranes, an even lower rate of resistant was developed when bacteria were treated with liposomal FFAs. This may explain why LipoLLA did not provoke drug resistance of H. pylori during the treatment period.
Conclusions
In this study, we demonstrated the bactericidal activities of LipoLLA and free LLA in treating H. pylori infection. Using H. pylori SS1 as a model strain, we showed that LipoLLA and free LLA were effective in killing both spiral and coccoid forms of the bacteria. Using a metronidazole-resistant strain of H. pylori and seven clinical isolates, we further demonstrated that LipoLLA eradicated all strains of the bacteria regardless of their resistance status to metronidazole. Furthermore, under our experimental conditions, bacteria did not develop noticeable drug resistance when cultured with LipoLLA at various sub-bactericidal concentrations, whereas they acquired resistance to both metronidazole and free LLA at comparable concentrations. While both LipoLLA and free LLA were tested in parallel for their bactericidal activities against H. pylori in this study, we expect that LipoLLA holds greater potential to be translated as a new therapeutic option to H. pylori infection. This is because of the poor water-solubility of LLA, which needs to be pre-dissolved in organic solvents prior to uses. In addition, using LipoLLA to treat H. pyloriinfection is expected to less likely induce bacterial drug resistance because of the liposome-bacterial membrane fusion mechanism, through which the drug molecules are directly introduced to the bacterial membranes rather than intracellular compartments. Moreover, our approach of using LLA-loaded liposomal nanoformulation to treat H. pylori infection bridges the accumulated knowledge of antimicrobial activities of FFAs and the fast emerging field of nanotechnology. As a result, LipoLLA developed in this study would allow us to implement numerous nanotechnology-based strategies, particularly those established to overcome mucosal barrier and gastric conditions,35–39 for more advanced drug delivery to treat H. pylori infection.
Acknowledgement
We are very grateful to Drs. Douglas Berg and Dangeruta Kersulyte for providing H. pylori strains and for sequencing their genomes. This work is supported by the University of California, San Diego (faculty startup funds) and the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number R01DK095168.
References
- 1.Hatakeyama M, Brzozowski T. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2006;11:14–20. doi: 10.1111/j.1478-405X.2006.00424.x. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez CA, Agudo A. Carcinogenesis, prevention and early detection of gastric cancer: Where we are and where we should go. Int. J. Cancer. 2012;130:745–753. doi: 10.1002/ijc.26430. [DOI] [PubMed] [Google Scholar]
- 3.Gisbert JP, Pajares JM. Treatment of Helicobacter pylori infection: the past and the future. Eur. J. Intern. Med. 2010;21:357–359. doi: 10.1016/j.ejim.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Paoluzi OA, Visconti E, Andrei F, Tosti C, Lionetti R, Grasso E, Ranaldi R, Stroppa I, Pallone F. Ten and eight-day sequential therapy in comparison to standard triple therapy for eradicating Helicobacter pylori infection: a randomized controlled study on efficacy and tolerability. J. Clin. Gastroenterol. 2010;44:261–266. doi: 10.1097/MCG.0b013e3181acebef. [DOI] [PubMed] [Google Scholar]
- 5.Yamaoka Y, Graham DY, Lu H. Should triple therapy for Helicobacter pylori infection be abandoned as no longer effective? US Gastroenterology. 2008;4:65–67. [Google Scholar]
- 6.Megraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Njume C, Afolayan AJ, Ndip RN. An overview of antimicrobial resistance and the future of medicinal plants in the treatment of Helicobacter pylori infections. Afr. J. Pharm. Pharmacol. 2009;3:685–699. [Google Scholar]
- 8.O'Connor A, Gisbert JP, McNamara D, O'Morain C. Treatment of Helicobacter pylori Infection 2011. Helicobacter. 2011;16:53–58. doi: 10.1111/j.1523-5378.2011.00881.x. [DOI] [PubMed] [Google Scholar]
- 9.Desbois AP, Smith VJ. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010;85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 10.Petschow BW, Batema RP, Ford LL. Susceptibility of Helicobacter pylori to bactericidal properties of medium-chain monoglycerides and free fatty acids. Antimicrob. Agents Chemother. 1996;40:302–306. doi: 10.1128/aac.40.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prajapati HN, Dalrymple DM, Serajuddin ATM. A comparative evaluation of mono-, di- and triglyceride of medium chain fatty acids by lipid/surfactant/water phase diagram, solubility determination and dispersion testing for application in pharmaceutical dosage form development. Pharm. Res. 2012;29:285–305. doi: 10.1007/s11095-011-0541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang D, Pornpattananangkul D, Nakatsuji T, Chan M, Carson D, Huang CM, Zhang L. The antimicrobial activity of liposomal lauric acids against Propionibacterium acnes. Biomaterials. 2009;30:6035–6040. doi: 10.1016/j.biomaterials.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CM, Chen CH, Pornpattananangkul D, Zhang L, Chan M, Hsieh MF. Eradication of drug resistant Staphylococcus aureus by liposomal oleic acids. Biomaterials. 2011;32:214–221. doi: 10.1016/j.biomaterials.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh AJ, Kwon YJ. Nanoantibiotics: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release. 2011;156:128–145. doi: 10.1016/j.jconrel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Bhardwaj V, Hariharan S, Bala I, Lamprecht A, Kumar N, Panchagnula R, Kumar MNVR. Pharmaceutical aspects of polymeric nanoparticles for oral drug delivery. J. Biomed. Nanotech. 2005;1:235–258. [Google Scholar]
- 16.Roger E, Lagarce F, Garcion E, Benoit J-P. Biopharmaceutical parameters to consider in order to alter the fate of nanocarriers after oral delivery. Nanomedicine. 2010;5:287–306. doi: 10.2217/nnm.09.110. [DOI] [PubMed] [Google Scholar]
- 17.Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 18.Obonyo M, Sabet M, Cole SP, Ebmeyer J, Uematsu S, Akira S, Guiney DG. Deficiencies of myeloid differentiation factor 88, toll-like receptor 2 (TLR2), or TLR4 produce specific defects in macrophage cytokine secretion induced by Helicobacter pylori. Infect. Immun. 2007;75:2408–2414. doi: 10.1128/IAI.01794-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong JY, Berg DE. Mouse-colonizing Helicobacter pylori SS1 is unusually susceptible to metronidazole due to two complementary reductase activities. Antimicrob. Agents Chemother. 2000;44:3127–3132. doi: 10.1128/aac.44.11.3127-3132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsugawa H, Suzuki H, Nakagawa I, Nishizawa T, Saito Y, Suematsu M, Hibi T. Alpha-ketoglutarate oxidoreductase, an essential salvage enzyme of energy metabolism, in coccoid form of Helicobacter pylori. Biochem. Biophys. Res. Commun. 2008;376:46–51. doi: 10.1016/j.bbrc.2008.08.078. [DOI] [PubMed] [Google Scholar]
- 21.Al-Nasiry S, Geusens N, Hanssens M, Luyten C, Pijnenborg R. The use of Alamar Blue assay for quantitative analysis of viability, migration and invasion of choriocarcinoma cells. Hum. Reprod. 2007;22:1304–1309. doi: 10.1093/humrep/dem011. [DOI] [PubMed] [Google Scholar]
- 22.Thompson LJ, Danon SJ, Wilson JE, O'Rourke JL, Salama NR, Falkow S, Mitchell H, Lee A. Chronic Helicobacter pylori infection with Syndey Strain 1 and a newly identified mouse-adapted strain (Sydney Strain 2000) in C57BL/6 and BALB/c mice. Infect. Immun. 2004;72:4668–4679. doi: 10.1128/IAI.72.8.4668-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 24.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 25.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen LP, Rasmussen L. Helicobacter pylori - coccoid forms and biofilm formation. FEMS Immunol. Med. Microbiol. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 27.She FF, Su DH, Lin JY, Zhou LY. Virulence and potential pathogenicity of coccoid Helicobacter pylori induced by antibiotics. World J. Gastroenterol. 2001;7:254–258. doi: 10.3748/wjg.v7.i2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Megraud F, Trimoulet P, Lamouliatte H, Boyanova L. Bactericidal effect of amoxicillin on Helicobacter pylori in an in vitro model using epithelial cells. Antimicrob. Agents Chemother. 1991;35:869–872. doi: 10.1128/aac.35.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canton R, de Argila CM, de Rafael L, Baquero F. Antimicrobial resistance in Helicobacter pylori. Rev. Med. Microbiol. 2001;12:47–61. [Google Scholar]
- 30.Co E-MA, Schiller NL. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 2006;50:4174–4176. doi: 10.1128/AAC.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Zwet AA, Thijs JC, de Graaf B. Explanations for high rates of eradication with triple therapy using metronidazole in patients harboring metronidazole-resistant Helicobacter pylori strains. Antimicrob. Agents Chemother. 1995;39:250–252. doi: 10.1128/aac.39.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weel JF, van der Hulst RW, Gerrits Y, Tytgat GN, van der Ende A, Dankert J. Heterogeneity in susceptibility to metronidazole among Helicobacter pylori isolates from patients with gastritis or peptic ulcer disease. J. Clin. Microbiol. 1996;34:2158–2162. doi: 10.1128/jcm.34.9.2158-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun CQ, O'Connor CJ, Roberton AM. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2003;36:9–17. doi: 10.1016/S0928-8244(03)00008-7. [DOI] [PubMed] [Google Scholar]
- 34.Knapp HR, Melly MA. Bactericidal effects of polyunsaturated fatty acids. J. Infect. Dis. 1986;154:84–94. doi: 10.1093/infdis/154.1.84. [DOI] [PubMed] [Google Scholar]
- 35.Lai SK, Wang Y-Y, Hanes J. Mucus-penetrating nanoparticles for drug and gene delivery to mucosal tissues. Adv. Drug Deliv. Rev. 2009;61:158–171. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, Chan JM, Farokhzad OC. pH-responsive nanoparticles for drug delivery. Mol. Pharm. 2010;7:1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Lai SK, Wang YY, Zhong WX, Happe C, Zhang M, Fu J, Hanes J. Biodegradable nanoparticles composed entirely of safe materials that rapidly penetrate human mucus. Angew. Chem. Int. Ed. 2011;50:2597–2600. doi: 10.1002/anie.201006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai SK, Wang YY, Hida K, Cone R, Hanes J. Nanoparticles reveal that human cervicovaginal mucus is riddled with pores larger than viruses. Proc. Natl. Acad. Sci. USA. 2010;107:598–603. doi: 10.1073/pnas.0911748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang BC, Dawson M, Lai SK, Wang YY, Suk JS, Yang M, Zeitlin P, Boyle MP, Fu J, Hanes J. Biodegradable polymer nanoparticles that rapidly penetrate the human mucus barrier. Proc. Natl. Acad. Sci. USA. 2009;106:19268–19273. doi: 10.1073/pnas.0905998106. [DOI] [PMC free article] [PubMed] [Google Scholar]