Abstract
Inflammation has long been implicated in secondary tissue damage after spinal cord injury (SCI). Our previous studies of inflammatory gene expression in rats after SCI revealed two temporally correlated clusters: the first was expressed early after injury and the second was up-regulated later, with peak expression at 1–2 weeks and persistent up-regulation through 6 months. To further address the role of inflammation after SCI, we examined inflammatory genes in a second species, mice, through 28 days after SCI. Using anchor gene clustering analysis, we found similar expression patterns for both the acute and chronic gene clusters previously identified after rat SCI. The acute group returned to normal expression levels by 7 days post-injury. The chronic group, which included C1qB, p22phox and galectin-3, showed peak expression at 7 days and remained up-regulated through 28 days. Immunohistochemistry and western blot analysis showed that the protein expression of these genes was consistent with the mRNA expression. Further exploration of the role of one of these genes, galectin-3, suggests that galectin-3 may contribute to secondary injury. In summary, our findings extend our prior gene profiling data by demonstrating the chronic expression of a cluster of microglial associated inflammatory genes after SCI in mice. Moreover, by demonstrating that inhibition of one such factor improves recovery, the findings suggest that such chronic up-regulation of inflammatory processes may contribute to secondary tissue damage after SCI, and that there may be a broader therapeutic window for neuroprotection than generally accepted.
Keywords: inflammation, microarray, microglia, motor function, NADPH oxidase, spinal cord contusion
Introduction
Traumatic spinal cord injury (SCI) induces tissue damage and cell death, leading to inflammation, including activation of local microglia and invasion of blood-borne immune cells(Beattie, 2004; Demjen et al., 2004; Dusart and Schwab, 1994; Fitch et al., 1999; Fleming et al., 2006; Popovich et al., 2002; Stirling et al., 2004; Teng et al., 2004). Along with this cellular activation and infiltration, there is a significant increase in inflammatory gene and protein expression. Microarray analysis is a useful tool to explore the gene expression alterations occurring after SCI, and several studies have demonstrated its utility for discovering novel genes in the spinal cord after trauma (Aimone et al., 2004; Carmel et al., 2001; Di Giovanni et al., 2003; Ghasemlou et al., 2010; Gris et al., 2003; Hashimoto et al., 2004; Tachibana et al., 2002; Zhang et al., 2004). In the rat SCI contusion model, we have shown that genes associated with inflammation, including those expressed primarily by microglia/macrophages, are strongly up-regulated immediately after injury and remain up-regulated for months (Byrnes et al., 2006; Byrnes et al., 2011).
Our earlier work investigated the delayed up-regulation of selected inflammatory genes, including C1qB, galectin-3 and p22PHOX, peaking at 7–14 days after SCI in the rat contusion model (Byrnes et al., 2006) and persisting to at least 6 months (Byrnes et al., 2011). Whether a similar pattern occurs in the mouse contusion SCI model is unknown, as there are substantial pathobiological differences in the spinal cord’s response trauma between the two species. For example, SCI in mice causes accumulation of a dense fibrous connective tissue at the lesion site (Bilgen et al., 2007), whereas rats develop cystic cavities, extending rostral and caudal to the initial injury site following SCI (Tang et al., 2003). However, similar inflammatory and astroglial activity resulting in glial scar formation and secondary tissue damage around the lesion site have been observed in rats and mice (Byrnes et al., 2010; Silver and Miller, 2004; Tang et al., 2003), suggestive of similarities in gene expression patterns across the two species.
Moreover, it is unclear if the chronically up-regulated genes have a functional significance in the pathophysiology of SCI. For example, galectin-3 is known to be involved in inflammatory responses (Hsu et al., 2000; Jiang et al., 2009). Its inhibition with the carbohydrate binding blocker modified citrus pectin (MCP) can reduce several markers of inflammation, including nitric oxide secretion and COX2 expression (Chen et al., 2006). However, to date, no study has evaluated the potential of galectin-3 inhibition on functional recovery after SCI.
The goal of this work was to examine the inflammatory gene expression profile after a moderate contusion SCI in the mouse and to explore the functional significance of one of the chronically expressed genes, galectin-3. Using microarray data from 30 minutes to 28 days post-injury, we now show that the acute and sub-acute expression profiles for microglial associated inflammatory genes parallel previous reports in the rat. In addition, we show that these genes may have a detrimental effect on spinal cord recovery, as inhibition of one of them, galectin-3, increases white matter sparing and improves BMS scores in mice after moderate SCI. These data indicate that chronic inflammation contributes to tissue damage after mouse contusion SCI.
2. Results
2.1 Evidence of chronic inflammatory gene expression after SCI
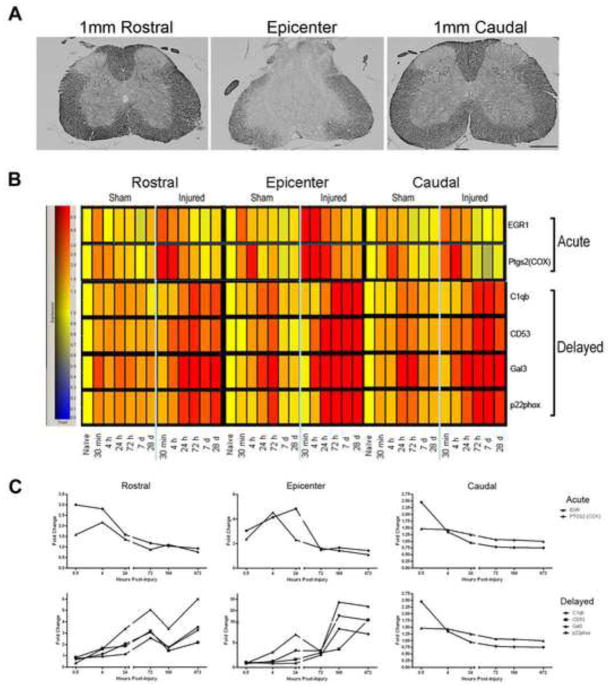
In order to confirm the extent of injury, sections from the epicenter as well as 1mm rostral and 1mm caudal to the injury site were stained using eriochrome cyanine R at 28 days after injury (Figure 1A). The spared white matter is shown in darker gray and is clearly reduced at the lesion epicenter.
Figure 1.
Inflammation-related gene expression in spinal cord injured mouse tissue. (A) Eriochrome cyanine R stained spinal cord tissue from the epicenter, 1mm rostral, and 1mm caudal to the injury site 28 days after injury demonstrate the severity of the moderate injury (bar = 500μm). Microarray data results are shown in a heat map (B) and graphical form (C). Time post-injury and group/location are represented on the y-axis; specific genes are indicated on the x-axis. Gene expression of injured and sham-injured samples is shown for the lesion epicenter, rostral and caudal after moderate injury. Note the increase in expression at 30 min post-injury in the acute-expression cluster, and the more delayed increase in intensity in the delayed-expression cluster compared to that in the sham-injured group. Cool colors represent decreased expression and warmer colors indicate higher expression relative to naïve controls (yellow). These same genes are graphed in (C) to demonstrate the fold expression change over naïve from 30 minutes to 28 days post-injury. Note the similar pattern of expression within the group as well as between regions (epicenter, rostral, caudal). Note also that expression of genes at the epicenter in the delayed-expression cluster remained up-regulated (above a 2 fold increase) through 28 days post-injury.
Two gene clusters were identified in the microarray analysis: an acute and a delayed group (Fig. 1B, C). Anchor gene analysis revealed 4 genes that had expression profiles that were temporally correlated at a level of 0.99 within the acute group (Table 1). This cluster was up-regulated between 30 minutes and 24 hours post-injury. Within the delayed expression group, the expression of 64 genes was temporally correlated at a level of 0.99 with the anchors C1qB and galectin-3 (Table 1). The delayed cluster, which includes the genes C1qB, CD53, galectin-3, and p22phox, demonstrated up-regulation at 72 hours (3 days) with sustained increases through 672 hrs (28 days). These expression profiles were consistent at both the epicenter and, to a lesser extent, rostral and caudal to the epicenter.
Table 1.
Gene clusters using the Ptgs2, C1qB and Lgals3 anchors (bolded), with a correlation of at least 0.99.
| Common Name | Genebank Number | Description |
|---|---|---|
| Ptgs2 | M94967 | prostaglandin-endoperoxide synthase 2 |
| Il1b | BC011437 | interleukin 1 beta |
| Cxcl2 | NM_009140 | chemokine (C-X-C motif) ligand 2 |
| Cxcl1 | NM_008176 | chemokine (C-X-C motif) ligand 1 |
| EGR1 | NM_007913 | Early growth response 1 |
| C1qb | NM_009777 | complement component 1, q subcomponent, beta polypeptide |
| Laptm5 | BB264849 | lysosomal-associated protein transmembrane 5 |
| Slamf9 | NM_029612 | SLAM family member 9 |
| C1qg | NM_007574 | complement component 1, q subcomponent, gamma polypeptide |
| Ctsh | NM_007801 | cathepsin H |
| Ppgb | NM_008906 | protective protein for beta-galactosidase |
| C1qa | NM_007572 | complement component 1, q subcomponent, alpha polypeptide |
| Igsf7 | AF251705 | immunoglobulin superfamily, member 7 |
| Lair1 | AK017222 | DNA segment, Chr 7, Brigham & Women’s Genetics 0421 expressed |
| Hexb | NM_010422 | hexosaminidase B |
| Itgb5 | NM_010580 | integrin beta 5 |
| Ctsz | NM_022325 | |
| Gns | BB445684 | glucosamine (N-acetyl)-6-sulfatase |
| Unc93b | BC018388 | unc-93 homolog B (C. elegans) |
| Abca1 | BB144704 | ATP-binding cassette, sub-family A (ABC1), member 1 |
| Mpeg1 | L20315 | macrophage expressed gene 1 |
| Tyrobp | NM_011662 | TYRO protein tyrosine kinase binding protein |
| Creg | BC027426 | cellular repressor of E1A-stimulated genes |
| Ctsd | NM_009983 | cathepsin D |
| Naglu | NM_013792 | alpha-N-acetyl glucosaminidase (Sanfilippodisease IIIB) |
| Grn | M86736 | Granulin |
| Lyn | M64608 | Yamaguchi sarcoma viral (v-yes-1) oncogenehomolog |
| Pik3ap1 | BI684288 | phosphoinositide-3-kinase adaptor protein 1 |
| C3ar1 | NM_009779 | complement component 3a receptor 1 |
| Fcgr2b | M14216 | Fc receptor, IgG, low affinity IIb |
| Clecsf5 | NM_021364 | C-type (calcium dependent, carbohydrate-recognition domain) lectin, superfamily member 5 |
| Lcp2 | BC006948 | lymphocyte cytosolic protein 2 |
| Lgals3 | X16834 | lectin, galactose binding, soluble 3 |
| Fer1l3 | BI555209 | fer-1-like 3, myoferlin (C. elegans) |
| Rgs19 | BC003838 | regulator of G-protein signaling 19 |
| Sh3glb1 | AV005520 | SH3-domain GRB2-like B1 (endophilin) |
| Cln5 | AV315220 | ceroid-lipofuscinosis, neuronal 5 |
| Lcp1 | NM_008879 | lymphocyte cytosolic protein 1 |
| Gnpda1 | NM_011937 | glucosamine-6-phosphate deaminase 1 |
| Syngr2 | BC004829 | synaptogyrin 2 |
| Asc | BG084230 | apoptosis-associated speck-like protein containing a CARD |
| P2ry6 | BC027331 | Pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| Gcnt1 | AK017462 | glucosaminyl (N-acetyl) transferase 1, core 2 |
| Capg | NM_007599 | capping protein (actin filament), gelsolin-like |
| Ly96 | NM_016923 | lymphocyte antigen 96 |
| Fes | BG867327 | feline sarcoma oncogene |
| B4galt1 | NM_022305 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| Cyba | AK018713 | cytochrome b-245, alpha polypeptide |
| Il10rb | NM_008349 | interleukin 10 receptor, beta |
| Fcer1g | NM_010185 | Fc receptor, IgE, high affinity I, gamma polypeptide |
| Pscd4 | AK010908 | pleckstrin homology, Sec7 and coiled/coil domains 4 |
| Mafb | AW412521 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| Apobec1 | BC003792 | Apolipoprotein B editing complex 1 |
| Cd68 | BC021637 | CD68 antigen |
| Ifi203 | AI607873 | interferon-activatable protein |
| Gngt2 | AK010554 | guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| Apbb1ip | BC023110 | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| Kdt1 | U13371 | kidney cell line derived transcript 1 |
| Myo1f | AK021181 | myosin IF |
| Slc11a1 | NM_013612 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| Lyzs | AW208566 | Lysozyme |
| Tcirg1 | NM_016921 | T-cell, immune regulator 1 |
| Cd53 | NM_007651 | CD53 antigen |
| Slc7a7 | NM_011405 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| Hcph | NM_013545 | hemopoietic cell phosphatase |
| Inpp5d | U39203 | inositol polyphosphate-5-phosphatase D |
| Anxa4 | NM_013471 | annexin A4 |
| Guca1a | NM_008189 | guanylatecyclase activator 1a (retina) |
| Cklfsf3 | NM_024217 | chemokine-like factor super family 3 |
| Tgfbr2 | BG793483 | transforming growth factor, beta receptor II |
| Igsf6 | NM_030691 | immunoglobulin superfamily, member 6 |
2.2 Chronic inflammatory protein expression confirmation
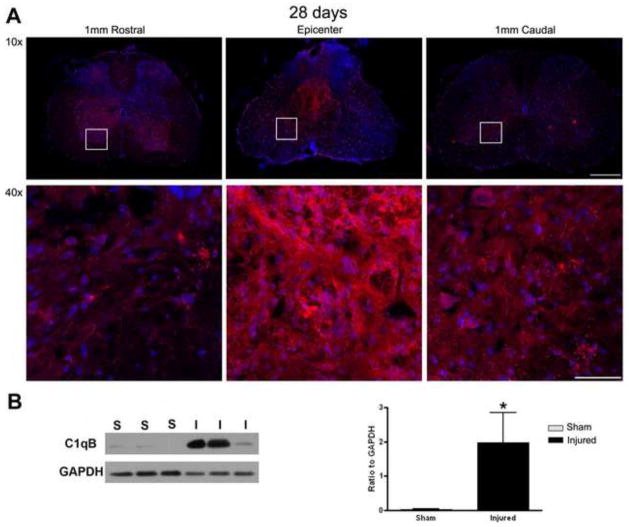
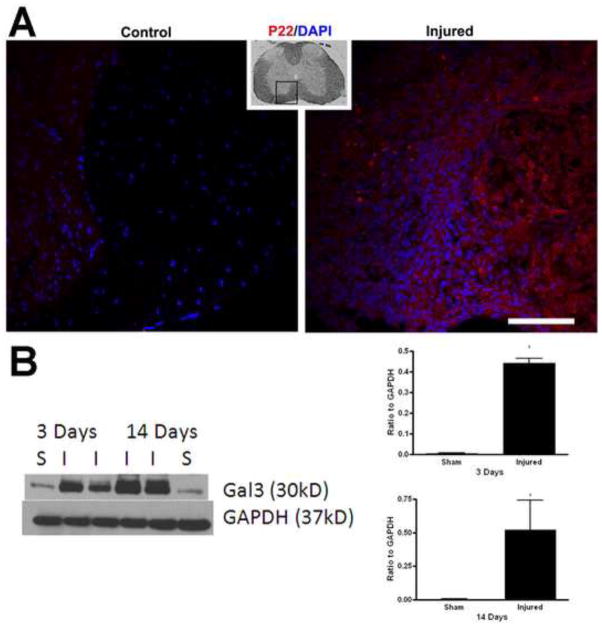
In order to confirm the gene expression data, we investigated the protein expression of a selection of genes. C1qB protein was expressed at the epicenter as well as 1mm caudal and rostral to the injury site at 28 days after injury (Fig. 2A). Western blot also showed a significant increase of this protein in injured spinal cord as compared to sham at 28 days after injury (Fig. 2B). The protein expression level was also evaluated for p22phox at 28 days after injury, as shown by immunohistochemistry (Fig. 3A). Finally, western blot analysis demonstrated that galectin-3 protein was significantly up-regulated as early as 3 days post-injury (Fig. 3B) which continued through at least 14 days post-injury (Fig. 3B).
Figure 2.
Confirmation of C1qB expression at the protein level at 28 days post-injury. (A) Sections from the epicenter, 1mm rostral, and 1mm caudal to the injury site were stained using C1qB antibody (red) and DAPI (blue) and demonstrate marked C1qB immunolabeling, particularly in the epicenter region. White boxes represent areas of enlarged images in 40X. 10X images bar= 500μm. 40X images bar= 100μm. Western blot analysis was also performed on sham and injured spinal cords. (B) Data were normalized to GAPDH expression and demonstrate a significant increase in C1qB protein. *p<0.05, Student’s t-test. Bars represent mean +/− SEM.
Figure 3.
Confirmation of p22PHOXand galectin-3protein expression. (A) Spinal cord tissue from sham and injured mice was obtained at 28 days post-injury and processed for p22PHOX immunolabeling (red), with a DAPI counterstain (blue), demonstrating a marked increase in staining within injured tissue. Small inset is a low mag image of the spinal cord, demonstrating the region represented in the larger images. Bar= 50μm. (B)Western blots were performed on sham (S) and injured (I) spinal cords at 3 and 14 days. Significant increases (*p<0.05, Student’s t-test) were observed at all time points, with protein expression normalized to GAPDH level, which was performed on the same blot after stripping. Bars represent mean +/− SEM.
2.3 Galectin-3 plays a role in SCI responses
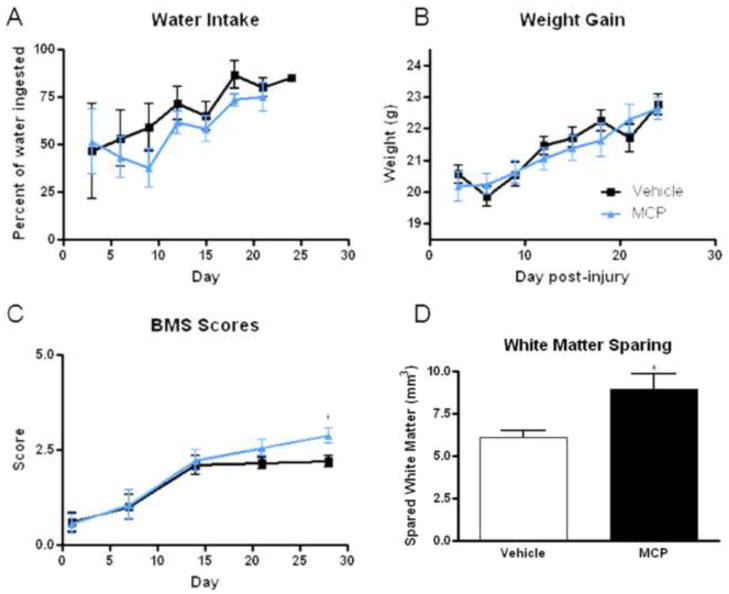
To further investigate the role of galectin-3 after SCI, spinal cord injured mice were given access to water containing 1% MCP or control water (plain distilled water) immediately after injury. Addition of MCP to the water had no significant effect on water consumption or weight gain (Fig. 4A and B). However, MCP addition did have a significant effect on motor function after injury. Vehicle-treated mice achieved a score of 2.1 +/− 0.2 by 14 days post-injury, and maintained this score through day 28, representing achievement of extensive ankle movement, but no other recovery. MCP-treated mice, on the other hand, had a score of 2.2 +/− 0.3 by day 14, and increased to a score of 2.9 +/− 0.2 by day 28 post-injury, a significant increase over vehicle treated mice (p<0.05, Repeated Measures ANOVA, Fig. 4C). In the MCP group, over half (5 of 9) of the mice were able to achieve plantar placement with support and occasional stepping, while none (0 of 10) of the vehicle treated mice achieved this function.
Figure 4.
The effect of galectin-3 inhibition by MCP administration in the water.(A, B) To ensure that addition of MCP to the water did not negatively affect responses, water intake and weight gain were measured and found to be similar between vehicle and MCP treated mice. (C) Functional recovery was measured using the BMS score at 1, 7, 14, 21 and 28 days post-injury, and demonstrates a significant (*p<0.05, Repeated Measures ANOVA) improvement in score with MCP treatment at day 28. (D) White matter sparing was quantified using unbiased stereology of eriochrome C stained spinal cord sections and the data show a significant (*p<0.05, Student’s t-test) difference between vehicle and MCP-treated mice 28 days post-injury. Bars represent mean +/− SEM.
At 28 days post-injury, spinal cords were removed and processed to assess white matter sparing. Quantitation of white matter sparing using unbiased stereology showed that MCP-treated mice had a significantly greater amount of white matter sparing than vehicle-treated mice 28 days post-injury (Fig. 4D).
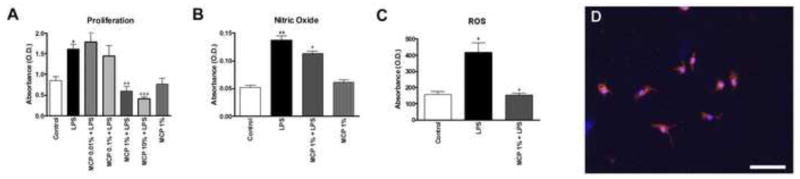
To examine the mechanism of the beneficial effects of galectin-3 inhibition in the SCI model, a microglial cell culture model was studied in which microglia were exposed to MCP 1 hour prior to a lipopolysaccharide (LPS) challenge. At 24 hours after the LPS challenge, microglia demonstrated significant increases in proliferation (Fig. 5A), nitric oxide (NO) production (Fig. 5B), and ROS release (Fig. 5C). Pre-treatment with MCP significantly reduced these responses in a dose-dependent manner, with a maximal inhibition at 1%. MCP alone had no effect on any outcome measure (Fig 5A–C).
Figure 5.
Effects of MCP in microglial culture. Primary microglia cells were exposed to MCP 1 hour prior to an LPS challenge. At 24 hours after the LPS challenge (A) proliferation, (B) NO production, and (C) ROS release were measured and quantified. (A) MCP resulted in a dose dependent reduction in microglial responses to LPS. *p<0.05, **p<0.01 vs. control; +p<0.05, ++p<0.01; +++p<0.001 vs. LPS; One-Way ANOVA. Bars represent mean +/− SEM. Microglia in all cultures were found to be 92.9% pure, as measured by percentage of cells positive for the microglial marker Iba1 (red, D). DAPI+ nuclei are blue. Bar = 50μm.
3. Discussion
Here we provide evidence for chronic inflammation in the spinal cord after a moderate contusion injury in mice. This inflammatory response at the gene expression level is markedly similar to that seen in rats (Byrnes et al., 2006; Byrnes et al., 2011), and supports the idea of a conserved response to injury across species. Moreover, we demonstrate that expression at the mRNA and protein level of the delayed expression group is sustained for at least 1 month after injury. In addition, we show that one of these genes, galectin-3, may contribute to post-traumatic tissue damage and associated neurological dysfunction.
In our previous work in rats, two clusters of genes were identified: an acute-expression cluster that demonstrated early up-regulation and down-regulation over the first 24 hours to 7 days post-injury, and a delayed-expression cluster that demonstrated up-regulation later than 24 hours and sustained up-regulation through 6 months post-injury. Many of the same genes were identified and confirmed at the protein level in mice, including C1qB, galectin-3 and p22PHOX. This may reflect the similarities in cellular composition of the lesion after SCI in rats and mice (Byrnes et al., 2010). In particular, the microglia/macrophage invasion and concentration around the lesion was markedly similar between the two species, despite significant differences in lesion evolution and cavity formation.
Although the role of many of these genes in the injured spinal cord is currently unknown or unclear (Byrnes et al., 2006), some have been implicated in the pathophysiological effects of chronic inflammation after CNS injury. For example, gp91PHOX, also referred to as NOX2, is a cell membrane-bound component of NADPH oxidase. This enzyme has been investigated as a primary source of ROS after injury, and recent studies have begun to investigate the effects of inhibiting this enzyme (Byrnes et al., 2011).
A 2010 proteomics study also identified galectin-3 as a chronically up-regulated protein after CNS injury (Yan et al., 2010), consistent with our current data. This protein, also known as MAC-2, is expressed primarily in microglia, but its role is currently unclear. Some studies of galectin-3 in stroke or other injury models have suggested that lack of this protein in knockout animal models or cells results in impaired recovery or function (Lalancette-Hebert et al., 2007). Others have shown that galectin-3 plays a significant role in phagocytosis (Caberoy et al., 2011; Olah et al., 2011; Reichert and Rotshenker, 1999), which may play a role during both development and injury (Rigato et al., 2011). Alternatively, stimulation of microglia with the classical activator LPS induces an increase in secretion of galectin-3, which may have cytokine-like properties (Liu et al., 1995). Furthermore, galectin-3 mutation has been found to induce NADPH oxidase activity, resulting in ROS production (Fernandez et al., 2005). This is also supported by our current in vitro studies, in which blocking microglial galectin-3 activity reduced ROS production (Fig.5). In our SCI model, we found that pharmacological inhibition of galectin-3 activity with MCP significantly improved functional recovery and white matter sparing. This may be a result of the potent anti-inflammatory effect of MCP on galectin-3 cytokine-like activities, which we demonstrated as well in our in vitro microglial model.
In conclusion, we now demonstrate that, similar to rat, mouse SCI results in a chronic up-regulation of microglial-related genes. While, structurally, human SCI is more similar to rat SCI (Byrnes et al., 2010; Potter and Saifuddin, 2003), similarities in the gene expression profiles between the two species suggest genomic conservation and indicates the importance of these genes in the spinal cord’s response to injury. These data provide strong support for the need for future research into chronic inflammation after SCI.
4. Experimental Procedure
4.1 Spinal Cord Injury
Contusion spinal cord injury was performed in adult male C57Bl/6 mice. Mice (20 – 25g) were anesthetized with isoflurane (induction: 4%, maintenance: 1.5%) and moderate injury was induced using a weight drop method, in which a 1g weight was dropped from 30mm onto an impounder positioned on the exposed spinal cord at vertebral level T9 as previously described (Pajoohesh-Ganji et al., 2010). The weight drop method is a traditionally accepted model of contusion SCI that was chosen to mimic the rat model previously studied in our laboratory (Byrnes et al., 2006; Byrnes et al., 2011). With a moderate injury, less than 15% mortality was noted, mostly within the first few hours to days after injury. Two mice were removed from the study after a recovery of 11 – 14 days due to autophagia. With normal care, including manual bladder expression for 7 – 10 days after injury and Cell-sorb bedding, most mice survived to the required time points with no complications. Sham injured animals underwent the same experimental procedures and received a laminectomy without weight drop. All experiments complied fully with the principles set forth in the “Guide for the Care and Use of Laboratory Animals” prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Resources, National Research Council (DHEW pub. No. (NIH) 85–23, 2985) and were approved by the Georgetown University IACUC.
4.2 Expression Profiling
Spinal cords were analyzed at the site of impact and from adjacent rostral and caudal regions (4 mm in length per region) at 0.5, 4, 24, 72 h and 7 and 28 days after injury (n=3/per group), sham-injury (n=2 per group), or from naïve mice (n=2). Four mice were pooled for each individual n, for a total of 12 mice for each injury time point.
Expression profiling was performed as described previously (Byrnes et al., 2006; Di Giovanni et al., 2005). Briefly, total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA), and quality was assessed via spectrophotometry and gel electrophoresis. Five μg of total RNA was amplified (one-cycle) and hybridized to Affymetrix 430 2.0 mouse arrays following standard manufacturer’s protocols (Affymetrix, Santa Clara, CA).
4.3 Microarray Analysis
Quality control methods were previously published (Di Giovanni et al., 2003). Samples fulfilled the following quality control measures: cRNA fold changes between 5 to 10, scaling factor from 0.3–1.5, percentage of “present” (P) calls from 40–55%, average signal intensity levels between 900–1100, housekeeping genes and internal probe set controls showed > 80% present calls, consistent values and 5′/3′ ratios were < 3. Experimental normalization, data filtering and statistical analysis of gene expression profiles were generated with dCHIP and GeneSpring software (Agilent Technologies, Santa Clara, CA). No arrays were detected as project outliers by the dCHIP algorithm. Sham vs injured groups and gene differences were compared with Welch ANOVA/t-tests with p values <0.05 considered significant, as previously described (Kerr and Churchill, 2001).
Anchor gene analysis was performed using genes known to be associated with post-injury inflammation: C1qB, Lgals3 (galectin-3) and PTGS2 (Byrnes et al., 2006). Inflammatory gene clusters were defined as all genes that were significantly different from controls and temporally expressed at a correlation of ≥0.99 with the anchors (nucleators). Original raw microarray data are publicly available on the GEO omnibus (www.ncbi.nlm.nih.gov).
4.4 Western Blot
At 72 hours, 7 days, 14 days and 28 days post-injury, 4 moderate-contusion injured and 2 sham injured mice per time point were anesthetized (100 mg/kg sodium pentobarbital, I.P.) and decapitated. A 5mm section of the spinal cord (approximately 50 mg of tissue weight) centered at the lesion epicenter, T-9, was dissected, and immediately frozen on dry ice and western blot was performed as described previously (Byrnes et al., 2006). Briefly, tissue was homogenized in RIPA buffer and centrifuged to isolate protein. Twenty-five μg of protein were run in an SDS polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane. The blot was then probed with antibodies against C1q (1:200; US Biologicals, Swampscott, MA), galectin-3 (1:1000; Abcam, Cambridge, MA), and p22PHOX (1:200; Santa Cruz Biotechnology, Santa Cruz, CA). Immune complexes were detected with appropriate secondary antibodies (HRP secondaries, Fisher Scientific, Pittsburgh, PA; 1:2000) and chemiluminescence reagents (Pierce, Rockford, IL). GAPDH was used as a control for gel loading and protein transfer. Scion Image Analysis (http://www.scioncorp.com/) was used to assess pixel density of resultant blots for comparison between sham-injured and injured spinal cord tissue.
4.5 Immunohistochemistry
At 14 and 28 days post-injury, 4 moderate-contusion injured and 2 sham injured mice per time point were anesthetized (100 mg/kg sodium pentobarbital, I.P.) and intracardially perfused with 10 ml of 0.9% saline followed by 50 ml of 10% buffered formalin. A 1 cm section of the spinal cord centered at the lesion epicenter, T-9, was dissected, post-fixed in 10% buffered formalin overnight and cryoprotected in 30% sucrose for 48 hours. Standard fluorescent immunocytochemistry on serial, 20 μm thick coronal sections was performed as described previously (Byrnes et al., 2006).
Antibodies included Iba1 (1:100, Wako, Richmond, VA), C1q (1:500Abcam, Cambridge, MA) and p22phox (1:50, Santa Cruz). Appropriate secondary antibodies linked to fluorophores (1:2000; Invitrogen, Grand Island, NY) were incubated with tissue sections for 1 hour at room temperature. Slides were coverslipped using mounting media with DAPI, a counterstain for nuclei (Vector Labs, Burlingame, CA). To ensure accurate and specific staining, negative controls were used in which the primary antibody was not applied. Immunofluorescence was detected using confocal microscopy or an AxioPlan Zeiss Microscopy system (Carl Zeiss, Inc., Thornwood, NY). Immunolabeling was quantified as needed as previously described (Donnelly et al., 2009).
4.6 Water Administration
MCP was made as previously described (Nangia-Makker et al., 2002) and diluted to 1% in distilled water. MCP treated water or normal distilled water (vehicle) was administered to C57Bl/6 mice in their normal drinking water bottles beginning immediately after injury. Water was changed every 3 days and water volume consumed was measured.
4.7 Behavioral Testing
C57Bl/6 mice receiving MCP or vehicle-treated water (n = 10/group) were tested for hindlimb functional deficits at 1, 7, 14, 21, and 28after moderate SCI. Hindlimb locomotor recovery was assessed in an open field using the BMS previously described in detail (Basso et al., 2006). This scale ranges from 0, indicating complete paralysis, to 9, indicating normal movement of the hindlimbs. Performance of the left and right hindlimbs was averaged in order to obtain the BMS score.
4.8 Histology
Quantitative assessment of white matter sparing was performed using the Cavalieri method of unbiased stereology, as previously described (Byrnes et al., 2007; Iannotti et al., 2004). Briefly, a 1 cm section of the spinal cord centered at the lesion epicenter was cut at 20μm and every 50th section of the 1 cm spinal cord block, with a random starting section, was processed with a standard eriochrome cyanine R staining protocol for histological analysis. Volume estimations were obtained from at least 10 randomly selected sections spanning the 1 cm centered around the lesion site.
4.9 Microglial Cultures
Primary microglial cells were obtained from post-natal day 2 C57Bl6 mouse pups and cultured as previously described (Byrnes et al., 2006; Tamashiro et al., 2012). Briefly, the whole brain was carefully dissected and homogenized in L15 media (Gibco, Carlsbad, CA). Mixed glial cultures were incubated for 8–10 days at 37°C with 5% CO2 in Dulbecco’s modified eagle media (Gibco) with 10% fetal calf serum (Hyclone, Logan, UT), 1% L-glutamine (Gibco), 1% sodium pyruvate (Gibco), and 1% Pen/Strep (Fisher, Pittsburgh, PA). After the initial incubation, the cells were shaken for 1 h at 100 rpm and at 37°C to allow microglia to detach from the astrocyte monolayer within flasks. Detached microglia were collected and replated as purified cultures with greater than 92% purity (Fig. 5D).
4.10 Microglial Proliferation and Viability
Proliferation of microglia in 96-wellplates was assessed using the MTS assay (MTS tetrazolium compound; Cell Titer 96Aqueous One Solution, Promega, Madison, WI) according to the manufacturer’s protocol.
4.11 Nitric Oxide Production
NO production was assayed using the Griess Reagent Assay (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions.
4.12 ROS Detection
Intracellular reactive oxygen species (ROS) production was assessed at 24 h after stimulation by measuring the oxidation of 5 (and 6)-chloromethyl-20, 70-dichlorodihydrofluoresceindiacetate-acetyl ester (CM-H2DCFDA; Molecular Probes, Eugene, OR). Media from microglia plated into 96-well plates was aspirated and replaced with warmed PBS. CM-H2DCFDA (10 μM) was added to microglia and incubated for 45 min. Fluorescence was measured using excitation and emission wavelengths of490 and 535 nm, respectively.
4.13 Statistical Analysis
Quantitative data are presented as mean +/− standard error of the mean. Lesion volume, western blot, and immunohistochemical data were obtained by an investigator blinded to treatment group. All data were analyzed using Student’s t test or one-way ANOVA, where appropriate. Functional testing data were analyzed using Repeated Measures ANOVA. All statistical tests were performed using the GraphPad Prism Program, Version 5.0 for Windows (GraphPad Software, San Diego, CA). A p value < 0.05 was considered statistically significant.
Highlights.
Inflammatory gene expression after SCI was profiled using microarray in mice.
There is delayed and persistent up-regulation of inflammatory genes after mouse SCI
The delayed expression group includes NADPH oxidase components and galectin-3
The protein expression of these genes is consistent with the gene expression
Inhibition of galectin-3 improves motor function and white matter sparing after SCI
Acknowledgments
This work was supported by pilot funding from the National Capital Area Rehabilitation Research Network (KB) and National Institutes of Health (NIH) Grant number R01NS054221-06 (AIF). The authors would like to thank the technical support of Dr. Jorge Garay, Nicole Hockenbury, and Yujia Zhao in this work.
Abbreviations
- MCP
modified citrus pectin
- NO
nitric oxide
- LPS
lipopolysaccharide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ahdeah Pajoohesh-Ganji, Email: ahdeah@gwu.edu.
Susan M. Knoblach, Email: sknoblach@cnmcresearch.org.
Alan I. Faden, Email: afaden@anes.umm.edu.
Kimberly R. Byrnes, Email: kbyrnes@usuhs.mil.
References
- Aimone JB, et al. Spatial and temporal gene expression profiling of the contused rat spinal cord. Exp Neurol. 2004;189:204–21. doi: 10.1016/j.expneurol.2004.05.042. [DOI] [PubMed] [Google Scholar]
- Basso DM, et al. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J Neurotrauma. 2006;23:635–59. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- Beattie MS. Inflammation and apoptosis: linked therapeutic targets in spinal cord injury. Trends Mol Med. 2004;10:580–3. doi: 10.1016/j.molmed.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Bilgen M, et al. Longitudinal magnetic resonance imaging of spinal cord injury in mouse: changes in signal patterns associated with the inflammatory response. Magn Reson Imaging. 2007;25:657–64. doi: 10.1016/j.mri.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Byrnes KR, et al. Expression of two temporally distinct microglia-related gene clusters after spinal cord injury. Glia. 2006;53:420–33. doi: 10.1002/glia.20295. [DOI] [PubMed] [Google Scholar]
- Byrnes KR, et al. Cell cycle activation contributes to post-mitotic cell death and secondary damage after spinal cord injury. Brain. 2007;130:2977–92. doi: 10.1093/brain/awm179. [DOI] [PubMed] [Google Scholar]
- Byrnes KR, Fricke ST, Faden AI. Neuropathological differences between rats and mice after spinal cord injury. J Magn Res Imag. 2010;32:836–46. doi: 10.1002/jmri.22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes KR, et al. Delayed inflammatory mRNA and protein expression after spinal cord injury. J Neuroinflammation. 2011 doi: 10.1186/1742-2094-8-130. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, et al. Galectin-3 is a new MerTK-specific eat-me signal. J Cell Physiol. 2011 doi: 10.1002/jcp.22955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel JB, et al. Gene expression profiling of acute spinal cord injury reveals spreading inflammatory signals and neuron loss. Physiol Genomics. 2001;7:201–13. doi: 10.1152/physiolgenomics.00074.2001. [DOI] [PubMed] [Google Scholar]
- Chen CH, et al. Suppression of endotoxin-induced proinflammatory responses by citrus pectin through blocking LPS signaling pathways. Biochem Pharmacol. 2006;72:1001–9. doi: 10.1016/j.bcp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Demjen D, et al. Neutralization of CD95 ligand promotes regeneration and functional recovery after spinal cord injury. Nat Med. 2004;10:389–95. doi: 10.1038/nm1007. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, et al. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann Neurol. 2003;53:454–68. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, et al. Neuronal plasticity after spinal cord injury: identification of a gene cluster driving neurite outgrowth. Faseb J. 2005;19:153–4. doi: 10.1096/fj.04-2694fje. [DOI] [PubMed] [Google Scholar]
- Donnelly DJ, et al. An efficient and reproducible method for quantifying macrophages in different experimental models of central nervous system pathology. J Neurosci Methods. 2009;181:36–44. doi: 10.1016/j.jneumeth.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusart I, Schwab ME. Secondary cell death and the inflammatory reaction after dorsal hemisection of the rat spinal cord. Eur J Neurosci. 1994;6:712–24. doi: 10.1111/j.1460-9568.1994.tb00983.x. [DOI] [PubMed] [Google Scholar]
- Fernandez GC, et al. Galectin-3 and soluble fibrinogen act in concert to modulate neutrophil activation and survival: involvement of alternative MAPK pathways. Glycobiology. 2005;15:519–27. doi: 10.1093/glycob/cwi026. [DOI] [PubMed] [Google Scholar]
- Fitch MT, et al. Cellular and molecular mechanisms of glial scarring and progressive cavitation: In vivo and in vitro analysis of inflammation-induced secondary injury after cns trauma. J Neurosci. 1999;19:8182–8198. doi: 10.1523/JNEUROSCI.19-19-08182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JC, et al. The cellular inflammatory response in human spinal cords after injury. Brain. 2006;129:3249–69. doi: 10.1093/brain/awl296. [DOI] [PubMed] [Google Scholar]
- Ghasemlou N, et al. Mitogen-activated protein kinase-activated protein kinase 2 (MK2) contributes to secondary damage after spinal cord injury. J Neurosci. 2010;30:13750–9. doi: 10.1523/JNEUROSCI.2998-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gris P, et al. Differential gene expression profiles in embryonic, adult-injured and adult-uninjured rat spinal cords. Mol Cell Neurosci. 2003;24:555–67. doi: 10.1016/s1044-7431(03)00211-2. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, et al. Gene expression profiling of cathepsin D, metallothioneins-1 and -2, osteopontin, and tenascin-C in a mouse spinal cord injury model by cDNA microarray analysis. Acta Neuropathol (Berl) 2004 doi: 10.1007/s00401-004-0926-z. [DOI] [PubMed] [Google Scholar]
- Hsu DK, et al. Targeted disruption of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am J Pathol. 2000;156:1073–83. doi: 10.1016/S0002-9440(10)64975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti C, et al. A neuroprotective role of glial cell line-derived neurotrophic factor following moderate spinal cord contusion injury. Exp Neurol. 2004;189:317–32. doi: 10.1016/j.expneurol.2004.05.033. [DOI] [PubMed] [Google Scholar]
- Jiang HR, et al. Galectin-3 deficiency reduces the severity of experimental autoimmune encephalomyelitis. J Immunol. 2009;182:1167–73. doi: 10.4049/jimmunol.182.2.1167. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet Res. 2001;77:123–8. doi: 10.1017/s0016672301005055. [DOI] [PubMed] [Google Scholar]
- Lalancette-Hebert M, et al. Selective ablation of proliferating microglial cells exacerbates ischemic injury in the brain. J Neurosci. 2007;27:2596–605. doi: 10.1523/JNEUROSCI.5360-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FT, et al. Expression and function of galectin-3, a beta-galactoside-binding lectin, in human monocytes and macrophages. Am J Pathol. 1995;147:1016–28. [PMC free article] [PubMed] [Google Scholar]
- Nangia-Makker P, et al. Inhibition of human cancer cell growth and metastasis in nude mice by oral intake of modified citrus pectin. J Natl Cancer Inst. 2002;94:1854–62. doi: 10.1093/jnci/94.24.1854. [DOI] [PubMed] [Google Scholar]
- Olah M, et al. Identification of a microglia phenotype supportive of remyelination. Glia. 2011 doi: 10.1002/glia.21266. [DOI] [PubMed] [Google Scholar]
- Pajoohesh-Ganji A, et al. A combined scoring method to assess behavioral recovery after mouse spinal cord injury. J Neurosci Res. 2010 doi: 10.1016/j.neures.2010.02.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovich PG, et al. The neuropathological and behavioral consequences of intraspinal microglial/macrophage activation. J Neuropathol Exp Neurol. 2002;61:623–33. doi: 10.1093/jnen/61.7.623. [DOI] [PubMed] [Google Scholar]
- Potter K, Saifuddin A. Pictorial review: MRI of chronic spinal cord injury. Br J Radiol. 2003;76:347–52. doi: 10.1259/bjr/11881183. [DOI] [PubMed] [Google Scholar]
- Reichert F, Rotshenker S. Galectin-3/MAC-2 in experimental allergic encephalomyelitis. Exp Neurol. 1999;160:508–14. doi: 10.1006/exnr.1999.7229. [DOI] [PubMed] [Google Scholar]
- Rigato C, et al. Pattern of invasion of the embryonic mouse spinal cord by microglial cells at the time of the onset of functional neuronal networks. Glia. 2011;59:675–95. doi: 10.1002/glia.21140. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–56. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- Stirling DP, et al. Minocycline treatment reduces delayed oligodendrocyte death, attenuates axonal dieback, and improves functional outcome after spinal cord injury. J Neurosci. 2004;24:2182–90. doi: 10.1523/JNEUROSCI.5275-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T, Noguchi K, Ruda MA. Analysis of gene expression following spinal cord injury in rat using complementary DNA microarray. Neurosci Lett. 2002;327:133–7. doi: 10.1016/s0304-3940(02)00375-0. [DOI] [PubMed] [Google Scholar]
- Tamashiro TT, Dalgard CL, Byrnes KR. Primary microglia isolation from mixed glial cell cultures of neonatal rat brain tissue. J Vis Exp. 2012 doi: 10.3791/3814. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Davies JE, Davies SJ. Changes in distribution, cell associations, and protein expression levels of NG2, neurocan, phosphacan, brevican, versican V2, and tenascin-C during acute to chronic maturation of spinal cord scar tissue. J Neurosci Res. 2003;71:427–44. doi: 10.1002/jnr.10523. [DOI] [PubMed] [Google Scholar]
- Teng YD, et al. Minocycline inhibits contusion-triggered mitochondrial cytochrome c release and mitigates functional deficits after spinal cord injury. Proc Natl Acad Sci U S A. 2004;101:3071–6. doi: 10.1073/pnas.0306239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, et al. Proteomic profiling of proteins in rat spinal cord induced by contusion injury. Neurochem Int. 2010;56:971–83. doi: 10.1016/j.neuint.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang KH, et al. Differential gene expression after complete spinal cord transection in adult rats: an analysis focused on a subchronic post-injury stage. Neuroscience. 2004;128:375–88. doi: 10.1016/j.neuroscience.2004.07.008. [DOI] [PubMed] [Google Scholar]