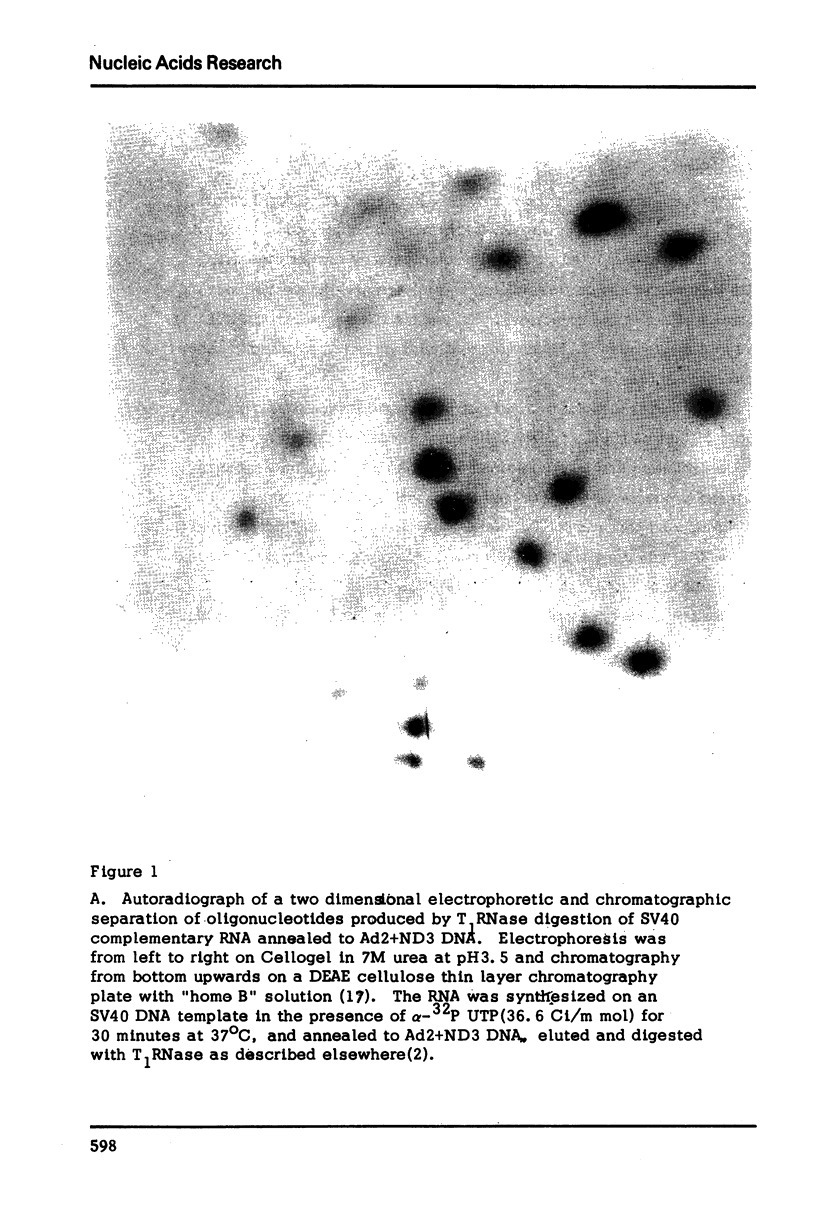
Abstract
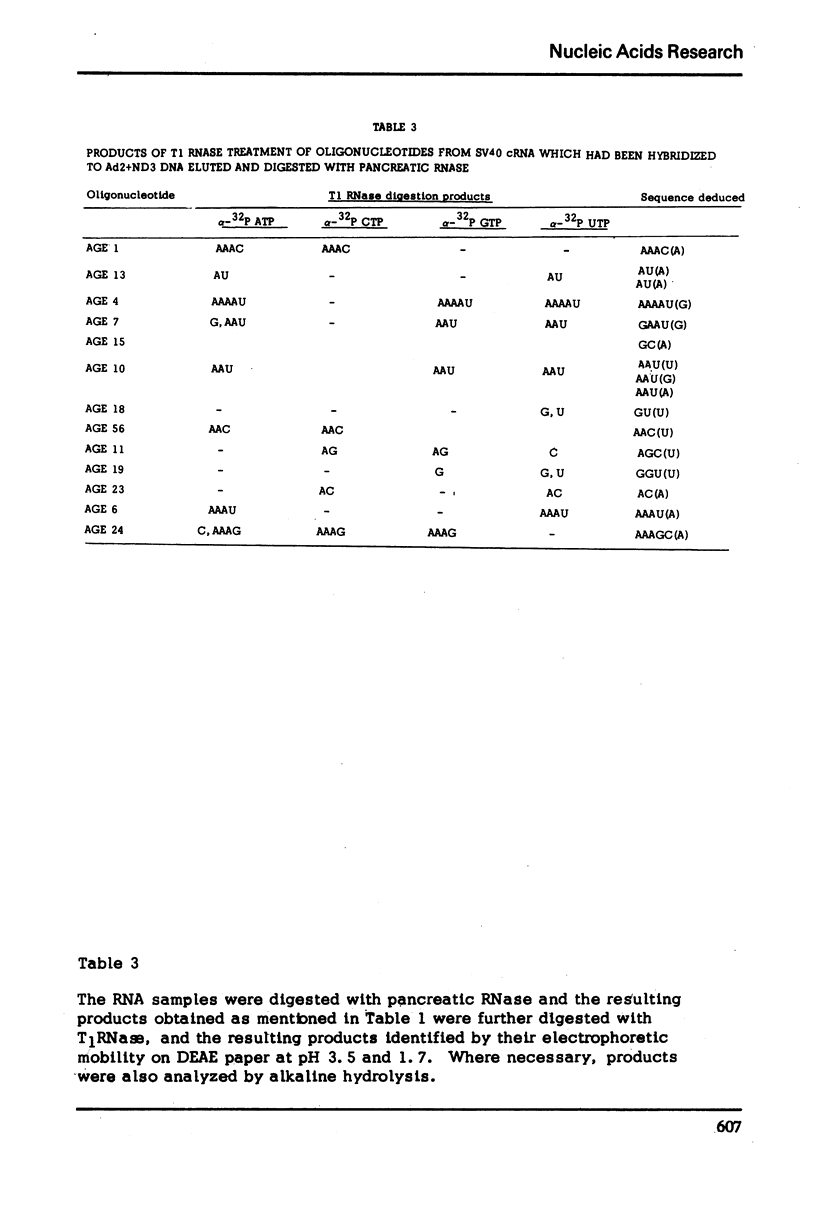
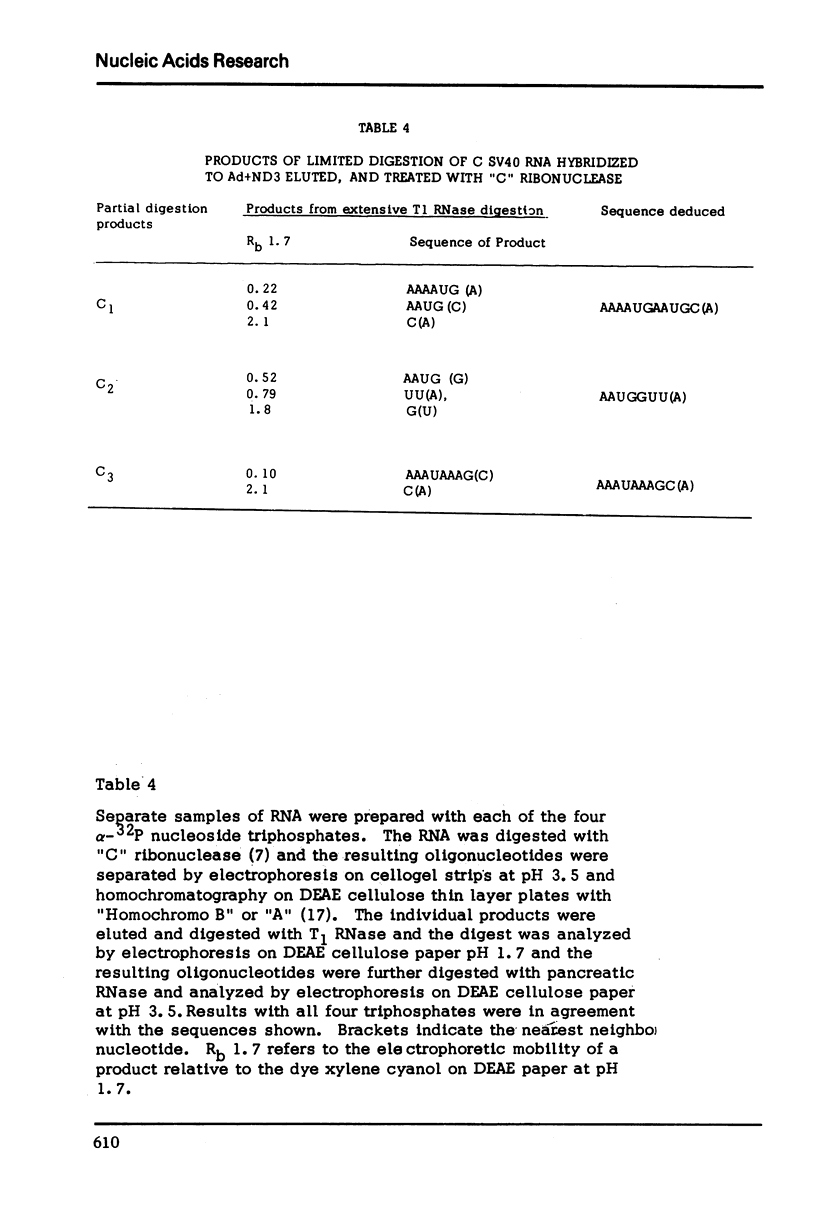
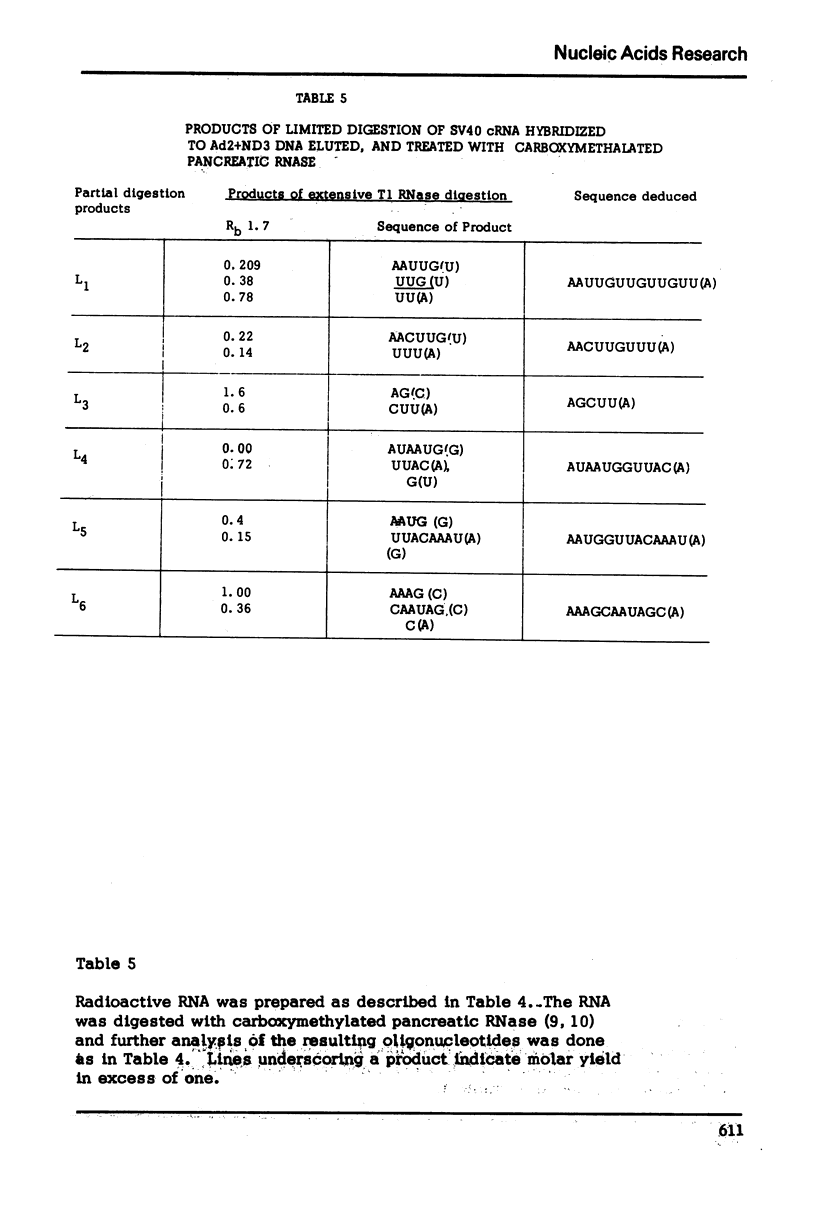
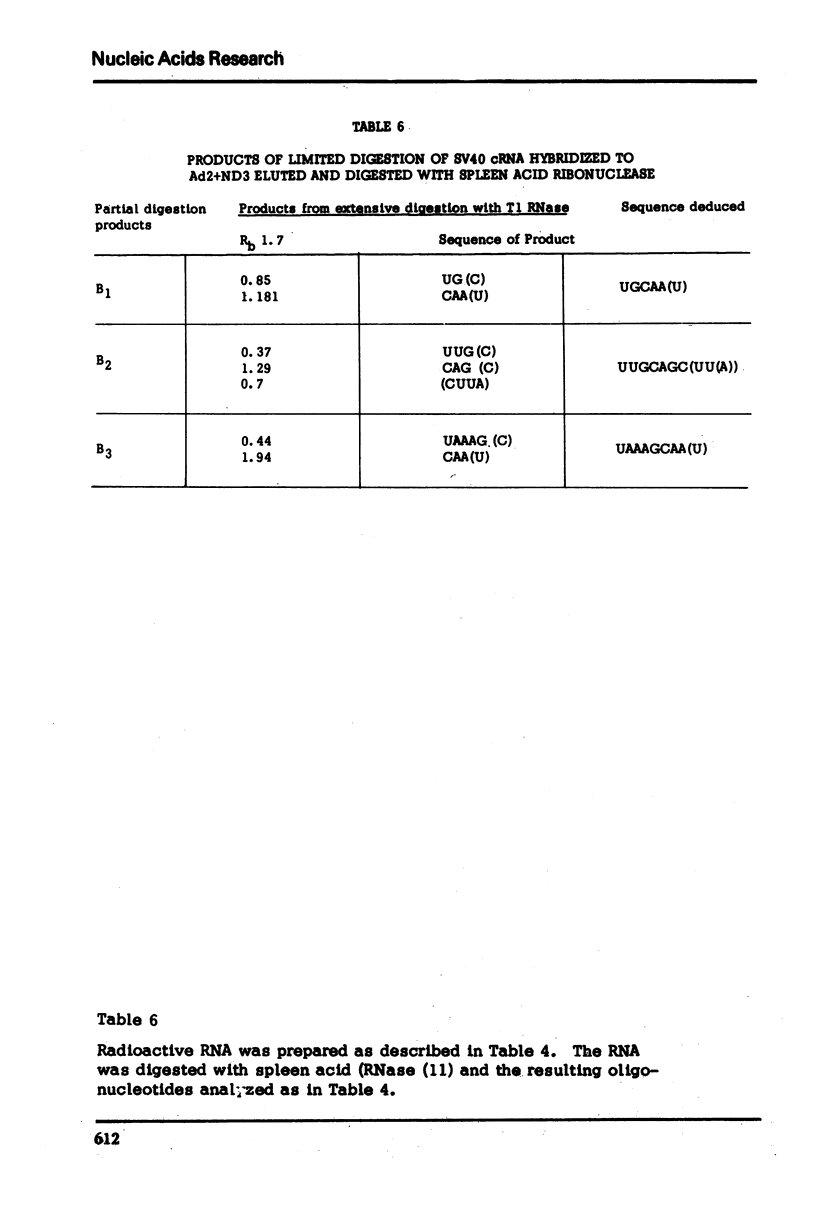
The nucleotide sequence of the RNA transcript from the “early” (E) strand of SV40 DNA immediately preceding the preferred E. coli RNA polymerase start site is G-(A-A-A-C, -A-U-)-A-A-A-A-U-G-A-A-U-G-C-A-A-U-U-G-U-U-G-U-U-G-U-U-A-A-C-U-U-G-U-U-U-A-U-U-G-C-A-G-C-U-U-A-U-A-A-U-G-G-U-U-A-C-Ap. The last nucleotide of the sequence is the first nucleotide transcribed by E. coli RNA polymerase from the “E” strand. The DNA template contains a palindrome of 17 residues that includes the Hemophilus influenza restriction endonuclease cleavage site G-T-T-A-A-Cp. The DNA which gives this transcript lies very close to one end of SV40 DNA segment in the Adeno-SV40 hybrid virus Ad2+ND3 and appears to contain sufficient untranscribed information to specify the E. coli RNA polymerase start.
Full text
PDF


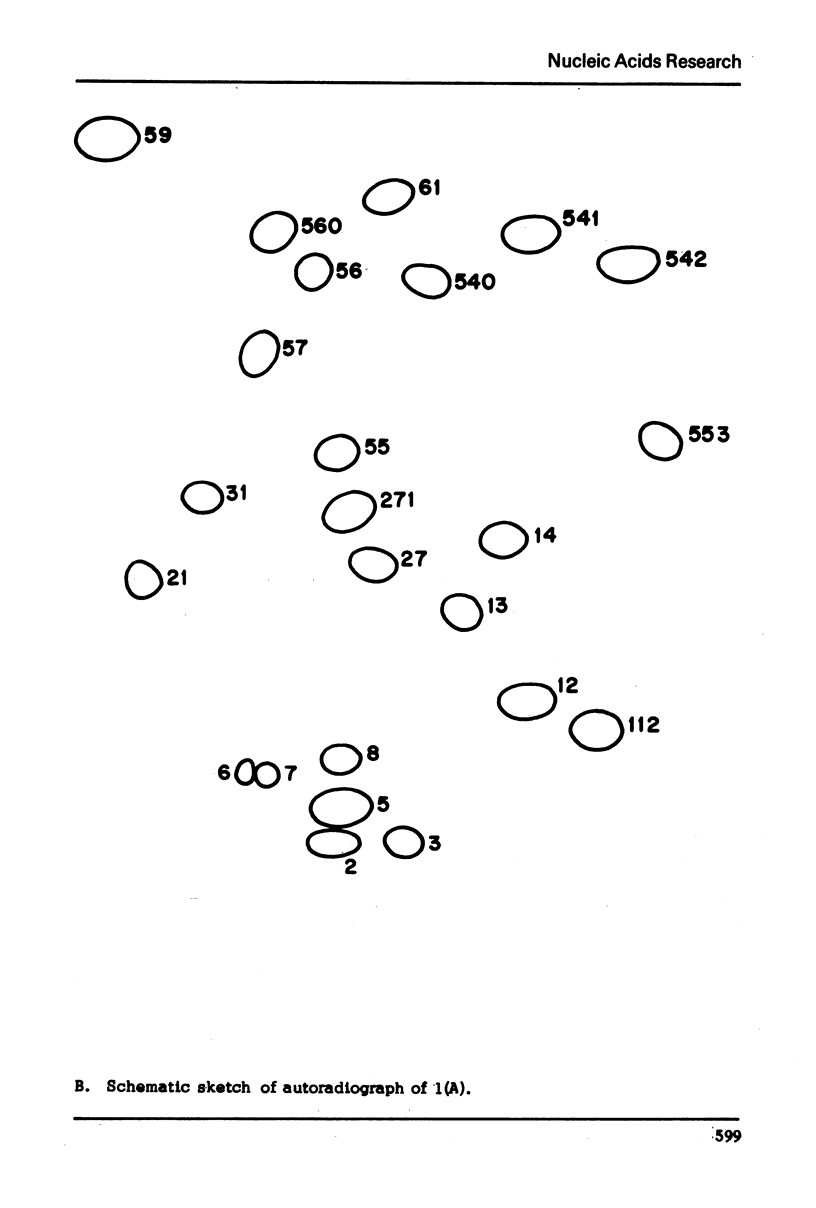


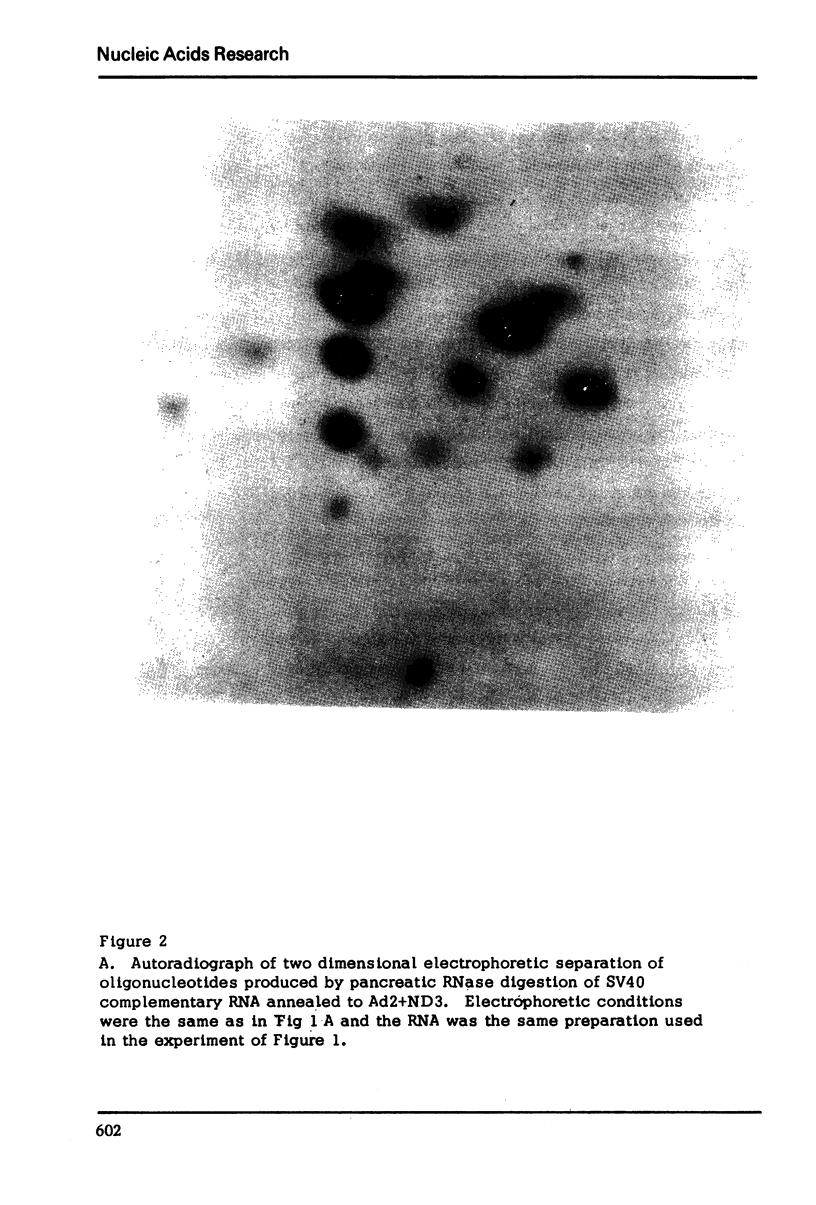
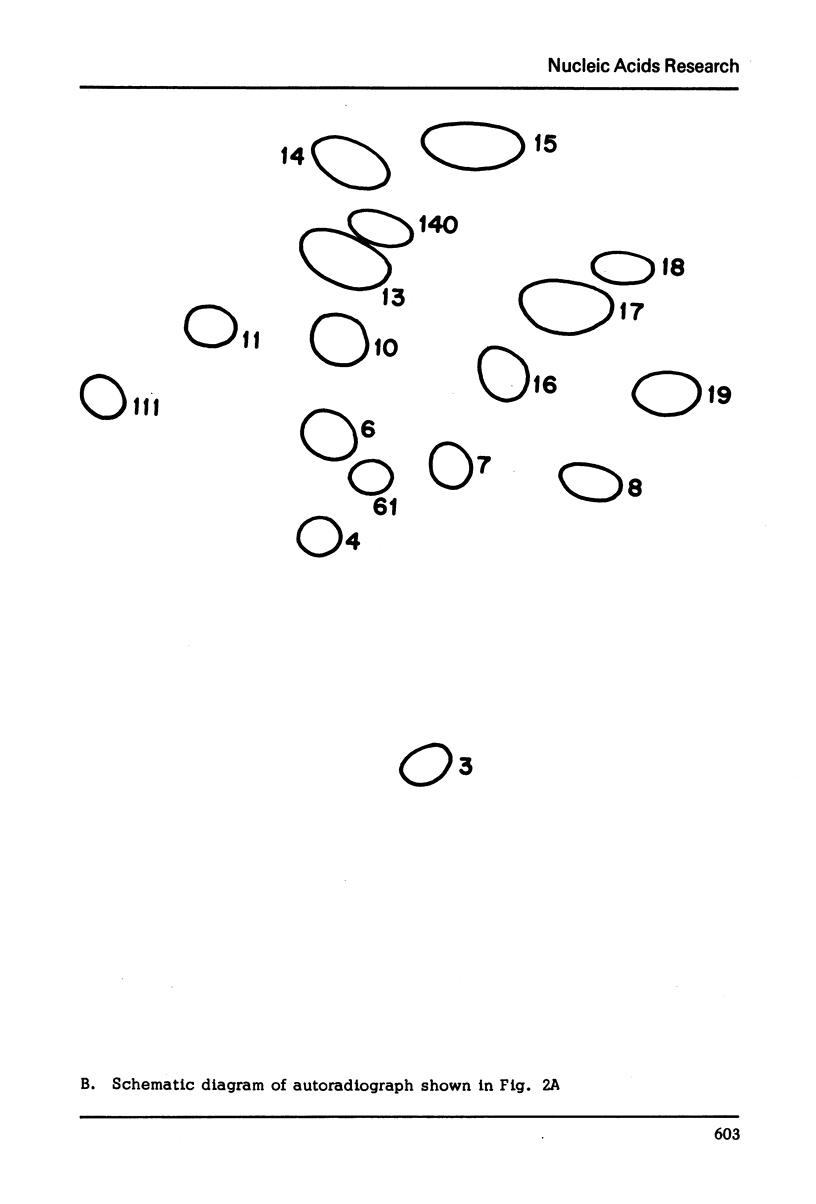
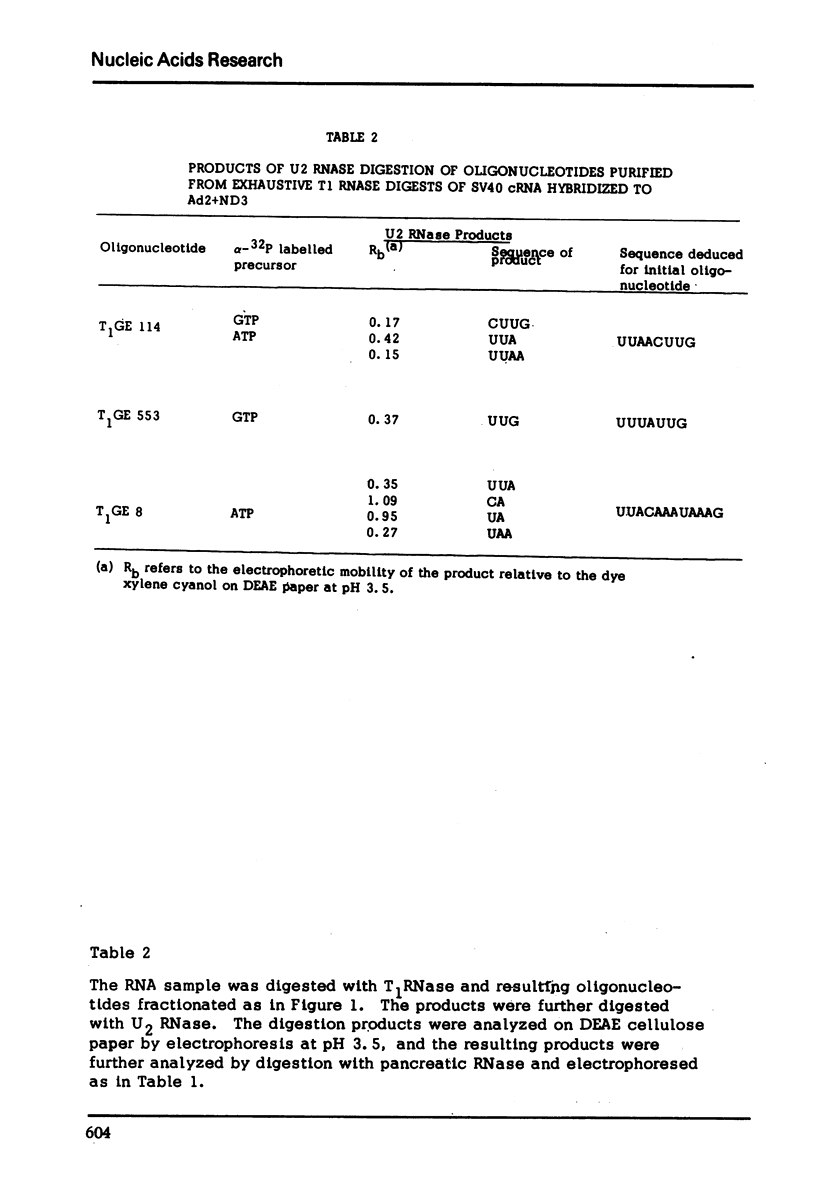



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Contreras R., Fiers W. A new method for partial digestion useful for sequence analysis of polynucleotides. FEBS Lett. 1971 Sep 1;16(4):281–283. doi: 10.1016/0014-5793(71)80370-8. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Goldstein J. Digestion of ribonucleic acid by an alkylated ribonuclease. J Mol Biol. 1967 Apr 14;25(1):123–130. doi: 10.1016/0022-2836(67)90283-5. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Levy C. C., Goldman P. Residue specificity of a ribonuclease which hydrolyzes polycytidylic acid. J Biol Chem. 1970 Jun;245(12):3257–3262. [PubMed] [Google Scholar]
- Lewis A. M., Jr, Levine A. S., Crumpacker C. S., Levin M. J., Samaha R. J., Henry P. H. Studies of nondefective adenovirus 2-simian virus 40 hybrid viruses. V. Isolation of additional hybrids which differ in their simian virus 40-specific biological properties. J Virol. 1973 May;11(5):655–664. doi: 10.1128/jvi.11.5.655-664.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta C. A., Levy C. C., Weissman S. M., Varricchio F. Preferred sites of digestion of a ribonuclease from Enterobacter sp. in the sequence analysis of Bacillus stearothermophilus 5S ribonucleic acid. Biochemistry. 1973 Jul 17;12(15):2901–2904. doi: 10.1021/bi00739a020. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Westphal H. SV40 DNA strand selection by Escherichia coli RNA polymerase. J Mol Biol. 1970 Jun 14;50(2):407–420. doi: 10.1016/0022-2836(70)90201-9. [DOI] [PubMed] [Google Scholar]
- Zain B. S., Dhar R., Weissman S. M., Lebowitz P., Lewis A. M., Jr Preferred site for initiation of RNA transcription by Escherichia coli RNA polymerase within the simian virus 40 DNA segment of the nondefective adenovirus-simian virus 40 hybrid viruses Ad2 + ND 1 and Ad2 + ND 3 . J Virol. 1973 May;11(5):682–693. doi: 10.1128/jvi.11.5.682-693.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]