Abstract
In a synchronized photoautotrophic culture of Chlamydomonas reinhardtii, cell size, cell number, and the averaged starch content were determined throughout the light-dark cycle. For single-cell analyses, the relative cellular starch was quantified by measuring the second harmonic generation (SHG). In destained cells, amylopectin essentially represents the only biophotonic structure. As revealed by various validation procedures, SHG signal intensities are a reliable relative measure of the cellular starch content. During photosynthesis-driven starch biosynthesis, synchronized Chlamydomonas cells possess an unexpected cell-to-cell diversity both in size and starch content, but the starch-related heterogeneity largely exceeds that of size. The cellular volume, starch content, and amount of starch/cell volume obey lognormal distributions. Starch degradation was initiated by inhibiting the photosynthetic electron transport in illuminated cells or by darkening. Under both conditions, the averaged rate of starch degradation is almost constant, but it is higher in illuminated than in darkened cells. At the single-cell level, rates of starch degradation largely differ but are unrelated to the initial cellular starch content. A rate equation describing the cellular starch degradation is presented. SHG-based three-dimensional reconstructions of Chlamydomonas cells containing starch granules are shown.
Introduction
Systems biology aims at modeling complex cellular and/or intercellular processes. Modeling is based on large sets of quantitative empirical data, such as the levels of metabolites, lipids, proteins, and transcripts. Validity of modeling largely depends on the quality of the empirical data. Because of the limited sensitivity of methods applied, empirical studies are frequently forced to use large cell collections. Thus, data obtained are average values that do not necessarily reflect individual cells, especially if the cell collection analyzed is heterogeneous.
Synchronization is known to increase homogeneity of a cell culture. Synchrony has been achieved for highly diverse cell or tissue types, such as prokaryotes (1), eukaryotic algae (2,3), plant cells and tissues (4,5), lower eukaryotes (6), mammalian cells (7,8), and even eukaryotic parasites of mammalian cells (9,10). A wide range of synchronization methods has been applied (8,10,11).
When photoautotrophic cells are grown under appropriate conditions in a synthetic medium lacking any reduced carbon, continuous synchrony often is achieved by a strict light-dark cycle (total length, 24 h) and dilution to a constant cell density at the beginning of each cycle using fresh culture medium (12). Under these conditions, each cell has permanent access to all nutrients (including CO2) and, averaged over time, is exposed to the same illumination.
Most eukaryotic unicellular algae undergo multiple cell divisions. The number, N, of cells formed by a single mother cell is defined by the equation , where n is the number of divisions before the release of offspring. In synchronized algal cultures, the averaged cell number increases often 10- to 24-fold (13), but these values do not reflect cell division of any real cell. Why cells form daughter cells below or above the average number of offspring is unknown. As each cell faces essentially equal external conditions, some diversity has to be assumed, but demonstration requires single-cell analyses and exceptionally sensitive methods.
Several nonlinear microscopical approaches are promising tools that, due to the high sensitivity of these instruments, may even allow high spatiotemporal resolution. Second harmonic generation (SHG) laser scanning microscopy has been used for biological imaging (14). SHG is a nonlinear optical process in which two photons possessing the same wavelength interact with the target and thereby are converted to a single photon whose energy is exactly twice that of each incident photon. SHG signals originate from a strong light field interacting with a noncentrosymmetric structure of biomolecules. The optical properties of SHG are characterized by the second-order tensor, , reflecting structural features of the target(s) (15,16). Photons generated by SHG are largely emitted in the direction of the incident photons (17).
Typically, short laser pulses are applied in the near-infrared region not absorbed by most biological samples. SHG signals are generated only near the focal point of a tightly focused laser beam, and therefore, a high local resolution is achieved. As SHG does not include molecular energy absorption, no photodamage is likely to occur (18). SHG signals have been obtained from a wide range of biological materials possessing a high intra- or intermolecular order. Both large molecules and complexes of macromolecules are potential targets, but macromolecules lacking any extended centrosymmetry or crystallinity (such as most soluble proteins) remain undetected unless SHG labels are introduced (19–21).
In mammalian tissues, SHG signals have been obtained from materials such as collagen (22,23). Starch and cellulose are known to be the most prominent SHG sources in plants (24–29). As a quantitative relative measure, SHG signal intensities appear to correlate with both the square of illumination intensity and starch amount (16,18). Thus, the optical device used for SHG measurements needs to be as stable as possible.
Native starch mainly consists of two types of polyglucans, amylose and amylopectin. Amylose comprises various essentially unbranched α-1,4-glucans. Amylopectin is usually the qualitatively and quantitatively dominant constituent defining most of the structural features of native starch. Due to the relatively high degree of branching, amylopectin is, to some extent, similar to glycogen, the most common storage polysaccharide in animals and other heterotrophic organisms (for review, see Roach et al. (30)). However, as opposed to glycogen, branching points occur as intramolecular clusters, permitting neighboring helices to form double helices that are strictly ordered and define the starch allomorph (for review, see Pérez and Bertoft (31)). Native starch exists as water-insoluble granules, which in green algae and higher plants are formed inside the plastid. The internal granule structure is largely uniform, evolutionarily conserved, and consists of alternating layers of highly ordered (unbranched double helices) and less ordered (helices enriched in branching points) materials. The highly ordered regions constitute biophotonic structures generating strong SHG signals (16,32).
Using SHG measurements, we have analyzed content and metabolism of starch in a synchronized Chlamydomonas culture at the single-cell level. In illuminated cells, starch accumulation is diverse and does not correlate with cell size. Likewise, the rate of mobilization is heterogeneous and does not reflect cellular starch levels.
Materials and Methods
Biological materials
Chlamydomonas reinhardtii strain no. CC-1690 wild-type mt+ (Sager 21gr) was obtained from the Chlamydomonas Resource Center, University of Minnesota, St. Paul, Minnesota.
Preculture and synchronization
Chlamydomonas cells were precultured under axenic conditions for 5 days at room temperature in a medium containing five macrocompounds, following the procedure of Sueoka (33), and trace elements modified according to Kuhl and Lorenzen (12). Subsequently, cells were synchronized under axenic conditions in the same medium (34°C; 12 h light/12 h dark; continuous agitation by air containing 2% (v/v) CO2, dilution to 7 × 105 cells ml−1 at the end of each dark period; illumination inside the cell culture, ∼900 and 550 μmol photons m−2 s−1 at the beginning and end, respectively, of the light period). For details, see the Supporting Material.
Ammonium measurement
Extracellular ammonium content was determined using the NANOCOLOR ammonium test (Macherey-Nagel, Düren, Germany) according to manufacturer instructions.
Quantification of cell number and cell size
Cell number and size were determined using a Beckman Coulter Counter MULTISIZER 3 (Beckman Coulter, Krefeld, Germany). Aliquots of the cell suspensions were directly diluted with the Isoton II electrolyte (Beckman) according to the instructions of the manufacturer. Measurements (based on at least 2 × 104 cells) were completed within 1 min after the addition of Isoton II. The accumulated cell volume, meaning the sum of all cellular volumes in 1 ml cell suspension, is given as relative units, setting the value at the onset of the light phase to unity. The accumulated cell volume divided by the number of cells/ml is the averaged cellular volume (procedure A).
Enzymatic starch quantification
Cells (2 ml suspension each) were centrifuged (5 min at 5000 × g), frozen in liquid N2, and stored at −80°C until use. After resuspension in 1 ml 80% (v/v) ethanol and ultrasonification (4 × 8 s each; 25% maximum power; Sonopuls, Bandelin Electronic, Berlin, Germany), homogenates were heated (10 min at 80°C) and centrifuged (10 min at 20,000 × g; 4°C), and the pellets were lyophilized. The pellets were washed with H2O (1 ml each) and then resuspended in 200 μl 0.2 N KOH each and heated (1 h at 95°C). After the addition of 35.2 μl each of 1 N acetic acid, samples were centrifuged (10 min at 10,000 × g). Aliquots of the supernatants (200 μl each) were mixed with 30 μl amyloglucosidase (Starch Assay Kit; R-Biopharm, Darmstadt, Germany) and incubated overnight at 55°C. Starch-derived glucose was enzymatically quantified (34) at 340 nm using a microplate reader (30°C; GENios, Tecan; Crailsheim, Germany).
Commercial starches
Three native maize starches (wild-type, Amioca, and Hylon VII) were provided by National Starch (Bridgewater, NJ). Amioca consists essentially of amylopectin. Hylon is a so-called high-amylose corn starch (35,36). Starches (∼100 mg each) were suspended in water (2 ml each). After washing with water (five times), suspensions were directly used for SHG measurements. For enzymatic starch quantification, the washed starch granules were further treated twice with acetone (2 ml each) and lyophilized. Aliquots (10 mg each) of the dried pellets were hydrolyzed in 0.7 N HCl (3 h at 95°C). After centrifugation, aliquots of the supernatants were used for the enzymatic glucose assay.
Fixation and destaining of the cells
Aliquots of the cell suspensions (10 ml each) were mixed with ethanol (final concentration 70% (v/v)). After 1 h at 4°C, cells were stored at −20°C for up to 5 days. Before optical analyses, the fixed cells were washed twice with H2O. Destained cells (as well as isolated starch granules) were immobilized using glass slides pretreated with Vectabond (Vectabond Laboratories, Burlingame, CA) and were covered with water (1.5 ml each). Slides were stored in a cell chamber (Minicem, Jenlab, Jena, Germany).
Vectabond modifies the surface of the glass slides and is removed before the addition of the targets. We did not notice any alteration of the targets when measured repeatedly over an extended period of time.
Optical device
The laser scanning SHG device (see Fig. S2 in the Supporting Material) consists of an inverted confocal microscope (TE2000), a scanning unit (ECLIPSE C1+; Nikon, Düsseldorf, Germany), and a Ti:Sapphire femtosecond laser (Tsunami, Spectra Physics, Darmstadt, Germany) aircoupled to the scanning unit. The laser emitted pulse trains at a central wavelength of 808 nm with an average power of 500 mW (pulse width, ∼120 fs; repetition rate, 80 MHz). The pulse width was measured behind the laser system using an autocorrelator (Type 409-08, Spectra Physics). Linearly polarized laser light was converted to circularly polarized light by a quarter-wave plate placed closely to the objective. Circular polarization was measured at an accuracy of >95% using a glan-laser polarizer and a polarization-insensitive detector (thermal power sensor S302C, Thorlabs, Dachau, Germany).
An oil immersion objective (40×; Nikon CFI Plan Fluor) with a numerical aperture (NA) of 1.3 was applied. SHG signals in the forward direction (that of the incoming light) were collected using a long-distance condenser (Nikon; NA 0.52). SHG signals were separated from the laser light by both a short-pass and a band-pass filter (BP 405/10 nm; both Semrock SP-750, Rochester, NY). The same filter set was applied for backward signals. Forward SHG signals were monitored using a photomultiplier tube (PMT) and a preamplifier unit (R1463, C7319; Hamamatsu, Herrsching am See, Germany). Voltage signals were synchronized to the focal position of the laser by using a controller of the Eclipse C1+ scanner unit. To ensure constancy, both the averaged laser power and the center wavelength (Spectrometer OSM2-400DUV, Newport, Irvine, CA) were measured throughout the experiments. The average laser power, measured behind the objective, was 10 mW (error ±2%, S302C; Thorlabs). SHG signals were normalized using a reference SHG signal derived from a LBO crystal.
SHG measurements and image analysis
The size of the scanning field was 200 × 200 μm (1024 × 1024 pixels). Pixel dwell time was 2.64 μs and step width in the z-direction was 0.5 μm. The raw image data obtained with the microscope software (Nikon) were analyzed by a routine written in MATLAB (The MathWorks, Natick, MA). For raw data reading, the Java tool loci tools was used (37). Single cells were identified in every slice. Subsequently, the stacks were used to build the holistic cell. For each cell, the volume (procedure B), the summation of SHG signals, and the cellular SHG signals based on cell volume were determined. For relative starch quantification, the procedure of Zhuo et al. (16) was used. The total starch amount/cell is referred to as relative cellular starch content (RCSC). The RCSC, based on the volume of the respective cell (determined by procedure B), is defined as cellular starch density (CSD). When calculating CSD, the volume of all destained cells was corrected for shrinking (see Results). Therefore, the values of starch density can be directly compared with the averaged cellular starch content determined by the enzyme-based starch assay.
Results
Synchronized Chlamydomonas cells
Photoautotrophic vegetative cells of C. reinhardtii strain CC-1690 wild-type mt+ were synchronized at a high productivity that is not further increased when synchronization is performed at 30°C or 32°C (data not shown). Ammonium consumption is restricted to the light period. After 12 h of growth, the medium still contains ∼75% of the initial NH4Cl content.
Before cell division, a single chloroplast occupies most of the volume of the Chlamydomonas cell. Multiple cell divisions require a coordination of DNA replication, nucleus amplification, and chloroplast fragmentation. Under standard conditions, cell divisions are restricted to the late light phase (data not shown). Release of daughter cells starts at the end of the light period and is essentially completed after 4 h in the dark period. It results in a roughly 12-fold increase in cell number/suspension volume (Fig. 1 A). The accumulated cell volume (normalized to the onset of the light period) also increases. Cells grow throughout the light period (Fig. 1 B), but in darkness, the accumulated cell volume slightly decreases.
Figure 1.
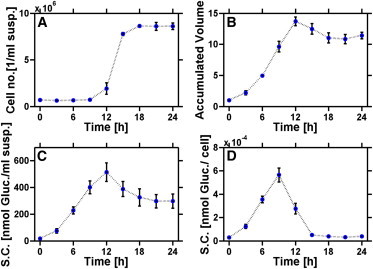
Synchronized vegetative cells of Chlamydomonas reinhardtii strain CC-1960. Cell number based on suspension volume (A), relative accumulated cell volume (B), averaged starch content based on suspension volume (C), and averaged cellular starch content (D) are plotted against time of the light-dark cycle (12 h light, 12 h darkness; standard conditions). Means of three independently performed experiments ± SD are given (n = 3).
The averaged starch levels, based on suspension volume or per cell, increase during the entire light period and exceed cellular growth (Fig. 1, B–D). In darkness, the starch content/cell massively decreases, and after 6 h darkness, the starting level of the next light-dark cycle is reached (Fig. 1 D). Decrease is caused by two processes: first, darkened cells degrade starch to maintain metabolic and biosynthetic processes, but this process is largely restricted to the first half of the dark period (Fig. 1 C). Second, the residual starch granules of the mother cell are distributed among the offspring during multiple cell divisions (Fig. 1 D).
Validation of the relative starch quantification by nonlinear microscopy
Because of the phase-matching conditions, SHG signals propagate in the direction of the incoming light (17). However, in cells, the propagation of the SHG signals could be more complex due to secondary effects, i.e., scattering (38). Therefore, scattering was tested by monitoring signals in the backward direction using destained Chlamydomonas cells. For this purpose, both the photomultiplier voltage and the integration time were adjusted to obtain an appropriate signal/noise ratio. The backward SHG signals obtained were ∼250-fold weaker compared to the forward-signal intensities. Furthermore, starch exhibits birefringence, which will lead to a decreasing SHG signal with increasing penetration depth of laser light, as also occurs with scattering of the laser light. These processes are of minor relevance in this study (for details, see the Supporting Material).
Starch analyses presented here are facilitated by features of the Chlamydomonas cells. During the first half of the light period, the unicellular alga is relatively small (50–1000 μm3; Fig. 2). Furthermore, each cell is surrounded by a large liquid volume. Photoautotrophic Chlamydomonas cells contain relatively low amounts of starch existing as several distinct starch granules. The volume of a single starch granule is more than two orders of magnitude below that of a cell. The total volume of the granules/cell does not exceed 30% of the cell volume. Therefore, SHG measurements are largely unaffected by birefringence, absorbance, and/or scattering processes inherent to optical analyses of compact tissues from mammals or higher plants consisting of several compact cell layers (see Supporting Material). Nevertheless, SHG signal intensities were validated as a relative measure of the cellular starch content by using several approaches enumerated below.
Figure 2.
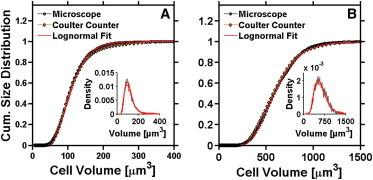
Size distribution of living and destained Chlamydomonas cells. Cell-size distributions were determined for living cells using the Coulter Counter (procedure A). Size distribution of the destained cells was measured by microscopical analyses (procedure B). Cells were measured at 0 h (A) and after 6 h illumination (B). (Insets) Size distribution of living cells and corrected distribution of destained cells.
SHG signal intensities reflect structural features of amylopectin
Three maize starches were analyzed: wild-type, waxy (largely amylose-free), and high-amylose (amylose-extender) starch. The latter contains a less frequently branched amylopectin (35,36) and represents the B-type allomorph, whereas the two others (and that from Chlamydomonas) are A-type allomorph (39–41). An altered starch allomorph has also been observed in so-called high-amylose starches from other plant species (42). SHG signal intensities from at least 300 individual starch granules were analyzed according to the procedure of Zhuo et al. (16). The sum of the signal intensities was divided by the number of particles to yield an average SHG intensity/granule. Based on light microscopical inspection, the three starches possess essentially equal size distributions.
Based on equal dry weight, hydrolysates of the three starch samples contain almost the same amount of glucosyl residues, but SHG signal intensities vary: waxy (high-amylopectin) starch is most efficient, and intensities from amylose-extender starch are lower than those from wild-type granules (Table 1). Thus, the highly ordered regions within the amylopectin are the actual source of SHG signals. The less frequently branched amylopectin of extender starch is a less efficient source (16,25,43).
Table 1.
Starch quantification by SHG signals and enzymatic starch determination using granules from wild-type maize and two mutants
| WT | Waxy | Extender | |
|---|---|---|---|
| SHG Signals (relative units) | 68 ± 3.2 | 100 ± 5 | 37.8 ± 3.6 |
| Glucose content (μmol glucose/10 mg DW) | 51.2 ± 2.7 | 50.2 ± 0.7 | 49.7 ± 3.6 |
Waxy starch consists essentially of amylopectin. Extender is a so-called high-amylose starch (see text). For glucose quantification, average values plus SD are given (n = 5). SHG signals were summed up for at least 300 particles each. WT, wild-type; DW, dry weight.
SHG signal intensities in intact and destained Chlamydomonas cells
In intact cells, SHG signals were almost undetectable because of the strong absorbance by endogenous pigments (data not shown). By contrast, high SHG signal intensities were measurable using destained cells. Treatment with ethanol (final concentration 70% (v/v)) is a simple procedure offering several advantages: Metabolic processes are immediately quenched, and starch remains unchanged (see below). Furthermore, destained cells are stable for several days at 4°C.
Destaining does not noticeably alter optical features of starch, as ensured by control experiments. Native starch granules from wild maize were suspended in water and immobilized. SHG signal intensities derived from the same starch granules were monitored before and after equilibration with 70% (v/v) ethanol and were essentially unchanged (>98% identity).
The destaining procedure inevitably reduces cell size by ∼30%, as evidenced when comparing data obtained using the Coulter counter technique (procedure A) with those obtained using microscopical analyses (procedure B). At the onset of illumination, native Chlamydomonas cells have a wide size distribution. The mean value and the width are ∼114 μm3 and 97 μm3, respectively (Fig. 2 A, inset, and Table S2). After 6 h illumination, the size distribution is shifted to a mean value of 570 μm3 with a width of 440 μm3 (Fig. 2 B, inset, and Table S2). When corrected, the size distribution of destained cells coincides with that of the living cells (Fig. 2). Similarly, the Kolmogorow-Smirnow test reveals that the corrected size distributions of destained and living cells coincide (probability at least 99%). Thus, cell-size distributions are not noticeably altered by destaining (Fig. 2).
Relative starch quantification by SHG signals matches enzymatic starch determination
To validate SHG signal intensities as a relative in situ measure of starch, aliquots of the same suspension of Chlamydomonas cells were used for both SHG signal intensity measurements and an established enzyme-based starch assay. In the first series of experiments, starch accumulation was followed during 6 h illumination.
SHG signal intensities of individual cells were collected for a large number of cells, and mean values were calculated. For the enzyme-based assay, native starch was isolated from ∼1.4 × 106 cells, and after hydrolysis, starch-derived glucose was photometrically determined. Both procedures result in a similar starch accumulation (Fig. 3 A).
Figure 3.
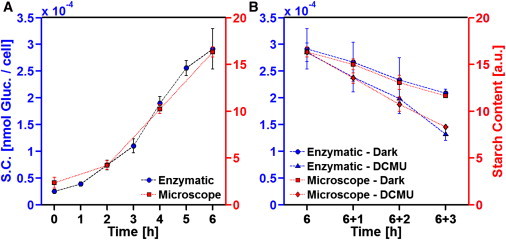
Starch accumulation (A) and degradation (B) in synchronized Chlamydomonas cells. (A) Averaged cellular starch content during 6 h illumination as determined by microscopic analysis (A; square, right axis) or by the enzyme-based assay (A; circle, left axis). Average of two independently performed experiments ± SD (n = 2) are given. (B) After 6 h illumination, starch degradation was initiated by the addition of DCMU (B; triangle and diamond) or by darkening (B; circle and square). The averaged cellular starch content was determined by microscopical analysis (B; diamond and square; right axis) or by the enzyme-based assay (B; cirlce and triangle; left axis). Means of two independently performed experiments ± SD are given (n = 2).
Subsequently, starch degradation was followed after 6 h illumination. Degradation was initiated by either adding 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; continuous illumination) or darkening of the cell suspsension. DCMU is a potent inhibitor of the photosynthetic electron transport chain. Both conditions resulted in essentially constant rates of starch degradation, but mobilization is consistently faster during DCMU treatment. Furthermore, the enzymatic starch quantification and the nonlinear microscopic approach essentially coincide (Fig. 3 B).
The data shown in Fig. 3 have important implications. The biochemical starch quantification measures the total amount of starch-derived glucosyl residues irrespective of any physical order of glucan chains within starch granules. The latter, however, is the essential cause of the SHG signals. The graphs obtained for the biochemical starch assay and the nonlinear optical approach are similar during both starch accumulation and starch degradation (Fig. 3). Because of the similarity, several conclusions can be drawn: First, during starch biosynthesis and degradation, granule features such as the amylopectin/amylose ratio or the frequency of branchings, remain largely unchanged. Second, unwanted optical processes, such as light scattering or birefringence, do not significantly affect the measurements of SHG signal intensities, as we do not see a systematic increase in deviation at higher cellular starch levels. Third, in destained Chlamydomonas cells, amylopectin is essentially the only relevant biophotonic structure. This is a safe conclusion, as during the light-dependent growth, essentially all cellular structures are amplified, but the measured SHG signal intensities are not affected. Furthermore, during prolonged darkening of living cells, both enzymatic starch quantification and SHG signal intensities decrease to the limit of detection and exhibit similar time dependency (data not shown).
Starch metabolism exhibits a large cell-to-cell diversity
Both at the onset of the light period and after 6 h illumination, the RCSC plotted against the cellular volume varies largely, and the starch-related cell-to-cell diversity exceeds that of the cell size (Fig. 4). Similarly, the scatter plot of the CSD against the cellular volume indicates a high heterogeneity (Fig. 4). At the onset of illumination, the starch-related cellular heterogeneity may even be underestimated, as the automatic cell identification is unable to identify cells lacking any detectable SHG signals.
Figure 4.
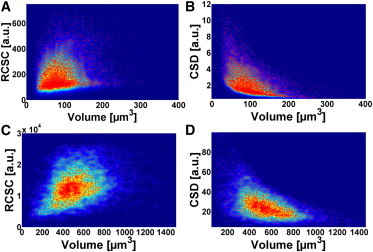
Scatter plots of the RCSC and CSD as determined by single-cell analyses. (A and B) RCSC and CSD, respectively, before the beginning of the light period. (C and D) RCSC and CSD, respectively, after 6 h illumination. For A and B, the number of cells analyzed was 7300; for C and D, the number measured was 4300 cells. Chlamydomonas cells were synchronized using the standard procedure. Warm and cold colors represent high and low densities, respectively.
During photosynthesis-driven growth, larger cells appear to accumulate less starch compared to smaller ones (Fig. 4, B and D). For a more detailed statistical analysis, we applied the Spearman formalism to test for a potential correlation between the cellular size and the relative cellular starch content. The Spearman coefficient ranges from 0 (no correlation) to an absolute value of 1 (high correlation; see Table S1). The coefficient between RCSC and cellular volume ranges from 0.08 to 0.30, thus confirming that there is no obvious correlation. As CSD includes the cellular volume, the absolute value of the correlation coefficient between CSD and cellular volume increases as expected (0.37–0.67).
When considering the cellular starch content in the population, we consistently observe lognormally distributed CSD. In a general form, a log normal distribution is given by
| (1) |
The distribution density, , is determined by the two parameters σ and μ. Using σ and μ., we obtain the standard deviation (SD) and the mean value of the lognormal distribution. The standard deviation is given by
| (2) |
whereas the expected value (arithmetic mean) reads
| (3) |
At least 4000 cells were studied for both synthesis and degradation of starch. CSD (Figs. 5 and 6 A, Fig. S3 A, and Table S3) and cellular volume (Fig. 2 and Table S2) were fitted by a lognormal distribution, and both the mean value and the width were determined.
Figure 5.
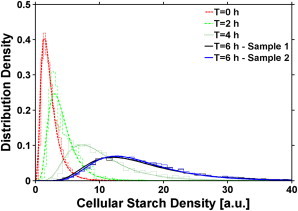
PDF of the CSD during photosynthesis-driven starch accumulation. Synchronized Chlamydomonas cells (standard conditions) were harvested at intervals during 6 h illumination. With increasing time, the PDF broadens and the mean values shift to higher values.
Figure 6.
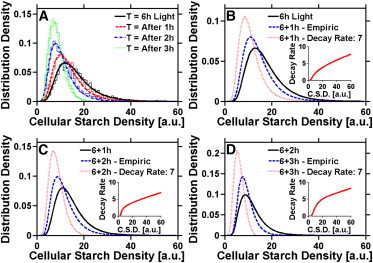
Single-cell analyses of the DCMU-induced starch degradation. (A) PDF of the CSD during starch degradation exhibits a lognormal distribution. Unlike starch accumulation, the mean value decreased with time, and a more narrow distribution was observed. (B–D) Theoretical and empirical analyses after 1 (B), 2 (C), and 3 h (D) of starch degradation. Black, CSD at the onset of the degradation (i.e., after 6 h illumination); red, distribution obtained by Eq. 4 assuming a constant degradation rate; blue, distribution obtained by using the degradation rate obtained for each interval (see inset).
Starch degradation at the single-cell level described by a rate equation
The transition from photosynthesis-driven starch biosynthesis to starch degradation largely affects the central carbon metabolism. Starch degradation induced by DCMU (Fig. 6) and by darkness (Fig. S3) is described by the simple rate equation below (Eq. 4). Based on the cellular starch content, cells were subdivided into N equidistant subpopulations (Figs. 5 and 6 A and Fig. S3 A). The term denotes the occupation number of the different subpopulations, where k indicates the range of values of cellular starch densities. The change of the occupation numbers between two consecutive time points is described by the rate function β. We emphasize that in general the degradation rate is dependent on the cellular starch content, resulting in . The master rate equation (Eq. 4) describes the regrouping of subpopulations during starch degradation:
| (4) |
By using two consecutive time points, can be determined. We consistently observed that the rate of degradation increases with the increasing CSD. The starch degradation rate is higher in DCMU-treated cells (Fig. 6, B–D, inset), than for darkened cells (Fig. S3, B–D, inset). Thus, the synchronized Chlamydomonas cells have a significant cell-to-cell heterogeneity in the rate of starch degradation.
Discussion
In this communication, SHG signal measurements have been used to study starch metabolism at the single-cell level. The nonlinear optical method was validated by several approaches. First, we quantified starch using an established enzyme-based assay and compared the results with those obtained by SHG signal measurements (Table 1 and Fig. 3). Both unrelated methods lead to largely coinciding results, and therefore, significant errors of the optical approach can be excluded. More specifically, we measured three commercial starch samples (wild-type, waxy, and amylose-extender) using both techniques. Based on equal glucose and dry weight contents, the three starches result in different SHG signal intensities (Table 1). From these data we conclude that the highly ordered regions of the wild-type and high-amylopectin starch are the essential SHG source. Amylose-extender starch is less branched, and the lower branching frequency is associated with a lowered efficiency of SHG. By contrast, the actual starch allomorph is of minor relevance. As the total starch-related glucose content and the SHG signal intensities essentially coincide in synchronized Chlamydomonas cells, it is safe to conclude that within the 6-h light period, the physical order of starch granules remains largely unchanged.
Similarly, under the conditions used, scattering of SHG does not cause any significant deviation between the enzyme-based and the optical data (Fig. 3). Within the Chlamydomonas cells, starch granules appear to be randomly distributed in the single chloroplast (Fig. S4). When performing a three-dimensional imaging of single cells, we noticed that starch particles are detected with similar sensitivity irrespective of their distance from neighboring granules (Fig. S4 and Fig. S5). This may, however, not be valid for extremely starch-rich cells, as observed under prolonged N or P deficiency. Finally, we provide clear evidence that under the conditions used, starch granules contain essentially the only biophotonic structure.
Using the nonlinear optical approach described above, a large cell-to-cell diversity was observed in a synchronized photoautotrophic culture of C. reinhardtii. A similar heterogeneity has also been observed using other Chlamydomonas strains, including cell-wall-deficient mutants (data not shown). Recloning of the Chlamydomonas strains did not diminish heterogeneity (data not shown). Therefore, the features described here are likely to be more common in isogenic unicellular algae.
Cell size and RCSC were studied in more detail. We focused on the first half of the light period. During this part of the cell cycle, no plastid or cell division is detectable, and therefore, heterogeneity is strictly attributed to an undivided organelle and/or cell. Furthermore, during this period of time, the cellular starch levels are relatively low (Figs. 1 and 3).
Both the probabilty density function (PDF) of cell size and the CSD of the synchronized Chlamydomonas culture describe a lognormal distribution (Figs. 5 and 6 A and Fig. S3 A). Importantly, RCSC and cell volume are not strictly correlated (Fig. 4 and Table S1).
Mechanistically, cell-to-cell diversity is difficult to explain. However, single-cell analyses of bacterial cells clearly reveal that isogenic daughter cells are far from being uniform. There are several reasons for diversity, most of which are functional despite genetic identity (44), including stochasticity at the levels of transcription, partitioning of transcripts, and partitioning of soluble proteins. The copy numbers of many proteins and transcripts vary largely, and for almost all protein species, the cellular copy numbers follow a gamma distribution (45). When analyzing eukaryotic algal cells, the most simple (and, perhaps, most likely) explanation for the heterogeneity of the synchronized Chlamydomonas cells is that the multiple divisions considerably expand the cell-to-cell diversity inherent in the binary division of rodlike bacteria.
When analyzing the interdependency of photosynthesis and growth, diversity has important implications: First, each cell cycle is initiated with a heterogeneous cell population. Second, despite the fact that all cells of the culture are likely to grow under essentially equal external conditions, there is a wide variation in the number of daughter cells produced by mother cells, with a range of 2–32 ((12,13), M. Sandmann, unpublished data). At the single-cell level, the number of offspring equals that of successive DNA replications and that of the divisions of the nucleus, chloroplast, and cell. Even in a synchronized culture, these closely coordinated processes differ quantitatively (and, presumably, temporarily). The factor(s) specifying the complex cell divisions remain(s) to be identified.
Starch biosynthesis covers two distinct biochemical processes, i.e., the enlargement of existing granules and the de novo formation of particles. Initiation of starch-granule formation deviates from the common path of glycogen de novo biosynthesis, but many biochemical details are not yet clear (46–48). Enlargement of granules proceeds by several closely interconnected reactions, such as the elongation of glucan chains, the conversion of linear chains into branched polyglucans, the local interaction of suitably sized α-glucans yielding double helices, and the arrangement of double helices to give a defined starch allomorph. In rapidly growing systems, such as Chlamydomonas cells, both the number and size of starch granules increase.
As opposed to rapidly growing Chlamydomonas cells, mesophyll cells of mature leaves from Arabidopsis thaliana possess a largely constant starch-granule number throughout the light-dark cycle, although the total amount of leaf starch exhibits an ∼40-fold diurnal fluctuation. Thus, starch metabolism proceeds largely (if not exclusively) by alternating periods of increasing and decreasing granule size, but the starch content/chloroplast varies widely (49).
Unlike Arabidopsis mesophyll cells, nondividing Chlamydomonas cells possess a single chloroplast, and therefore, the varying CSD (Fig. 5) indicates a heterogeneity that is related to a single compartment. RCSC and cell size are not strictly correlated (Fig. 4 and Table S1). Currently, it is not known whether the different cellular starch contents reflect largely altered intracellular carbon fluxes or different rates of photosynthetic CO2 fixation. For methodological reasons, the latter are difficult to estimate at the single-cell level.
A more detailed analysis of starch degradation was performed using cells that undergo a transition to starch degradation after 6 h of photosynthesis. Two extreme metabolic processes can be hypothesized: First, at the single-cell level, the rate of starch mobilization is uniform and totally unrelated to the cellular starch content. Mechanistically, this type of degradation could be reflected by a sequential mode in which the chloroplast contains (at least) two granule pools, i.e., a metabolically active and an inactive granule pool. The former defines the rate of starch degradation, but the cellular starch content is determined by the sum of the two pools. The alternative hypothesis is that the rate of starch mobilization and the cellular starch content are strictly correlated. Mechanistically, this mode is consistent with a highly synchronized degradation of the entire cellular starch pool but other explanations cannot be excluded.
The data presented in this communication (Fig. 6 and Fig. S3) support neither of the two modes; rather, they point to a more complex mode of degradation that requires largely unknown regulatory mechanisms. Within a single chloroplast, these mechanisms enable the cell to initiate the degradation of distinct starch granules and to define variable rates of mobilization. The data presented here have clear implications for any models that are exclusively based on bulk methods. If so, metabolic complexity is largely underestimated.
Acknowledgments
The authors are grateful to Henrike Brust for her involvement in experiments during the very early state of this study. The authors thank Ms. Carola Kuhn and Dr. Mahdi Hejazi for experimental support.
Financial support within the Systems Biology Project (GoFORSYS) by the Bundes Ministerium für Bildung und Forschung to R.M. and M.S. is gratefully acknowledged.
Footnotes
Michael Sandmann's present address is Institute of Chemistry, Department of Physical Chemistry, University of Potsdam, Potsdam-Golm, Germany.
Supporting Material
References
- 1.Ferullo D.J., Cooper D.L., Lovett S.T. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein E. Synchronous division in Chlamydomonas moewusii. Science. 1960;131:1528–1529. doi: 10.1126/science.131.3412.1528. [DOI] [PubMed] [Google Scholar]
- 3.Moriyama T., Terasawa K., Sato N. Characterization of cell-cycle-driven and light-driven gene expression in a synchronous culture system in the unicellular rhodophyte Cyanidioschyzon merolae. Microbiology. 2010;156:1730–1737. doi: 10.1099/mic.0.037754-0. [DOI] [PubMed] [Google Scholar]
- 4.Cools T., Iantcheva A., De Veylder L. A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J. 2010;64:705–714. doi: 10.1111/j.1365-313X.2010.04361.x. [DOI] [PubMed] [Google Scholar]
- 5.Riou-Khamlichi C., Huntley R., Murray J.A. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- 6.Hur J.Y., Park M.C., Park S.H. Synchronization of cell cycle of Saccharomyces cerevisiae by using a cell chip platform. Mol. Cells. 2011;32:483–488. doi: 10.1007/s10059-011-0174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schorl C., Sedivy J.M. Analysis of cell cycle phases and progression in cultured mammalian cells. Methods. 2007;41:143–150. doi: 10.1016/j.ymeth.2006.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thévoz P., Adams J.D., Soh H.T. Acoustophoretic synchronization of mammalian cells in microchannels. Anal. Chem. 2010;82:3094–3098. doi: 10.1021/ac100357u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radfar A., Méndez D., Bautista J.M. Synchronous culture of Plasmodium falciparum at high parasitemia levels. Nat. Protoc. 2009;4:1899–1915. doi: 10.1038/nprot.2009.198. [DOI] [PubMed] [Google Scholar]
- 10.Ranford-Cartwright L.C., Sinha A., Mwangi J.M. New synchronization method for Plasmodium falciparum. Malar. J. 2010;9:170. doi: 10.1186/1475-2875-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn S.-Y., Shin M.-Y., Park J.W. Magnetic separation: a highly effective method for synchronization of cultured erythrocytic Plasmodium falciparum. Parasitol. Res. 2008;102:1195–1200. doi: 10.1007/s00436-008-0893-8. [DOI] [PubMed] [Google Scholar]
- 12.Kuhl A., Lorenzen H. Handling and culturing of chlorella. In: Prescott D.M., editor. Methods in Cell Physiology. Academic Press; New York: 1964. pp. 159–166. [Google Scholar]
- 13.Vítová M., Bišová K., Cížková M. Chlamydomonas reinhardtii: duration of its cell cycle and phases at growth rates affected by light intensity. Planta. 2011;233:75–86. doi: 10.1007/s00425-010-1282-y. [DOI] [PubMed] [Google Scholar]
- 14.Zipfel W.R., Williams R.M., Webb W.W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 15.Gauderon R., Lukins P.B., Sheppard C.J.R. Optimization of second-harmonic generation microscopy. Micron. 2001;32:691–700. doi: 10.1016/s0968-4328(00)00066-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhuo Z.Y., Liao C.S., Chu S.W. Second harmonic generation imaging—a new method for unraveling molecular information of starch. J. Struct. Biol. 2010;171:88–94. doi: 10.1016/j.jsb.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Boyd R.W. 3rd ed. Elsevier; New York: 2008. Nonlinear Optics. [Google Scholar]
- 18.Sacconi L., Dombeck D.A., Webb W.W. Overcoming photodamage in second-harmonic generation microscopy: real-time optical recording of neuronal action potentials. Proc. Natl. Acad. Sci. USA. 2006;103:3124–3129. doi: 10.1073/pnas.0511338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gualtieri E.J., Guo F., Simpson G.J. Detection of membrane protein two-dimensional crystals in living cells. Biophys. J. 2011;100:207–214. doi: 10.1016/j.bpj.2010.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreaux L., Sandre O., Mertz J. Membrane imaging by second-harmonic generation microscopy. J. Opt. Soc. Am. B. 2000;17:1685–1694. [Google Scholar]
- 21.Salafsky J.S. 'SHG-labels' for detection of molecules by second harmonic generation. Chem. Phys. Lett. 2001;342:485–491. [Google Scholar]
- 22.Roth S., Freund I. Second harmonic generation in collagen. J. Chem. Phys. 1979;70:1637–1643. [Google Scholar]
- 23.Wang M., Reiser K.M., Knoesen A. Spectral moment invariant analysis of disorder in polarization-modulated second-harmonic-generation images obtained from collagen assemblies. J. Opt. Soc. Am. A –Opt. Image Sci. Vis. 2007;24:3573–3586. doi: 10.1364/josaa.24.003573. [DOI] [PubMed] [Google Scholar]
- 24.Brown R.M., Jr., Millard A.C., Campagnola P.J. Macromolecular structure of cellulose studied by second-harmonic generation imaging microscopy. Opt. Lett. 2003;28:2207–2209. doi: 10.1364/ol.28.002207. [DOI] [PubMed] [Google Scholar]
- 25.Cox G., Moreno N., Feijó J. Second-harmonic imaging of plant polysaccharides. J. Biomed. Opt. 2005;10:024013. doi: 10.1117/1.1896005. [DOI] [PubMed] [Google Scholar]
- 26.Mizutani G., Sonoda Y., Ushioda S. Detection of starch granules in a living plant by optical second harmonic microscopy. J. Lumin. 2000;87–9:824–826. [Google Scholar]
- 27.Nadiarnykh O., Lacomb R.B., Mohler W.A. Coherent and incoherent SHG in fibrillar cellulose matrices. Opt. Express. 2007;15:3348–3360. doi: 10.1364/oe.15.003348. [DOI] [PubMed] [Google Scholar]
- 28.Psilodimitrakopoulos S., Amat-Roldan I., Artigas D. Estimating the helical pitch angle of amylopectin in starch using polarization second harmonic generation microscopy. J. Opt. 2010;12:084007. [Google Scholar]
- 29.Reshak A.H. Second harmonic generation from thick leaves using the two-photon laser scanning microscope. Micron. 2009;40:455–462. doi: 10.1016/j.micron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Roach P.J., Depaoli-Roach A.A., Tagliabracci V.S. Glycogen and its metabolism: some new developments and old themes. Biochem. J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez S., Bertoft E. The molecular structures of starch components and their contribution to the architecture of starch granules: a comprehensive review. Starch/Stärke. 2010;62:389–420. [Google Scholar]
- 32.Anisha Thayil K.N., Gualda E.J., Loza-Alvarez P. Starch-based backwards SHG for in situ MEFISTO pulse characterization in multiphoton microscopy. J. Microsc. 2008;230:70–75. doi: 10.1111/j.1365-2818.2008.01956.x. [DOI] [PubMed] [Google Scholar]
- 33.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stitt M., Lilley R.M., Heldt H.W. Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol. 1989;174:518–552. [Google Scholar]
- 35.Liu F., Makhmoudova A., Tetlow I.J. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 2009;60:4423–4440. doi: 10.1093/jxb/erp297. [DOI] [PubMed] [Google Scholar]
- 36.Yao Y., Thompson D.B., Guiltinan M.J. Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiol. 2004;136:3515–3523. doi: 10.1104/pp.104.043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linkert M., Rueden C.T., Swedlow J.R. Metadata matters: access to image data in the real world. J. Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menzel R. 2nd ed. Springer; Berlin, Heidelberg: 2007. Photonics, Linear and Nonlinear Interactions of Laser Light and Matter. [Google Scholar]
- 39.Fettke J., Hejazi M., Steup M. Eukaryotic starch degradation: integration of plastidial and cytosolic pathways. J. Exp. Bot. 2009;60:2907–2922. doi: 10.1093/jxb/erp054. [DOI] [PubMed] [Google Scholar]
- 40.Gerard C., Colonna P., Planchot V. Amylolysis of maize mutant starches. J. Sci. Food Agric. 2001;81:1281–1287. [Google Scholar]
- 41.Hizukuri S. Relationship between the distribution of the chain-length of amylopectin and the crystalline-structure of starch granules. Carbohydr. Res. 1985;141:295–306. [Google Scholar]
- 42.Wei C., Xu B., Liu Q. C-type starch from high-amylose rice resistant starch granules modified by antisense RNA inhibition of starch branching enzyme. J. Agric. Food Chem. 2010;58:7383–7388. doi: 10.1021/jf100385m. [DOI] [PubMed] [Google Scholar]
- 43.Chu S.W., Chen I.H., Liu H.L. Nonlinear bio-photonic crystal effects revealed with multimodal nonlinear microscopy. J. Microsc. 2002;208:190–200. doi: 10.1046/j.1365-2818.2002.01081.x. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd-Price J., Lehtivaara M., Ribeiro A.S. Probabilistic RNA partitioning generates transient increases in the normalized variance of RNA numbers in synchronized populations of Escherichia coli. Mol. Biosyst. 2012;8:565–571. doi: 10.1039/c1mb05100h. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi Y., Choi P.J., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Hulst C., Mérida A. The priming of storage glucan synthesis from bacteria to plants: current knowledge and new developments. New Phytol. 2010;188:13–21. doi: 10.1111/j.1469-8137.2010.03361.x. [DOI] [PubMed] [Google Scholar]
- 47.Issoglio F.M., Carrizo M.E., Curtino J.A. Mechanisms of monomeric and dimeric glycogenin autoglucosylation. J. Biol. Chem. 2012;287:1955–1961. doi: 10.1074/jbc.M111.287813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeeman S.C., Tiessen A., Smith A.M. Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol. 2002;129:516–529. doi: 10.1104/pp.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crumpton-Taylor M., Grandison S., Smith A.M. Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol. 2012;158:905–916. doi: 10.1104/pp.111.186957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf M.J., Ruggles V.J., MacMasters M.M. Refractive indices of wheat starch granules at various moisture levels determined with an interference microscope. Biochim. Biophys. Acta. 1962;57:135–142. doi: 10.1016/0006-3002(62)91089-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


