Abstract
Conformational changes induced by ATP hydrolysis on actin are involved in the regulation of complex actin networks. Previous structural and biochemical data implicate the DNase I binding loop (D-loop) of actin in such nucleotide-dependent changes. Here, we investigated the structural and conformational states of the D-loop (in solution) using cysteine scanning mutagenesis and site-directed labeling. The reactivity of D-loop cysteine mutants toward acrylodan and the mobility of spin labels on these mutants do not show patterns of an α-helical structure in monomeric and filamentous actin, irrespective of the bound nucleotide. Upon transition from monomeric to filamentous actin, acrylodan emission spectra and electron paramagnetic resonance line shapes of labeled mutants are blue-shifted and more immobilized, respectively, with the central residues (residues 43–47) showing the most drastic changes. Moreover, complex electron paramagnetic resonance line shapes of spin-labeled mutants suggest several conformational states of the D-loop. Together with a new (to our knowledge) actin crystal structure that reveals the D-loop in a unique hairpin conformation, our data suggest that the D-loop equilibrates in F-actin among different conformational states irrespective of the nucleotide state of actin.
Introduction
Dynamic cycling between monomeric (globular (G)) and polymeric (filamentous (F)) actin, termed treadmilling, forms the basis of actin cytoskeleton’s remodeling (1). Actin-bound ATP is hydrolyzed mainly upon incorporation of G-actin into the filament, and after a delay, the inorganic phosphate is released (1). Consequently, the filament has an ATP-rich barbed end to which the monomers rapidly associate and an ADP-rich pointed end from which they rapidly dissociate. Numerous lines of evidence suggest that the ADP-bound filaments are more flexible (2,3) and less stable than ATP- and ADP-Pi filaments (4). The destabilization of ADP-F-actin is correlated with structural changes observed in subdomain 2 of both ADP-G-actin and ADP-F-actin, as detected by their increased proteolytic susceptibility (5–7) and fluorescent probes attached to residue 41 (8).
In addition to or through changes in the filament dynamics, ATP hydrolysis also regulates the interactions of actin with actin-binding proteins (ABPs). Intriguingly, many ABPs, including cofilin, thymosin β-4, profilin, and Arp2/3 complex, have different binding affinities toward ADP- and ATP-bound actin (reviewed in Paavilainen et al. (9)). This implies that actin undergoes conformational changes due to ATP hydrolysis. However, the nature of this structural transition is still under investigation.
A possible and attractive explanation for the differences between ATP- and ADP-bound actins emerged from the crystal structures of tetramethyl-rhodamine (TMR)-labeled (at cysteine 374) actin (10). The DNase I binding loop (D-loop) formed a short α-helix (residues 40–48) in the ADP-bound actin, but was disordered in the ATP-bound-TMR actin (11). This loop-to-helix switch could be the main conformational change between ATP- and ADP-actin. However, subsequent crystal structures of polymerization-impaired actin mutants locked the D-loop in a disordered conformation in both the ADP- and ATP-bound states (12), suggesting that the loop’s state may be influenced by local crystal contacts (12). The loop-to-helix transition in the D-loop was also observed in molecular dynamics (MD) simulations of ATP- and ADP-G-actin (13), but some simulations showed the helical state to be highly unstable in both nucleotide states (14,15). In recent studies, the helical D-loop was evaluated mainly in the filament structure (16) by coarse-grained analysis (17), MD (18), and metadynamics (19) simulations of F-actin trimers.
Sulfhydryl reactivity measurements have been extensively used to explore the secondary structures of membrane proteins such as ligand-gated ion channels (20,21), small-molecule transporters (22), and various receptors (23–26). The different reactivities of individual cysteines reflect their solvent accessibility and the deionization potential of each residue’s environment, which are a function of sequence position for α-helical or β-turn structures (27). Alternatively, one can assess the secondary structures of protein segments in solution by measuring the mobilities and accessibilities of the electron paramagnetic resonance (EPR) spin labels conjugated to them (28).
In this study, we investigated the solution structure of the D-loop in G- and F-actin in various nucleotide states using cysteine scanning mutagenesis and fluorescence and spin labeling of the mutant actins. Our results show that the D-loop is highly mobile and disordered in G-actin, irrespective of the bound nucleotide. However, the blue-shifted acrylodan (6-acryloyl-2-dimethylaminonaphthalene; Invitrogen, Eugene, OR) emission spectra and reduced spin-label mobilities on D-loop residues in F-actin point to their participation in interprotomer contacts, with the EPR spectra revealing the presence of structural substates. In addition, residue-dependent changes in the acrylodan labeling rates of F-actin in the ADP-, beryllium fluoride (BeFx)-, and phalloidin-stabilized filaments reveal site-specific differences in the D-loop that do not overlap with the predicted accessibility and pKa patterns for a helical D-loop in current filament models. Finally, we report crystallographic evidence of a new conformation of the D-loop. Altogether, our data suggest that the D-loop equilibrates among multiple conformational states regardless of the nucleotide and polymerization state of actin, and support the view of the polymorphic structural states of actin filaments (29,30).
Materials and Methods
Protein preparation and crystallization
Yeast actin mutant strains H40C/C374S (C40), Q41C/C374S (C41), G42C/C374S (C42), I43C/C374S (C43), M44C/C374S (C44), V45C/C374S (C45), G46C/C374S (C46), M47C/C374S (C47), G48C/C374A (C48), Q49C/C374A (C49), and K50C/C374A (C50) were described and characterized previously (31). Of these, mutants C40, C42, C43, C44, C45, C46, and C47 were erroneously reported to be in the C374A background (31). Yeast actin was purified on a DNase I affinity column as described previously (31) and kept in buffer A (10 mM HEPES (pH 7.4), 1 mM dithiothreitol (DTT), 0.2 mM ATP, and 0.2 mM CaCl2) on ice. To remove residual contaminants, actins used in acrylodan labeling reactions were subjected to cycling as described in the Supporting Material.
A chimera ABP was created by fusion of human gelsolin segment 1 (GS1), mouse WH2 domains of Cobl, and a pointed-end capping helix from thymosin β-4 (Supporting Material). The resulting DNA was cloned into pCold-1 vector. The chimera protein was expressed in Escherichia coli and purified as described in the Supporting Material. Fractions containing the full-length construct (apparent molecular mass on SDS-PAGE ∼ 38 kDa) and a product of its proteolytic degradation (∼34 kDa) were combined, dialyzed, mixed with excess of α-skeletal G-actin, and separated on a Superdex 75 column (Amersham Biosciences, Philadelphia, PA). Fractions containing actin and the cleaved chimera protein were used for crystallizations. Crystallization trays were set in a Mosquito crystallization robot with 2 μl drops containing a 1:1, 1:2, and 2:1 (v/v) mixture of protein solution and precipitant. Crystals were obtained in a precipitant solution of 10% (w/v) polyethyleneglycol 20000, 2% (v/v) dioxane, and 0.1 M bicine, pH 9.0, typically overnight. Data collection and structure determination were performed according to standard procedures (for details regarding the methods used, see Supporting Material and Table S1). The coordinates of the final model and the merged structure factors were deposited in the Protein Data Bank (PDB code 3TU5).
Preparation of actin in various nucleotide states
Mg-ATP-G-actin was prepared from Ca-G-actin by incubating it with 0.05 mM MgCl2 and 0.4 mM EGTA for 5 min. Mg-ADP-G-actin was prepared by incubating Ca-G-actin with 0.1 mM MgCl2, 0.4 mM EGTA, 0.4 mM ATP, 1 mM dextrose, and hexokinase (8 U/ml) for 30 min on ice.
Ca-G-actin was polymerized with 2.0 mM MgCl2 and 0.4 mM EGTA for 45 min, at 25°C. BeFx-F-actin (mimicking the ADP-Pi state of the filaments) was prepared by incubating F-actin with 0.1 mM BeCl2 and 5 mM NaF for 1 hr on ice. Phalloidin-stabilized actin filaments were prepared with a 1.1- to 1.2-fold molar excess of phalloidin over F-actin.
Fluorescence spectroscopy
To remove DTT, mutant G-actins were passed through a Sephadex G-50 column equilibrated with buffer A. Subsequently, various forms of G- and F-actins (5.0 μM) were prepared from DTT-free Ca-G-actin as described above. Before acrylodan reactivity experiments were conducted, the samples were incubated for 10 min at 25°C, but were kept on ice at all other times. Acrylodan labeling solution was prepared at 5.0 μM in dimethylformamide using a extinction coefficient of 20,000 at 391 nm (Molecular Probes Handbook). The pseudo-first-order labeling reactions were carried out at a 1:100 acrylodan/actin molar ratio and monitored via an increase in acrylodan emission at 465 nm, with the excitation wavelength set at 385 nm.
To record emission spectra of acrylodan-labeled actins, DTT-free G-actins were labeled with a 1.1- to 1.2-fold mole excess of acrylodan for 2.5 hr, at 24°C. The reactions were stopped with 1 mM DTT. Fluorescence emission spectra (400–650 nm) were recorded with the excitation wavelength set at 385 nm. All fluorescence experiments were carried out on a spectrofluorometer using FeliX32 Analysis, version 1.1 (Photon Technology International, Lawrenceville, NJ).
Site-directed spin labeling and EPR spectroscopy
Mutant G-actins were labeled as described previously (32), in buffer A without DTT, with a fivefold molar excess (over actin) of the nitroxide labeling reagent (MTSL [(1-oxy-2,2,5,5-tetramethylpyrrolinyl-3-methyl)methanethiosulfonate]) (33), creating the new R1 side chain or R1-bromo [(1-oxy-2,2,5,5-tetramethylpyrrolinyl-3-methyl-4-bromo)methanethiosulfonate]. After the unreacted reagent was removed, the sample was concentrated to 30 μM and various forms of actin were prepared. The X-band EPR spectra of 8.0 μL samples were recorded in glass capillaries using a Varian 109 spectrometer fitted with a loop gap resonator. The scan time was 30 s, with a field scan of 100 G, modulation amplitude of 1.0 G at 100 kHz, and an incident microwave power of 2.0 mW. Typically, 20–50 scans were averaged for each sample. EPR spectra were fitted to a simple isotropic model, assuming one or two spectral components, using the LabVIEW program Multicomponent (C. Altenbach, available by request).
Solvent-accessible surface area and pKa calculations of D-loop cysteine residues on actin models
To represent multiple D-loop conformers of different actin models, hybrid pdb files were created (Supporting Material). The solvent-accessible surface areas (SASA) of amino acid residues were calculated from the hybrid pdb coordinates using CCP4i (34) with a solvent probe radius of 1.4 Å. The same coordinates were uploaded to the PROPKA (35) website (http://propka.ki.ku.dk/) and the pKa values were calculated for each cysteine in the longitudinal barbed-end protomer of an actin trimer.
Results
To test for a possible transition of the D-loop of actin from a disordered state to α-helix in solution, we examined the secondary structure of this loop in G- and F-actins using previously characterized D-loop cysteine mutants (31) and spectroscopic methods. Specifically, both the reactivity of D-loop cysteines and the properties of fluorescent probes or EPR spin labels conjugated to these cysteines were analyzed for the presence of periodic patterns and compared with the predicted patterns of the SASA and pKa of D-loop residues in crystal structures of G-actin and the models of F-actin.
Cysteine mutagenesis and the characterization of D-loop mutants
All of the cysteine mutants of D-loop residues (a.a. 40–50) used in this study were used in previous work (31). As reported, mutations were done in wild-type (WT) yeast (Saccharomyces cerevisiae) actin gene, and with the exception of C44 (M44C/C374S), they all yielded viable colonies of yeast. Nucleotide (ε-ATP) exchange rates of all actin mutants were similar to the rate in WT yeast actin (31), indicating small (if any) changes in the nucleotide cleft of these actins. Almost all mutants (except for C40 and C50, which had a considerably longer lag phase than WT actin) had polymerization kinetics similar to that of WT actin (as detected by light scattering), suggesting minimal functional changes due to the mutations (31). The critical concentrations for polymerization of C40 and C50 actins were decreased with the fungal toxin phalloidin, which stabilizes actin filaments (36).
Chemical and spectroscopic scanning of D-loop in G-actin
EPR spectral line shapes of R1 side chains (R1 spin label attached to a cysteine) report on local protein environment and dynamics (on a nanosecond timescale (28,37)). An empirical parameter that can be extracted from an EPR spectrum is the inverse line width of the central resonance line (ΔH0−1) (38), which is proportional to the mobility of R1 and has been used in secondary structure predictions (37). We found that in Ca-G-actin, the mobility of R1 on all D-loop residue positions was exceptionally high, in agreement with ΔH0−1 values observed for surface loops (37,38) (Figs. 1 A and 2 A). To assess the magnitude of D-loop R1 side-chain mobility, we fitted the EPR spectra of Ca-ATP-G-actin to an isotropic model (a representative fit for C43R1 is given in Fig. 1 B) to obtain the correlation times for R1. For mutants C41, C43, and C45–C49, the spectra are a sum of two components, with rotational correlation times (τc) of 0.9 ± 0.2 ns and 2.9 ± 0.4 ns, and populations of ∼70% and 30%, respectively (Table S2). EPR spectra of the N- and C-terminal mutants C40 and C50 showed a single component with τc of 1.2 ns and 1.3 ns, respectively. To better resolve the origin of the high R1 mobility observed on the D-loop, we used a 4-bromo derivative of R1 that has internally restricted motions (39). This label also reports high mobility at the tested D-loop residues (see Fig. S1 for a representative spectrum for C45), showing that the observed mobility of R1 is not due to its side-chain motions, but results from D-loop backbone dynamics. Overall, the high spin-label mobility on all D-loop residues points to its disorder in Ca-ATP-G-actin.
Figure 1.
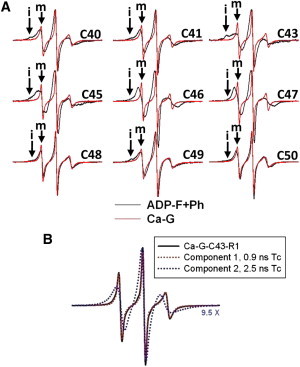
EPR spectra of R1-labeled D-loop mutants. (A) EPR line shapes of Ca-ATP-G (red traces) and phalloidin-stabilized Mg-ADP-F (black traces) actins. In each pair, the F-actin spectrum is scaled up to match the positive EPR central line height of the G-actin spectrum to emphasize the broader line shapes observed for F-actins. The scan width is 100 gauss. The mobile and immobile components of the spectra are labeled “m” and “i”, respectively. (B) Simulation of EPR spectrum for Ca-ATP bound C43R1-G-actin. The EPR spectra of monomeric C43R1-Ca-G-actin (black solid line) and individual components 1 (red dotted line overlaid with black solid line) and 2 (blue dotted line) of its nonlinear-least-squares fit are given with the corresponding rotational correlation times. Component 2 is scaled up 9.5 times to emphasize its EPR line shape.
Figure 2.
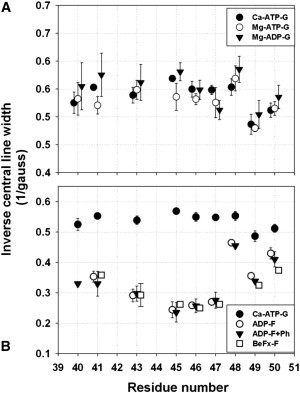
Mobility of the R1 side chain attached to D-loop residues (a.a. 40–50) in G- and F-actins in different nucleotide states. Mobility of R1 is measured by inverse central resonance line width (ΔH0−1) from the EPR spectra given in Fig. 1. ΔH0−1 values are shown for mutant G-actins bound to Ca-ATP (solid circles in A and B), Mg-ATP (open circles in A), and Mg-ADP (solid inverted triangles in A). ΔH0−1 values for F-actins in Mg-ADP-(open circles), Mg-ADP stabilized with phalloidin (solid inverted triangles), or Mg-BeFx (open squares) forms are given in B. Higher values of ΔH0−1 correspond to higher R1 mobility; values between 0.4and 0.6 are typical for R1 at surface exposed loops (38).
Nucleotide hydrolysis in monomeric actin was considered to induce a disorder→helix switch in the D-loop. In such a case, the mobility of R1 side chains on a helical D-loop of Mg-ADP-G actin would be lower (37), and the EPR spectra would reflect such a change. However, in contrast to this prediction, R1 mobility in Mg-ADP-bound mutant G-actins was slightly higher for many of the sequence positions than in Mg-ATP- and Ca-ATP- bound G-actins (Fig. 2 A). This small increase in mobility may be due to increased fluctuations of subdomain 2 upon Pi release. Clearly, the observed trend for ΔH0−1 values among D-loop positions was indicative of a disordered and mobile backbone structure, irrespective of the nucleotide state of actin. Likewise, the maximum emission wavelength of 1:1 actin/acrylodan-labeled Ca-G-actin mutants varied between 510 and 525 nm without a discernible helical pattern (i.e., reflecting a periodicity of 3.5 residues), consistent with a disordered D-loop in G-actin (Fig. 3 A and Table S3).
Figure 3.
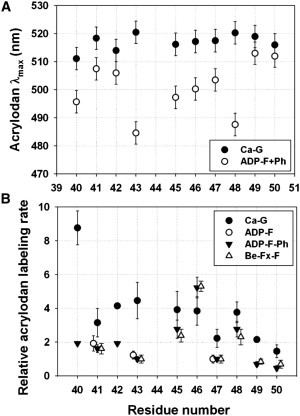
Cysteine scanning of D-loop mutants with acrylodan. (A) λmax of the fluorescence emission of acrylodan conjugated to D-loop cysteine mutants in G-actin (black circles) and F-actin (open circles) versus mutant cysteine sequence position. (B) Acrylodan-labeling rates of D-loop cysteine mutants in Ca-ATP-G-actin (black circles), BeFx-F-actin (open triangles), ADP-F-actin (open circles), and phalloidin-stabilized ADP-F-actin (inverted triangles) as a function of sequence position. Modifications by acrylodan were monitored via fluorescence increase at 462 nm. Rates of modification were determined by fitting experimental data to a single exponential rate equation. The resulting rate constants were normalized to the rate constant of C47 labeling in F-actin (0.0013 s−1).
When acrylodan emission scans or cysteine reactivity measurements were done with Mg-ATP-G or Mg-ADP-G actins, time-dependent fluorescence changes did not reach a stable plateau (data not shown). These changes result most likely from Mg-ATP-G actin forming polymerization-competent oligomeric species (40), whereas Mg-ADP-G-actin unfolds slowly after the loss of phosphate (8,41). It is also possible that the hydrophobic probe attached to the D-loop residues promotes G-actin transition to oligomeric species. Therefore, fluorescence measurements were not pursued any further with Mg-G-actins, and nucleotide-dependent changes in the D-loop of G-actin were studied only by EPR spectroscopy. EPR measurements allowed for faster data collection and did not involve potential complications due to the hydrophobicity of the acrylodan probe.
The reactivity of D-loop cysteines toward acrylodan was determined under pseudo-first-order reaction conditions (a representative plot for C43 modification by acrylodan is shown in Fig. S2). Acrylodan-labeling rates for individual D-loop mutants in Ca-ATP-G actin varied somewhat, but with no periodic pattern, indicating the absence of a regular secondary structure (Fig. 3 B). Notably, the reactivity of C40 was two- to fourfold higher, and that of C47, C49, and C50 was twofold lower compared with other D-loop mutants. Because reactivity is mainly a function of the accessibility and ionization state of the sulfhydryl, these two parameters should be taken into account in considering the observed reaction rates of D-loop substitution cysteines.
The accessibility and the deionization state of the thiols can be represented quantitatively by the SASA and pKa of a cysteine residue, respectively. However, both values depend strongly on protein conformation, and their predictions are based on a selected set of structural coordinates (34,35). Therefore, we created hybrid structure files using previously observed crystallographic and modeled conformers of the D-loop in the context of high-resolution Oda and Fujii F-actin models (42,43). Fig. 4 illustrates a single protomer from the Oda model with multiple D-loop conformers, including the one reported here for the crystal structure of actin in complex with gelsolin segment 1 (GS1) fused to a Cobl segment (PDB ID: 3TU5). It should be noted that only actin, GS1, and the first nine amino acids of Cobl WH2 domains are mapped in our structure (with an overall root mean-square deviation (RMSD) of 0.4 Å to a previous actin-GS1 structure; Fig. S3). Consequently, our structure merits attention only for its novel (to our knowledge) D-loop conformer.
Figure 4.
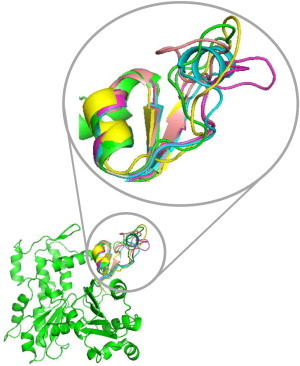
Structure of an actin protomer with various conformers of the D-loop. This figure was created using PYMOL to align subdomain 2 regions of actin monomer structures which show the D-loop in helical (1J6Z in blue), extended loop (3MFP in yellow and 2ZWH in green), and hairpin (1YAG in pink and 3TU5 (our new structure) in magenta) conformations, with a single protomer in Oda’s model of F-actin (2ZWH in green).
The predicted SASA values that were normalized (Table S4) to that of a cysteine with the highest SASA (C43, PDB: 3TU5) indicate that cysteines 49 and 50 have lower accessibility than other cysteines (considering multiple D-loop conformers), consistent with the up to twofold lower modification rates observed for these sites (Fig. 3 B). The same SASA analysis could not explain the two- to fourfold higher reactivity of C40. Therefore, we analyzed the predicted pKa values for this site (see Materials and Methods). We found that only a helical D-loop conformer has a low pKa value (∼5.0), i.e., high sulfhydryl reactivity, for C40 (Table S5). However, the higher reactivity of C40 is most likely due to the stabilization of a nucleophilic form of this cysteine’s side chain by the positively charged neighbor Arg-39, because the fraction of time that the D-loop spends as an α-helix must be relatively low considering our cysteine scanning evidence.
Chemical and spectroscopic scanning of D-loop in F-actin
The D-loop is known to form interprotomer contacts in F-actin and to regulate filament stability by its interactions with other structural elements (31,44). Therefore, R1 mobility is predicted to decrease upon formation of F-actin due to the restriction of nitroxide motions by tertiary interactions of D-loop residues with other protomers. As predicted, lower ΔH0−1 and the changes in EPR line shapes reveal a reduction of R1 mobility at all sites, in a site-specific manner (Figs. 1 A and 2 B). Qualitative and quantitative analyses of EPR spectra of R1 at positions 40, 41, 48, 49, and 50 show fast isotropic motions with a small population (∼10%) of highly immobilized components (denoted “i” for immobilized and “m” for mobile states in Fig. 1 A) in F-actin. This indicates that the main population of nitroxide spins at these positions retains substantial mobility while participating in F-actin contacts. On the other hand, C46R1 and C47R1 had quite similar EPR spectra demonstrating fast yet sterically hindered motions.
The central D-loop residues C43R1 and C45R1 had the most restricted EPR spectra (Fig. 1 A), indicating that this region may be more buried in the interprotomer space than other parts of the loop. The EPR spectra for C43R1 and C45R1 are a complex sum of immobilized and moderately restricted states (Fig. 1 A), which can be explained by the presence of multiple D-loop conformers. Indeed, when multiple conformers of D-loop are overlaid on the actin protomer in Oda’s F-actin model (42), states with varying degrees of restrictions are observed (Fig. 5). For example, the helical state may give rise to the most restricted component, whereas the hairpin or coils may lead to states with higher accessibility and mobility for C43R1. On the other hand, a helical loop would bring C45R1 to a state with average restrictions. The presence of multiple states in EPR spectrum may also arise from multiple rotamers of R1. As shown in a recent study (45), one can distinguish the possibilities of multiple loop conformers and rotamers by considering the effect of osmolytes on EPR spectra. Spectra resulting from multiple rotamers would be unchanged by the addition of osmolytes, whereas the R1 at a site undergoing conformational exchange would show increased fractions of immobilized components. As shown in Fig. S4, R1 at many sites shows clear osmolyte shifts upon addition of 1 M sucrose, suggesting that the D-loop might indeed undergo fast conformational exchange in F-actin.
Figure 5.
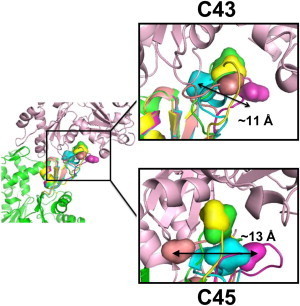
Structural substates of the D-loop are best described by the previously observed D-loop conformers. Variations in side-chain positions for C43 and C45 in different D-loop conformers are shown using Oda’s F-actin model. The color code and alignment of conformers are as described in the legend to Fig. 4. Multiple D-loop conformations are overlaid in the lower protomer (green), with C43 and C45 depicted by surface representation. The longest distance between alternative α carbon positions of C43 and C45 in D-loop conformers is given in angstroms. As shown here, individual conformers of the D-loop can selectively bury or expose a given side chain, providing an explanation for the complex EPR line shapes observed in Fig. 1A.
Notably, the EPR spectral shape and mobility of R1 attached to D-loop residues was unchanged (within error) between Mg-ADP-F and Mg-BeFx-F actin (see Fig. 2 B for mobility parameter). Because the latter is believed to represent the Mg-ADP-Pi-bound state of the filament, this indicates that the conformational fluctuations of individual D-loop residues in the filament are nucleotide independent.
Fluorescence emission spectra of acrylodan conjugated to D-loop mutants in F-actin had various degrees of blue shift and higher quantum yields compared with G-actin, indicating that the local environment of the probe is less polar at all sequence positions in F-actin (Fig. 3 A, Table S3, and Fig. S5). At residues 41, 42, 49, and 50, acrylodan shows smaller blue shifts (∼4–10 nm), suggesting that the D-loop ends participate less extensively in filament contacts. Acrylodan at positions 40, 43, and 45–48 in F-actin had 50–90% higher quantum yield and >15 nm blue shifts compared with the G-actin values, indicating probe burial due to filament contacts at these sites. The most dramatic blue shift (38 nm) in the emission spectra was observed for C43-acrylodan and C48-acrylodan (30 nm blue shift), with an accompanying 90% increase in maximum fluorescence relative to that in G-actin. Although the result for C43-acrylodan is in good agreement with the strongly immobilized states of C43-R1 observed in the EPR spectra, the C48-acrylodan result was less expected because the EPR spectrum revealed C48-R1 to be highly mobile and solvent-exposed in F-actin. Therefore, it is likely that the fluorescence result is influenced by acrylodan-driven hydrophobic interactions within the interprotomer space due to the tendency of hydrophobic fluorophores to be buried and shielded from the solvent.
Finally, acrylodan modification rates of D-loop mutants in F-actin (Fig. 3 B) did not correlate with the SASA and pKa patterns of D-loop residues modeled as a helix in current F-actin models. For example, as shown in Fig. S6, the trends of reactivity values deduced from the label incorporation rates do not overlap with the trends of predicted SASA values for current actin models incorporating a helical D-loop. As also shown in Table S6 and Table S7, the observed trends can be best explained by assuming multiple structural states of the D-loop. It should be noted that our experiments revealed site-specific differences among D-loop residues. As shown in Table S8, all mutants but C46 had an ∼2- to 5-fold slower labeling rate in F-actin than in G-actin, implying lower solvent exposure for these residues. On the other hand, the labeling rate of C46 was higher in F-actin than in G-actin, suggesting that the reactivity of this residue in F-actin is increased due to adjacent residues in the filament. Indeed, the pKa predicted for this cysteine in F-actin is lower in multiple conformers of D-loop compared with the value in G-actin (Table S5 and Table S7). Interestingly, the trend in the results of reactivity measurements was present in Mg-ADP-F, phalloidin-stabilized Mg-ADP-F, and Mg-BeFx-F (which mimics the Mg-ADP-Pi state) actins (Fig. 3 B), implying that individual D-loop residues are not strong conformational sensors of actin nucleotide hydrolysis in F-actin in these experiments.
Discussion
Our goal in this work was to clarify the structure of D-loop in G- and F-actins in solution via cysteine scanning mutagenesis and the use of spectroscopic methods to detect a sequence-dependent periodicity in short stretches of peptides. To that end, we used single cysteine mutants of D-loop and explored the reactivity of these cysteines and the properties of fluorescent and EPR spin labels attached to them. Taken together, our results provide evidence for multiple structural states of D-loop in monomeric and filamentous actin, and their relationship to the bound nucleotide. In addition, the properties of spin labels and acrylodan conjugated to the D-loop mutants demonstrate site-specific variations in the participation of D-loop residues in interprotomer contacts in F-actin.
D-loop is highly mobile and intrinsically disordered in G-actin
A helical D-loop has been used in several models of F-actin (29,46,47). Therefore, we searched for nucleotide-state-dependent structural changes in D-loop residues. Cysteine scans along the D-loop in Ca-ATP-G actin, with conjugates of R1 side chains and the fluorescent probe acrylodan, did not reveal any periodicity in the examined parameters (i.e., spin-label mobility, cysteine reactivity, and acrylodan emission parameters) as a function of sequence position. This indicates equivalent environment and solvent exposure of D-loop residues, i.e., dynamic structural disorder in the D-loop.
Our results showed only marginal differences in spin-label mobility (represented by ΔH0−1 in Fig. 2 A) and EPR spectral shapes (data not shown) among Ca-ATP-G, Mg-ATP-G, and Mg-ADP-G actin. A slower R1 mobility would be expected for the proposed helical D-loop fold of Mg-ADP-G actin than for a coil fold in Mg-ATP-G actin (16). In fact, the mobility of R1 side chain on Mg-ADP-G actin was slightly higher than for ATP-bound G-actins, perhaps due to the above-mentioned decreased stability of actin (8,41) and its subdomain 2 (4,5,48) upon phosphate release. Thus, our results are consistent with disordered D-loop states in both ATP- and ADP-bound G-actins (12).
D-loop assumes multiple structural states in F-actin
Crystal structures of G-actin have been solved >50 times (47). Strikingly, the D-loop electron density is missing, either completely or partially, in all but eight structures (actin-DNase I (49), β-actin-profilin (50), TMR-labeled ADP-actin (10), yeast actin-GS1 (51), WASP-actin-DNase I (52), actin-cytochalasin D (53), actin-twinfilin (54), and our most recently reported structure, actin-GS1-cobl (PDB: 3TU5); Fig. S3). The missing electron density is a characteristic of intrinsically disordered regions in proteins (55), which may assume a specific structure after making contacts with a binding partner (56). Our results and previous crystallographic observations suggest such a property for the D-loop in actin. Our analysis of lattice contacts in the eight resolved structures of D-loop-G-actin shows that in each case, 300–660 A2 of D-loop surface area is involved in crystal contacts. This supports the view that the D-loop is disordered in the absence of stabilizing side-chain interactions. The fact that distinct conformers of the D-loop involving α-helix, β-turns, and extended and compact hairpins are seen in the crystals further highlights the conformational plasticity of this loop.
Although the crystal lattice contacts in G-actin are not equivalent to those in F-actin, they are indicative of tertiary contacts that can be formed by D-loop residues in the interprotomer space in F-actin. Therefore, it has been reasonable to assume that ordered conformations of the D-loop, including a helical state, might exist within F-actin (16). Indeed, it was noted that a helical D-loop can be accommodated in the filament structure just as well as a coil (29,46,47). MD simulations and coarse-grained analysis (17,18) indicated that a helical D-loop might weaken the interactions between protomers, thereby resulting in a reduced persistence length of Mg-ADP-F actin. In addition, MD simulations using metadynamics algorithms suggested that folding of the D-loop into an α-helix is energetically unfavorable in Mg-ATP-F-actin as opposed to Mg-ADP-F-actin (19). In this study, the acrylodan-labeling reaction rates for D-loop residues in Mg-ADP-F, Mg-ADP-BeFx-F, and phalloidin stabilized Mg-ADP-F-actin filaments did not show helical periodicity (Fig. 3 B). Furthermore, site-specific labeling differences did not overlap with the trends projected from the SASA and pKa parameters calculated from actin models incorporating a helical D-loop (Fig. S6). However, if a helical D-loop were present in F-actin only a fraction of time, or in a fraction of F-actin, our reactivity scanning would not detect the presence of a helical state, and instead would report on the average of states, i.e., disorder. In addition to that, if the helical propensity of D-loop is indeed low, it is possible that cysteine substitutions could shift the equilibrium further toward the nonhelical states. Therefore, although our data cannot exclude the presence of a helical D-loop in F-actin, they suggest that it is not a predominant state.
Indeed, EPR data for many D-loop residues are suggestive of multiple R1 conformations, and the EPR spectral shapes are best understood by comparing D-loop conformers in a high-resolution model of F-actin (42). The structure overlay in Fig. 5 shows that a helical conformer of D-loop brings C43R1 to an immobilized state, whereas the hairpins may move it to higher mobility state that accounts for multiple spectral components (Fig. 1 A). In another example, the most restricted conformer of C45R1 would be represented by a structure observed in yeast actin-GS1 complex (1YAG), whereas the helical fold may result in an intermediate mobility level (Fig. 5). Therefore, our data indicate that the D-loop undergoes exchange among multiple conformational states. This is in line with a recent cryo-electron microscopy (cryo-EM) study in which the filaments were shown to exist in multiple states correlated with the conformations of subdomain 2 and the D-loop (29). In that work, the electron density for D-loop varied between compact and extended sizes, and was best described by helical and extended coils.
D-loop makes extensive contacts in actin filament
Actin filament models (42,57) and solution experiments including proteolysis (58) and chemical cross-linking (31,59) have shown that D-loop residues make essential contacts in the polymer. In this study, we observed a blue shift in the fluorescence of acrylodan on D-loop cysteines upon the G- to F-actin transition, indicating the burial of the probe in a more solvent-excluded environment (Fig. 3 A). Also, EPR spin-label mobilities were lower at all D-loop positions in F-actin than in G-actin (Fig. 2 B), implying reduced motions of the R1 side chains due to neighboring contacts. Overall, the best interpretation of our solution data is that all D-loop residues can be involved to a varying degree in actin interprotomer contacts or can be affected by neighbor contacts in a site-specific manner.
Three lines of evidence support the above conclusion. First, both acrylodan emission measurements and EPR data reveal that the central residues C43 and C45 are among the least solvent-accessible residues of the D-loop, suggesting their involvement in filament stabilization. Remarkably, mutation of methionine 44 to a cysteine was lethal to yeast cells, pointing to the importance of the hydrophobic central part of D-loop (31). Second, spin labels on C40 and C41 show relatively unrestricted motions in F-actin. Because N-terminal D-loop residues, and especially C40, were modeled to make important contacts with both the W-loop of an upper protomer and the H-loop from a lateral protomer (43), our spin-label mobility results show that these interactions must be very dynamic. A similar conclusion about dynamic interactions of C41 with the H-loop and C-terminus of adjacent actin protomers in F-actin was reached by Scoville et al. (32) in 2006. Lastly, based on R1 mobility and acrylodan emission properties, the C-terminal residues C49 and C50 appear to be highly solvent-exposed in the filament, implying that they make more transient bond contacts than other loop residues. This scenario provides a possible route for D-loop regulation by ABPs that bind to the side of the filament. Interestingly, a recent model of myosin-bound actin filaments suggests that myosin makes contacts with the C-terminal D-loop region of actin (60), and this is supported by biochemical evidence (61).
D-loop is a target for allosteric regulation of filament dynamics
Allosteric interactions between different structural elements in F-actin are well documented. The conformational coupling of the D-loop to the C-terminus, nucleotide-binding cleft, and N-terminus was previously shown in proteolysis (62,63) and cryo-EM (29) studies. Some of these interactions may actually arise from direct contacts among these structural elements. Most recently, the D-loop was proposed to extend to the proline-rich loop (a.a. 108–112) of actin upon outer domain rotation during ATP hydrolysis (47). In a recent study (31), we provided cross-linking evidence that all of the tested D-loop residues could approach (within a contact distance) other important loops of actin, including the H-loop, C-terminus, and W-loop. We showed that trapping these dynamic-contact-range states by disulfide bonds formed by cysteines engineered into these loops destabilized the filament in many cases. Although the cross-linking results are important for connecting the filament stability to the D-loop dynamics, they do not reveal the equilibrium distribution of this loop’s states. Our new (to our knowledge) results, which show the presence of multiple structural states of D-loop in F-actin, extend the current understanding of the conformational plasticity of this region of actin.
It has been known that the structure of yeast actin filaments is more dynamic than that of skeletal F-actin (64–66). This could imply that some differences in conformational states of D-loop in insect (12) and mammalian (10,11,13–15) skeletal actins, as well as yeast actin, are partially actin isoform specific. However, our previous molecular dynamics simulations of mammalian skeletal and yeast F-actins showed very similar dynamics in the proximities of D-loop residues to the other structurally connected interprotomer loops in these actins (31). Therefore, we expect that the actin-isoform-specific contributions to the observations presented here are rather low.
In summary, our solution experiments based on cysteine scanning mutagenesis reveal that the D-loop is disordered in G-actin in all nucleotide states and undergoes fast exchange among structural substates in F-actin. Our experimental data report on the average of such states and indicate that an α-helical D-loop can be present in F-actin for only a fraction of time. D-loop’s conformational plasticity and its shifts between different structural states, as shown here, are consistent with the cryo-EM documentation of structural polymorphism in F-actin (29,30), and provide an explanation for the essential role of D-loop in actin filament stability and function.
Acknowledgments
This work was supported by grants from the U.S. Public Health Service (GM-077190 to E.R. and EY 05216 to W.L.H.), and a Jules Stein Eye Institute core grant (EY00331) and Jules Stein Professor Endowment to W.L.H.
Footnotes
Dmitri S. Kudryashov’s present address is Department of Biochemistry, The Ohio State University, Columbus, OH.
Supporting Material
References
- 1.Carlier M.F. Measurement of Pi dissociation from actin filaments following ATP hydrolysis using a linked enzyme assay. Biochem. Biophys. Res. Commun. 1987;143:1069–1075. doi: 10.1016/0006-291x(87)90361-5. [DOI] [PubMed] [Google Scholar]
- 2.Isambert H., Venier P., Carlier M.F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 3.Orlova A., Egelman E.H. A conformational change in the actin subunit can change the flexibility of the actin filament. J. Mol. Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 4.Orlova A., Egelman E.H. Structural basis for the destabilization of F-actin by phosphate release following ATP hydrolysis. J. Mol. Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- 5.Muhlrad A., Cheung P., Reisler E. Dynamic properties of actin. Structural changes induced by beryllium fluoride. J. Biol. Chem. 1994;269:11852–11858. [PubMed] [Google Scholar]
- 6.Strzelecka-Gołaszewska H., Moraczewska J., Mossakowska M. Localization of the tightly bound divalent-cation-dependent and nucleotide-dependent conformation changes in G-actin using limited proteolytic digestion. Eur. J. Biochem. 1993;211:731–742. doi: 10.1111/j.1432-1033.1993.tb17603.x. [DOI] [PubMed] [Google Scholar]
- 7.Khaitlina S.Y., Strzelecka-Gołaszewska H. Role of the DNase-I-binding loop in dynamic properties of actin filament. Biophys. J. 2002;82:321–334. doi: 10.1016/S0006-3495(02)75397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moraczewska J., Wawro B., Strzelecka-Golaszewska H. Divalent cation-, nucleotide-, and polymerization-dependent changes in the conformation of subdomain 2 of actin. Biophys. J. 1999;77:373–385. doi: 10.1016/S0006-3495(99)76896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paavilainen V.O., Bertling E., Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Otterbein L.R., Graceffa P., Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 11.Graceffa P., Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 12.Rould M.A., Wan Q., Trybus K.M. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X., Diraviyam K., Sept D. Nucleotide effects on the structure and dynamics of actin. Biophys. J. 2007;93:1277–1283. doi: 10.1529/biophysj.107.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalhaimer P., Pollard T.D., Nolen B.J. Nucleotide-mediated conformational changes of monomeric actin and Arp3 studied by molecular dynamics simulations. J. Mol. Biol. 2008;376:166–183. doi: 10.1016/j.jmb.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Splettstoesser T., Noé F., Smith J.C. Nucleotide-dependence of G-actin conformation from multiple molecular dynamics simulations and observation of a putatively polymerization-competent superclosed state. Proteins. 2009;76:353–364. doi: 10.1002/prot.22350. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Chu J.W., Voth G.A. Allostery of actin filaments: molecular dynamics simulations and coarse-grained analysis. Proc. Natl. Acad. Sci. USA. 2005;102:13111–13116. doi: 10.1073/pnas.0503732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaendtner J., Lyman E., Voth G.A. Structure and dynamics of the actin filament. J. Mol. Biol. 2010;396:252–263. doi: 10.1016/j.jmb.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaendtner J., Branduardi D., Voth G.A. Nucleotide-dependent conformational states of actin. Proc. Natl. Acad. Sci. USA. 2009;106:12723–12728. doi: 10.1073/pnas.0902092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell J.G., Czajkowski C. Cysteine scanning mutagenesis: mapping binding sites of ligand-gated ion channels. In: Lajtha A., Johnson D., editors. Handbook of Neurochemistry and Molecular Neurobiology. Springer; New York: 2007. pp. 439–454. [Google Scholar]
- 21.Zichittella A.E., Shi H.G., Argüello J.M. Reactivity of cysteines in the transmembrane region of the Na, K-ATPase α subunit probed with Hg(2+) J. Membr. Biol. 2000;177:187–197. doi: 10.1007/s002320010002. [DOI] [PubMed] [Google Scholar]
- 22.Slotboom D.J., Konings W.N., Lolkema J.S. Cysteine-scanning mutagenesis reveals a highly amphipathic, pore-lining membrane-spanning helix in the glutamate transporter GltT. J. Biol. Chem. 2001;276:10775–10781. doi: 10.1074/jbc.M011064200. [DOI] [PubMed] [Google Scholar]
- 23.Cai K., Klein-Seetharaman J., Khorana H.G. Single-cysteine substitution mutants at amino acid positions 306-321 in rhodopsin, the sequence between the cytoplasmic end of helix VII and the palmitoylation sites: sulfhydryl reactivity and transducin activation reveal a tertiary structure. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 24.Klein-Seetharaman J., Hwa J., Khorana H.G. Single-cysteine substitution mutants at amino acid positions 55-75, the sequence connecting the cytoplasmic ends of helices I and II in rhodopsin: reactivity of the sulfhydryl groups and their derivatives identifies a tertiary structure that changes upon light-activation. Biochemistry. 1999;38:7938–7944. doi: 10.1021/bi990013t. [DOI] [PubMed] [Google Scholar]
- 25.Danielson M.A., Bass R.B., Falke J.J. Cysteine and disulfide scanning reveals a regulatory α-helix in the cytoplasmic domain of the aspartate receptor. J. Biol. Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bera A.K., Chatav M., Akabas M.H. GABA(A) receptor M2-M3 loop secondary structure and changes in accessibility during channel gating. J. Biol. Chem. 2002;277:43002–43010. doi: 10.1074/jbc.M206321200. [DOI] [PubMed] [Google Scholar]
- 27.Karlin A., Akabas M.H. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 28.Columbus L., Hubbell W.L. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 29.Galkin V.E., Orlova A., Egelman E.H. Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin V.E., Orlova A., Egelman E.H. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oztug Durer Z.A., Diraviyam K., Reisler E. F-actin structure destabilization and DNase I binding loop: fluctuations mutational cross-linking and electron microscopy analysis of loop states and effects on F-actin. J. Mol. Biol. 2010;395:544–557. doi: 10.1016/j.jmb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scoville D., Stamm J.D., Reisler E. Hydrophobic loop dynamics and actin filament stability. Biochemistry. 2006;45:13576–13584. doi: 10.1021/bi061229f. [DOI] [PubMed] [Google Scholar]
- 33.Altenbach C., Oh K.J., Hubbell W.L. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 34.Potterton E., Briggs P., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 36.Dancker P., Löw I., Wieland T. Interaction of actin with phalloidin: polymerization and stabilization of F-actin. Biochim. Biophys. Acta. 1975;400:407–414. doi: 10.1016/0005-2795(75)90196-8. [DOI] [PubMed] [Google Scholar]
- 37.Mchaourab H.S., Lietzow M.A., Hubbell W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 38.Czogalla A., Pieciul A., Sikorski A.F. Attaching a spin to a protein—site-directed spin labeling in structural biology. Acta Biochim. Pol. 2007;54:235–244. [PubMed] [Google Scholar]
- 39.Fleissner, M. R. 2007. X-ray structures of nitroxide side chains in proteins: a basis for interpreting distance measurements and dynamic studies by electron paramagnetic resonance. PhD thesis, University of California, Los Angeles.
- 40.Attri A.K., Lewis M.S., Korn E.D. The formation of actin oligomers studied by analytical ultracentrifugation. J. Biol. Chem. 1991;266:6815–6824. [PubMed] [Google Scholar]
- 41.Gershman L.C., Selden L.A., Estes J.E. Preparation and polymerization properties of monomeric ADP-actin. Biochim. Biophys. Acta. 1989;995:109–115. doi: 10.1016/0167-4838(89)90068-x. [DOI] [PubMed] [Google Scholar]
- 42.Oda T., Iwasa M., Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 43.Fujii T., Iwane A.H., Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 44.Reisler E., Egelman E.H. Actin structure and function: what we still do not understand. J. Biol. Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 45.López C.J., Fleissner M.R., Hubbell W.L. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes K.C., Angert I., Schröder R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 47.Murakami K., Yasunaga T., Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 48.Belmont L.D., Orlova A., Egelman E.H. A change in actin conformation associated with filament instability after Pi release. Proc. Natl. Acad. Sci. USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W., Mannherz H.G., Holmes K.C. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 50.Schutt C.E., Myslik J.C., Lindberg U. The structure of crystalline profilin-β-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 51.Vorobiev S., Strokopytov B., Almo S.C. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chereau D., Kerff F., Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair U.B., Joel P.B., Trybus K.M. Crystal structures of monomeric actin bound to cytochalasin D. J. Mol. Biol. 2008;384:848–864. doi: 10.1016/j.jmb.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paavilainen V.O., Oksanen E., Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunker A.K., Obradovic Z. The protein trinity—linking function and disorder. Nat. Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 56.Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmes K.C., Popp D., Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 58.Khaitlina S.Y., Moraczewska J., Strzelecka-Gołaszewska H. The actin/actin interactions involving the N-terminus of the DNase-I-binding loop are crucial for stabilization of the actin filament. Eur. J. Biochem. 1993;218:911–920. doi: 10.1111/j.1432-1033.1993.tb18447.x. [DOI] [PubMed] [Google Scholar]
- 59.Kim E., Wriggers W., Reisler E. Cross-linking constraints on F-actin structure. J. Mol. Biol. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 60.Lorenz M., Holmes K.C. The actin-myosin interface. Proc. Natl. Acad. Sci. USA. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oztug Durer Z.A., Kamal J.K., Reisler E. Myosin binding surface on actin probed by hydroxyl radical footprinting and site-directed labels. J. Mol. Biol. 2011;414:204–216. doi: 10.1016/j.jmb.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crosbie R.H., Miller C., Reisler E. Structural connectivity in actin: effect of C-terminal modifications on the properties of actin. Biophys. J. 1994;67:1957–1964. doi: 10.1016/S0006-3495(94)80678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strzelecka-Gołaszewska H., Mossakowska M., Nakayama H. Long-range conformational effects of proteolytic removal of the last three residues of actin. Biochem. J. 1995;307:527–534. doi: 10.1042/bj3070527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim E., Miller C.J., Reisler E. Polymerization and in vitro motility properties of yeast actin: a comparison with rabbit skeletal α-actin. Biochemistry. 1996;35:16566–16572. doi: 10.1021/bi9623892. [DOI] [PubMed] [Google Scholar]
- 65.Orlova A., Chen X., Egelman E.H. Modulation of yeast F-actin structure by a mutation in the nucleotide-binding cleft. J. Mol. Biol. 1997;271:235–243. doi: 10.1006/jmbi.1997.1163. [DOI] [PubMed] [Google Scholar]
- 66.Prochniewicz E., Thomas D.D. Differences in structural dynamics of muscle and yeast actin accompany differences in functional interactions with myosin. Biochemistry. 1999;38:14860–14867. doi: 10.1021/bi991343g. [DOI] [PubMed] [Google Scholar]
- 67.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Kudryashov D.S., Durer Z.A., Reisler E. Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin. Proc. Natl. Acad. Sci. USA. 2008;105:18537–18542. doi: 10.1073/pnas.0808082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laskowski R.A., Moss D.S., Thornton J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 71.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 72.McCoy A.J., Grosse-Kunstleve R.W., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 74.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr., Sweet R.M., editors. Macromolecular Crystallography, Part A. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 75.Winn M.D., Murshudov G.N., Papiz M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


