Abstract
A major challenge in neuroscience is to understand how universal behaviors, such as sensation, movement, cognition, and emotion, arise from the interactions of specific cells that are present within intricate neural networks in the brain. Dissection of such complex networks has typically relied on disturbing the activity of individual gene products, perturbing neuronal activities pharmacologically, or lesioning specific brain regions, to investigate the network’s response in a behavioral output. Though informative for many kinds of studies, these approaches are not sufficiently fine-tuned for examining the functionality of specific cells or cell classes in a spatially or temporally-restricted context. Recent advances in the field of optogenetics now enable researchers to monitor and manipulate the activity of genetically defined cell populations with the speed and precision uniquely afforded by light. Transgenic mice engineered to express optogenetic tools in a cell type-specific manner offer a powerful approach for examining the role of particular cells in discrete circuits in a defined and reproducible way. Not surprisingly then, recent years have seen substantial efforts directed towards generating transgenic mouse lines that express functionally relevant levels of optogenetic tools. In this chapter, we review the state of these efforts and consider aspects of the current technology that would benefit from additional improvement.
Keywords: transgenic mice, genetic manipulation, cell type, Cre, channelrhodopsin, halorhodopsin, archaerhodopsin, calcium indicator, voltage sensor
Introduction
Transgenic mice have been widely used in neuroscience research to facilitate the deciphering of gene and cellular functions. Perhaps the greatest advantage of using a transgenic approach in such studies is that cell population-restricted transgene expression can be achieved using specific promoters, and this restricted pattern of expression can be passed on to subsequent generations fairly reproducibly. In functional studies of the mouse brain, a variety of transgenic strategies have been used to inactivate or over-express particular genes, label specific cell populations or their subcellular compartments, and manipulate the activity or function of specific cell populations (Luo et al., 2008). For example, the strong, neuronally-restricted expression of fluorescent reporter in Thy1-EYFP mice has made possible studies of morphology, connectivity, electrophysiology, and mRNA content of a single neuron and has permitted long-term in vivo imaging of neurons (Feng et al., 2000; Micheva et al., 2010; Sugino et al., 2006). Further, using a strategy for combinatorial expression of fluorescent proteins, BrainBow mice have enabled the simultaneous mapping of projections and connectivity among multiple neurons (Livet et al., 2007). Given the wealth of information transgenic mice have yielded in past studies of neural circuits, it is not surprising that considerable efforts have been expended to establish lines in which the activity of populations of neurons can be both easily observed and reliably and reversibly manipulated.
One of the most exciting recent advances in experimental neuroscience has been the development of genetically encoded light-sensitive proteins, giving rise to the burgeoning field of optogenetics. In its broadest sense, optogenetic tools include both optical indicators of neuronal activity, such as genetically encoded calcium or voltage sensors, as well as optical actuators of neuronal activity, such as light-activated membrane channels and pumps. Although both types of tools are of intense interest to the neuroscience community, the latter group of molecules has been especially pursued given the opportunity they offer for being able to activate and inactivate particular neurons in live, behaving animals. Recent work incorporating three of these molecules, the neural-activating cation channel, channelrhodopsin-2 (ChR2) (Boyden et al., 2005; Nagel et al., 2003), the neural-silencing chloride transporter, halorhodopsin (NpHR) (Han and Boyden, 2007; Zhang et al., 2007), and the neural-silencing proton pump, archaerhodopsin (Arch) (Chow et al., 2010), has demonstrated the power of these tools to activate or silence neurons with unparalleled specificity and temporal precision on a millisecond scale. In addition, ChR2 has already been widely used in rodents to map circuits between defined neuronal populations.
Optogenetic actuators, such as ChR2, function by regulating the membrane potential of excitable cells. To generate sufficient membrane depolarization for light-induced action potentials, functional ChR2 protein must be expressed on the cell membrane at very high level or density due to the low single channel conductance. In the past, such high-level expression has routinely been achieved using strategies that rely on delivering high copy numbers of transgene to cells, such as by in utero electroporation or viral infection. With a few notable exceptions, it has proven more difficult to obtain transgenic mice that express these genetic tools both robustly and widely enough to allow for probing the functionality of a wide range of cell types. For example, although the Thy1 promoter directed sufficient ChR2 expression to investigate the cortical and olfactory circuits (Arenkiel et al., 2007; Wang et al., 2007), this promoter is sensitive to inhibitory positional effects when randomly integrated into the genome, and it lacks ubiquitous neuronal expression. Clearly, to exploit the full potential of current and future optogenetic tools for elucidating neural circuitry, transgenic lines need to be developed that will allow for high-level transgene expression in any specific cell type of interest. Recently developed Cre-dependent reporter mouse lines with the ability to robustly express a variety of opsins proffer great promise to fulfill this need.
Optogenetic indicators, such as genetically encoded calcium sensors (GECIs) (also called fluorescent calcium indicator proteins, FCIPs) and voltage sensitive fluorescent proteins (VSFPs), have had a longer history of development than the optogenetic actuators. In particular, a variety of GECIs have been engineered, based on combinations of different types of calcium-binding proteins and fluorescent proteins (Mank and Griesbeck, 2008). Major advantages of using genetically encoded optical sensors over synthetic indicators to monitor cell activity include their suitability for long-term tracking of particular cells over time, as well as their ability to target specific cell types or populations. Although recent work has improved upon the sensitivity and stability of early generation optical indicators, current versions of both GECIs and VSFPs still require very high-level expression to present changes in relative fluorescence at a sufficiently high signal-to-noise ratio (SNR). Thus, there is a continued need to improve both the SNR, and the sensitivity to sub-threshold and single spike-induced changes in calcium or voltage. Similar to optogenetic actuators, genetically encoded optical indicators are most commonly delivered through viral or DNA plasmid transduction in functional studies. The most recent versions of these indicators, such as GCaMP3 (Tian et al., 2009), possess greatly improved properties over earlier iterations, making the transgenic approach feasible for their application. Indeed, promising mouse lines that express these molecules have been developed and are currently being characterized.
In the following sections we will review common strategies for generating transgenic mice, discuss the significant progress that has been made over the past few years in developing transgenic lines that express optogenetic molecules to functional levels, and consider what improvements to current technologies are needed to allow transgenic lines to capitalize on the exceedingly powerful tools offered by optogenetics.
General transgenic approaches
There are two general strategies for expressing a transgene in a cell population-specific manner. The first is to express the transgene directly under a promoter that is active in only particular cell types. The second is to use a binary system, in which expression of the transgene is regulated by another ‘driver’ gene, whose own expression is controlled by a specific promoter.
Approaches based on a single transgenic line
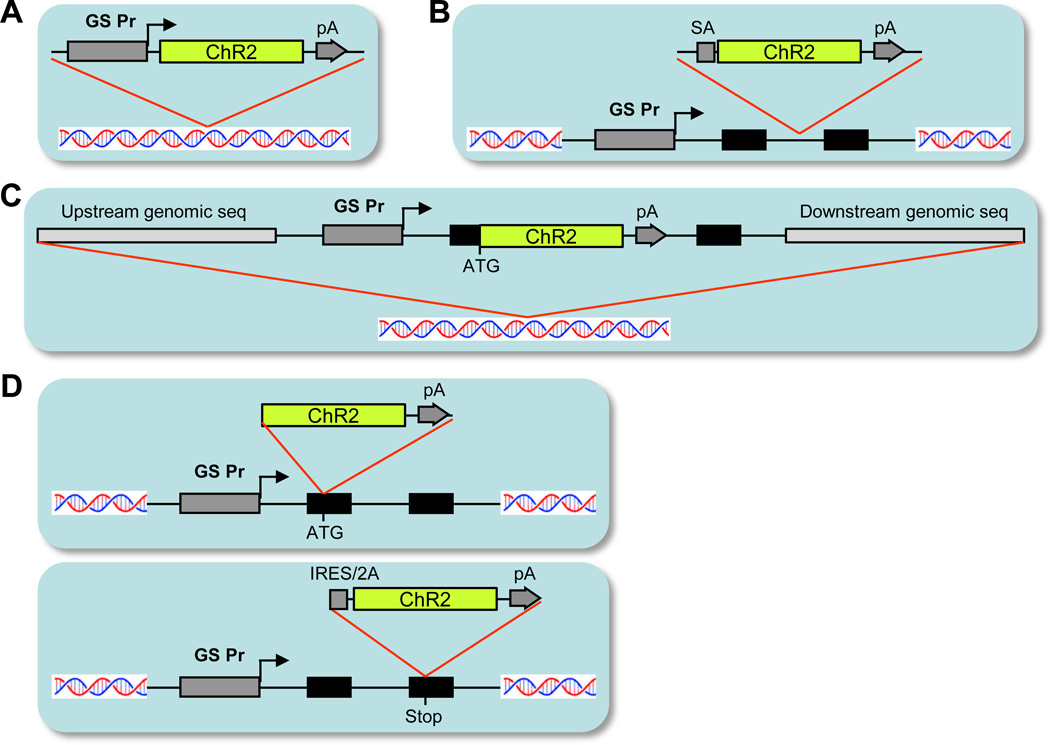
The single transgenic approach (Fig. 1) relies on one of several different methods to achieve promoter-specific transgene expression. In the simplest approach, a defined promoter that is active in a specific population of cells is directly assembled with the transgene of interest. Upon pronuclear injection into zygotic eggs, the transgenic construct randomly integrates into the mouse genome. Pronuclear injection of DNA plasmids often results in concatemerization of multiple copies of the transgenic construct and their co-integration into the same genomic locus, which can result in high-level expression of the transgene. In other instances, however, tandem arrays of co-integrated transgenes have been subject to silencing that is mediated by a heterochromatin-like complex (Henikoff, 1998). Examples of transgenic lines commonly used in neurobiology that were generated in this approach include: Thy1-YFP mice (Feng et al., 2000), Gad67-GFP mice (Ma et al., 2006), etc. A major limitation to the simple pronuclear injection of a promoter-transgene assembly is that for many genes, regulation of cell type-specific gene expression is poorly understood. Cis-acting enhancer elements that contribute to the specificity of expression can be located far away from the gene’s transcriptional start site and may not yet have been identified. In these instances, faithful recapitulation of a particular pattern of gene expression by a relatively short promoter surrounding the transcriptional start site may not be possible. Indeed, to date, there have been only a limited number of promoters successfully used to drive cell type-specific expression in neurons of transgenic mice. In addition, because these transgenes are randomly integrated, their expression can be greatly influenced by activating or inactivating positional effects, which may result in ectopic expression that is unrelated to the promoter in use or suppression of expression in relevant cells (sometimes in a mosaic manner). Although generally thought to be undesirable, positional effects and ectopic expression can occasionally direct novel and more restricted patterns of expression compared to that directed by the specific promoter.
Figure 1.
Single transgenic approaches to expressing optogenetic tools (using ChR2 as an example). (A) Conventional transgenic approach, in which an expression cassette contains a promoter and the transgene and is randomly integrated into the genome. GS Pr, gene-specific promoter. pA, polyA signal. (B) Gene trap approach, in which a promoterless cassette containing the transgene is randomly integrated into the genome, and the transgene expression is determined by a “trapped” nearby endogenous promoter. SA, splice acceptor. Black boxes indicate endogenous gene exons. (C) BAC transgenic approach, in which the transgene is inserted into the locus of the gene-of-interest contained within a BAC clone, and this BAC clone is randomly integrated into the genome. (D) Knock-in approach, in which the transgene is targeted to the endogenous locus of the gene-of-interest by homologous recombination. The targeting site can be either at the ATG start codon (upper panel), or at the STOP codon (lower panel).
A second method for attaining population-specific gene expression in transgenic mice is gene trapping. In this approach, a DNA cassette that contains an RNA splicing acceptor site, a promoterless transgene, and a polyadenylation sequence is introduced into mouse embryonic stem (ES) cells by transfection, viral infection, or transposition. Integration of the promoterless transgenic cassette into the genome is generally random, but when integrated into an intron of an expressed gene, the promoterless cassette is transcribed from the ‘trapped’ endogenous promoter. When mice are generated from a sufficiently large gene trap library (Nord et al., 2006), it is possible to obtain a variety of highly specific transgene expression patterns. On the other hand, many trapped events result in random, non-specific expression of the transgene, as the landing site may not be perfectly situated (e.g. too upstream, too downstream, in between genes, etc.) to capture the relevant, specific regulatory elements of a nearby gene. Therefore tremendous effort is needed to sort out the meaningful and useful specific patterns of expression from the random, non-specific ones.
In recent years, transgenic vectors based on bacterial artificial chromosomes (BACs) have become increasingly popular for attempting to generate patterns of restricted transgene expression. In this approach, an endogenous gene is selected as exhibiting the desired pattern of specific expression. A BAC clone that contains the selected endogenous gene near its center is identified, and a transgene of interest is inserted into the BAC, typically at the translation start site of the endogenous gene. Consequently, BAC constructs typically contain very large regions (~30–100 kb) of both 5’ and 3’ genomic sequences, which flank the specific promoter-linked transgene. As a result, they likely contain many of the cis-acting regulatory elements required to direct cell type-specific gene expression, increasing the probability that the transgene will be expressed in the same cells in which the ‘targeted’ promoter is normally active. The extremely long stretches of genomic sequence present in BAC vectors may also help buffer the transgene from activating or repressing positional effects encountered after random integration into the genome. In the large-scale BAC transgenic project, GENSAT (http://www.gensat.org/index.html), thousands of BAC-GFP mouse lines have been created in an effort to recapitulate the restricted expression patterns directed by hundreds of high-interest gene promoters (Gong et al., 2003). Characterization of GFP expression in these lines has revealed a variety of specific and nonspecific patterns, which may or may not faithfully recapitulate the endogenous gene’s expression pattern (often partially recapitulating), when compared with the Allen Mouse Brain Atlas gene expression database (http://www.brain-map.org/) (Lein et al., 2007), also as seen in a retinal cell type screening effort (Siegert et al., 2009). GFP labeling in some lines appears very restricted and is limited to a small population of either known or novel cell types. However, GFP expression in other GENSAT lines appears to be non-specific, demonstrating that significant variations in gene expression still occur when using the BAC transgenic strategy.
An alternative to the randomly-integrating transgenic or BAC transgenic approaches is the so-called site-specific transgenesis approach (Monetti et al., 2011; Tasic et al., 2011). In this approach, a docking site containing recombinase recognition site(s) (e.g. loxP, FRT, AttP, etc.) is created in a permissible genomic locus through homologous recombination, then a promoter-transgene vector or BAC-transgene vector of interest is integrated into the docking site through recombinase mediated cassette exchange (RMCE). The RMCE process can be carried out either in the ES cells or the embryos carrying the docking site through co-transfection or co-microinjection of the transgene vector and a recombinase vector. Commonly used genomic loci include the well-characterized Rosa26 and Hprt loci, as well as newly identified ones (Tasic et al., 2011). Site-specific transgenesis has the advantage of creating single-copy transgenics in a defined, consistent genomic locus, overcoming positional effect and multi-copy-induced gene silencing. On the other hand, its expression specificity is still dependent on the particular promoter fragment or BAC clone used.
Perhaps the most faithful method for generating single transgenic mice with promoter-specific gene expression is the knock-in approach. In this strategy, the transgene is inserted into a target gene’s genomic locus through homologous recombination. As a result, the transgene becomes located within the exact genomic context of the target gene, and transgene expression is controlled entirely by the target gene’s promoter and cis-acting regulatory elements. Depending on the precise experimental goals, transgenes can be knocked-in at either the target gene’s ATG start codon, for direct expression from a monocistronic transcript, or anywhere downstream of the start codon, where transgene expression is usually mediated by an IRES or 2A sequence from a bicistronic transcript. A drawback of the knock-in approach is that for transgenes inserted at the ATG, the endogenous gene locus is perturbed, leading to the disruption of the target gene’s transcription; this problem is theoretically avoided by use of an IRES or 2A sequence to place the transgene at the 3’ end of the endogenous gene. In either case, it is important to evaluate heterozygous mice for deleterious gene dosage effects, which could arise from having one wild-type and one altered allele. The utility of the knock-in approach is further limited by being inherently laborious. Each genetic marker or tool one would like to have expressed in a specific population requires isolation of a correct homologous recombination event in mouse ES cells, a task that has been difficult to realize when targeting some neuronal genes.
Regardless of the above method used to generate transgenic mice, the level and pattern of transgene expression depend tremendously on both the promoter linked to the transgene, either through design or through trapping, as well as on the genomic landscape the transgene integrates into, which may confer unpredictable transcriptional or epigenetic regulation. For this reason, transgene expression and functionality cannot be assumed; rather, they need to be carefully examined in each transgenic line.
The binary systems
The binary approach (Fig. 2) for expressing a transgene in a cell population-specific manner is based on the requirement of having two components, each essential for expression, in the same cell. Generally, these two components are brought into cells by the breeding of two separate mouse lines, which are often called driver and reporter lines. Driver lines express a master control ‘driver’ gene from a chosen specific promoter and are generated via any of the above-mentioned single transgenic approaches. Reporter lines carry the transgene of interest in a cassette, whose expression is regulated by driver gene activity. A major strength of the binary system is the flexibility conferred by having these two autonomous components combined to direct transgene expression. Assortments of unique driver and reporter lines can be generated and optimized independently of one another. Once cell type specificity is achieved in a collection of driver lines, and robust and functional expression of different genetic probes or tools is achieved in reporter lines, the lines can be merged in numerous combinations to direct a variety of cell type-specific genetic labeling methods and manipulations.
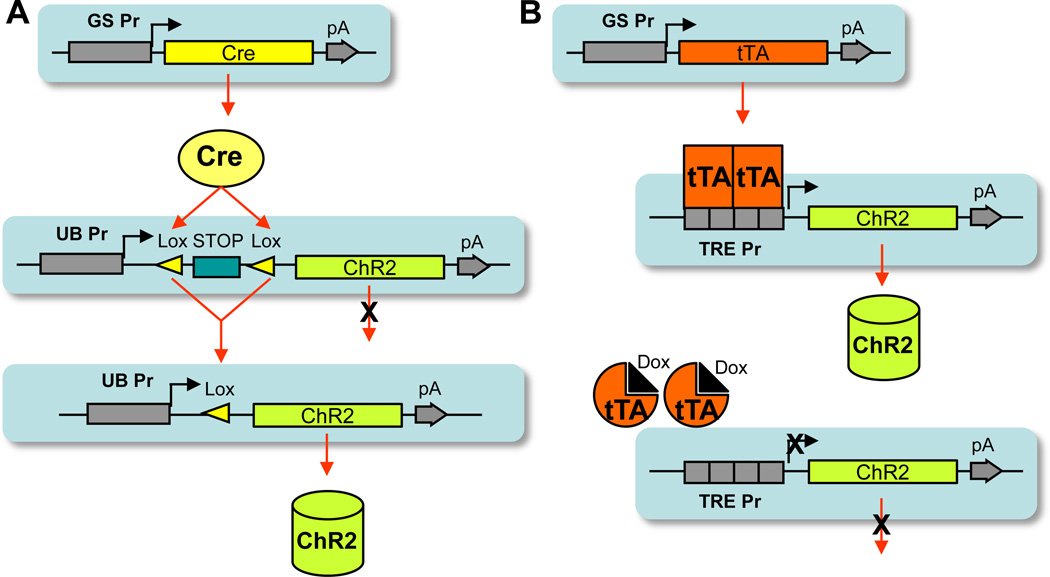
Figure 2.
The binary transgenic systems for expressing optogenetic tools (using ChR2 as an example). (A) The Cre/lox system, in which the driver line expresses Cre under the control of a gene-specific promoter, and the reporter line directs Cre-dependent expression of the transgene. Cre-mediated recombination between the two loxP sites deletes the STOP cassette and hence induces the transgene expression. UB Pr, ubiquitous promoter. (B) The Tet-inducible system, in which the driver line expresses tTA under the control of a gene-specific promoter, and the reporter line expresses the transgene under the TRE promoter. tTA binds to the TRE promoter (TRE Pr) to activate transcription of the transgene. Upon binding to tetracycline or doxycycline (Dox), tTA is released from the TRE promoter and transcription stops (bottom panel).
Binary expression systems established both in vitro and in vivo have generally incorporated one of two main kinds of driver genes: site-specific recombinases (SSRs) and transcriptional activators. The SSR Cre protein mediates recombination between loxP sites within DNA, and, due to being highly effective in mammalian cells, it is the most commonly used SSR in transgenic mice. When Cre functions as the driver in a binary system, transgene expression in the reporter line is generally initiated from a robust and ubiquitous promoter, but is then blocked by a loxP-flanked (floxed) transcriptional stop cassette located between the promoter and transgene (i.e., promoter-loxP-stop-loxP-transgene). Cell type-specific expression of the reporter occurs when Cre, produced in a limited cell population defined by its own linked promoter, mediates recombination between the two loxP sites within the reporter locus, resulting in deletion of the intervening stop cassette and activation of transgene expression. The Cre/lox recombinase system has proven extremely useful for manipulating the mouse genome in many ways, and it is used by researchers from a broad range of scientific disciplines. As a result, numerous Cre-driver mouse lines, generated in individual labs and through large-scale efforts (Gong et al., 2007; Madisen et al., 2010), have been established to drive specific gene expression in a variety of cell types or populations throughout the nervous system.
In binary approaches reliant on a transcriptional activator as the driver, the tetracycline/doxycycline-regulated tTA protein is the most commonly used activator for studies of neuronal function in the mouse brain. In this variation of the system, transgene expression in the reporter line is controlled by a TRE (Tet-regulated element)-containing promoter, which is activated by the binding of tTA dimers. In the presence of tetracycline or doxycycline, tTA dimers undergo a conformational change that prevents them from binding to the TRE. As a result, transgene expression stops. Although tTA-regulated strategies have been used successfully to direct cell type-restricted gene expression in mice, there are far fewer neuronally-specific tTA driver lines available than driver lines based on Cre recombinase. Further, in the lines that do exist, tTA has often been expressed from exceedingly strong neuronal promoters, such as αCaMKII (Mayford et al., 1996), NSE (Chen et al., 1998) and OMP (Yu et al., 2004). It remains unclear whether lower levels of tTA expression initiated from weaker promoters will be sufficient to stimulate TRE-dependent reporter expression efficiently.
Regardless of which type of driver gene is chosen, recombinase or transactivator, the binary approach requires the development of appropriate reporter lines for functionality. Ideally, the regulatable transgenic cassette within a reporter line would exhibit certain characteristics, such as having the capability being highly expressed in as many cell types as possible. An inherently strong, pancellular expression platform would be a powerful starting point from which to refine expression through driver-mediated regulation.
To date, the most commonly used locus for generating Cre-dependent responder mice is the Gt(ROSA)26Sor (Rosa26) locus, which has been shown to be a fairly permissive and ubiquitously expressed locus (Soriano, 1999). However, expression of fluorescent reporters (e.g., GFP) directly from the endogenous Rosa26 promoter (Srinivas et al., 2001) is poor in the adult mouse brain. Other Cre-dependent reporter lines have been made that include strong exogenous promoters, but since they have been integrated into random genomic loci [e.g., Z/EG (Novak et al., 2000) and BrainBow (Livet et al., 2007)], the reporters have generally not been universally expressed. The creation of the MADM (Zong et al., 2005) and mT/mG (Muzumdar et al., 2007) mice demonstrated an approach for achieving higher-level, universal expression by introducing a robust exogenous promoter into the Rosa26 locus. A further modification of this strategy, the incorporation of a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) into the reporter cassette, resulted in consistently higher expression of fluorescent proteins that efficiently label fine neuronal structures (Madisen et al., 2010).
The tTA-dependent reporter lines have been most commonly generated by random integration, which could often have positional effects. A so-called TIGRE locus, identified from a screen of hundreds of ES clones, showed reliable tTA-dependent expression with low basal activity and high inducibility (Zeng et al., 2008).
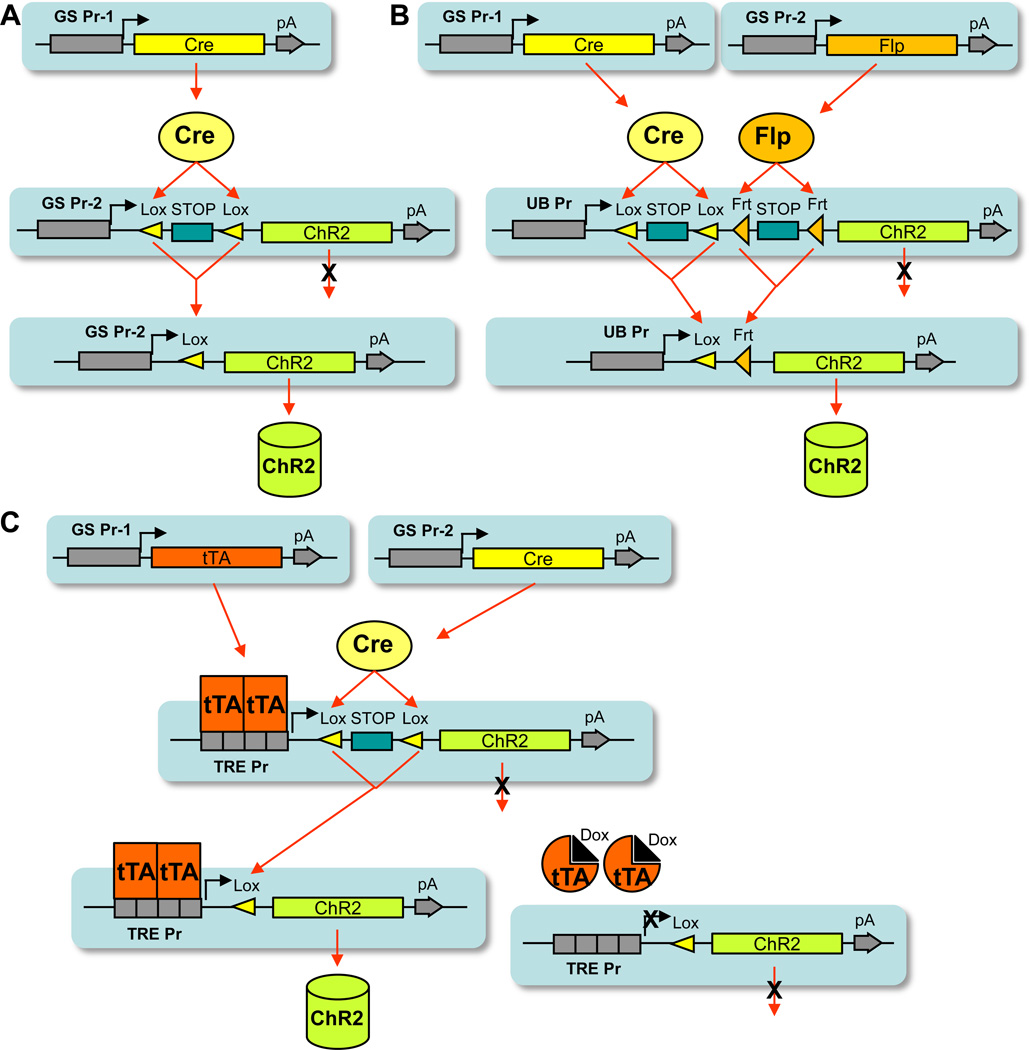
As described above, the pattern of driver gene expression largely dictates the specificity of transgene expression in the binary approach. Although Cre-driver lines are generally created using promoters or genomic loci of genes whose activity is strongest in particular kinds of cells, the promoters utilized are rarely restricted to a single neuronal population in one brain region. More frequently, the promoters are active in several brain regions in either similar or different types of neurons. As a result, it has been difficult to create driver lines that are altogether precise in marking a single cell type. Increased specificity of transgene expression may be achieved by employing an intersectional strategy of regulation. There are a variety of approaches (Fig. 3) that could be implemented in such a strategy, the simplest being substitution of the ubiquitous promoter, upstream of the floxed-stop cassette in the reporter line, with a cell type-specific promoter. Another possible approach incorporates the requirement for two different drivers, which are expressed in overlapping cell populations, to trigger reporter expression. For example, the Cre and Flp SSRs can be combined to control a ‘double reporter’ line that carries dual stop cassettes (promoter-loxP-stop-loxP-Frt-stop-Frt) (Farago et al., 2006). A similar approach can be taken using different kinds of driver genes, such as Cre and tTA. In this case, transgene expression in the ‘double reporter’ line would be regulated by a TRE-containing promoter in combination with a loxP-stop-loxP cassette. Expression of the two driver genes in these strategies could be regulated by two different cell type-specific promoters; alternatively, one of the two drivers could be delivered virally, in a region-specific manner.
Figure 3.
The intersectional approaches to expressing optogenetic tools (using ChR2 as an example) to higher specificity. (A) A simple intersectional approach, in which the driver line uses the gene-specific promoter 1 (GS Pr-1) and the reporter line uses the gene-specific promoter 2 (GS Pr-2). (B) A Cre/Flp dual recombinase intersectional approach, in which the Cre driver line uses gene-specific promoter 1, and the Flp driver line uses gene-specific promoter 2. The double reporter line is both Cre and Flp dependent. (C) A Cre/tTA intersectional approach, in which the tTA driver line uses gene-specific promoter 1, and the Cre driver line uses genespecific promoter 2. The double reporter uses the TRE promoter and is also Cre dependent. HZ - 23
Transgenic expression of optogenetic tools
Successful application of optogenetic tools to in vivo studies requires very high-level expression of these genes in the cells whose activity is to be manipulated. For this reason, investigators have typically expressed optogenetic proteins by methods that deliver high copy numbers of transgenes to target cells, such as with recombinant viral vectors, or by in utero electroporation (Zhang et al., 2010). Although these strategies can effectively achieve sufficient transgene expression for functional studies, they also have encumbering limitations. Both methods suffer from directing incomplete coverage and variable transgene expression across cells in targeted regions of a single animal. In addition, due to inherent variability between treated animal subjects, use of these methods often necessitates laborious validation of transgene expression for each animal, and still the comparison and interpretation of data between animals can be difficult. To circumvent these complications, it seems essential to develop transgenic lines that carry genetically-encoded actuators and indicators in cassettes whose expression is both tightly-regulatable and highly-inducible. The last ten years has seen considerable efforts put towards this task, and progress has been made. Yet, for a number of reasons, expression of optogenetic molecules at levels sufficient for function in transgenic mice remains challenging until recently. In the following sections, we will review the current state of transgenic mouse lines that express these genetically encoded light-responsive proteins.
Transgenic expression of optical actuators
Individual opsin molecules induce relatively small changes in membrane polarization following photostimulation. Because of this, initial attempts to develop mice that express effective levels of the optical activating molecule, channelrhodopsin-2, in specific neuronal cell populations were based on directly linking the ChR2 gene to very strong cell type-specific promoters, such as those from Thy1 (Arenkiel et al., 2007; Wang et al., 2007), Vglut2 (Hagglund et al., 2010), Omp (Dhawale et al., 2010) and Chat (Ren et al., 2011), or to the Tet-inducible promoter (Chuhma et al., 2011). Characterization of these mice revealed that membrane depolarization and spiking activity were evoked in predicted cell types following blue light stimulation. In contrast, expression of optical inhibitory molecules in mice has been more problematic, mainly due to protein aggregation and low current conductance (Chuhma et al., 2011; Zhao et al., 2008). To date, there has been only one report of functional NpHR expression in transgenic mice, using the Orexin promoter (Tsunematsu et al., 2011). Efforts to overcome limitations associated with early versions of the silencing opsins have led to the development of second- and third-generation optical silencing molecules, including eNpHR (Gradinaru et al., 2008), eNpHR3.0 (Gradinaru et al., 2010), various forms of Arch (Chow et al., 2010), and ArchT (Han et al., 2011). With greatly improved membrane expression and photoconductance, these reengineered opsins embody a major step toward reliable genetic silencing. Below we provide an overview of the published transgenic mouse lines expressing optical activators or silencers (see Table 1 for a summary).
Table 1.
Transgenic mouse lines expressing optical activators or silencers
| Name | Method of generation |
Promoter used | Cell type examined | Light power required to evoke spiking |
Peak photocurrent | Reference |
|---|---|---|---|---|---|---|
| Thy1-ChR2-EYFP (line 18) | Conventional transgenic | Thy1.2 | Cortical pyramidal neurons | 0.2 mW/mm2 | 500–600 pA under ~10 mW/mm2 light | (Wang et al., 2007) |
| MOB mitral cells | Unknown | ~200 pA | (Arenkiel et al., 2007) | |||
| Omp-ChR2-EYFP (line ORC-M) | Conventional transgenic | Omp | MOB mitral cells (postsynaptic to the ChR2-expressing OSNs) | <2 mW/mm2 to activate postsynaptic cells | Unknown | (Dhawale et al., 2010) |
| Vglut2-ChR2-YFP | BAC transgenic | Vglut2 | Spinal cord Vglut2-positive neurons | Unable to evoke spiking (35 mW/mm2 light only induces depolarization) | Unknown | (Hagglund et al., 2010) |
| Chat-ChR2-EYFP | BAC transgenic | Chat | MHb CHAT+ neurons | 20 mW/mm2 | ~500 pA under 20 mW/mm2 light | (Ren et al., 2011) |
| Mrgprd-ChR2-Venus | Knock-in | Mrgprd | DRG Mrgprd+ neurons | Unknown | Unknown | (Wang and Zylka, 2009) |
| BTR (bi-directional tetO promoter driven ChR2-mCherry on one side and HaloR-EGFP on the other side) (line BTR6) | Conventional transgenic, line BTR6 crossed to aCaMKII-tTA | tetO | Dorsal striatal medium spiny neurons | Unknown | ~350 pA for ChR2 (HaloR aggregated, not functional) | (Chuhma et al., 2011) |
| Thy1-NpHR-YFP | Conventional transgenic | Thy1.2 | Hippocampal or cortical pyramidal neurons (NpHR-YFP formed bright intracellular blebs) | 186 mW/mm2 to suppress spiking completely | ~20 pA under 23 mW/mm2 light ~−5 mV under 23 mW/mm2 light |
(Zhao et al., 2008) |
| Orexin-Halo-GFP | Conventional transgenic | Human prepro-orexin promoter | Hypothalamic orexin neurons | 4 mW light through objective lens to suppress spiking | ~6 pA under 4 mW light ~−11 mV under 4 mW light |
(Tsunematsu et al., 2011) |
| R26::ChR2(H134R)-EGFP | Knock-in to the Rosa26 locus | CAG promoter (with floxed-Neo-stop cassette) | Cortical interneurons | 2 mW laser light (20 ms duration) | ~190 pA under 2 mW, 20 ms laser light | (Katzel et al., 2011) |
| Ai27 (ChR2H134R-tdTomato) | Knock-in to the Rosa26 locus | CAG promoter (with floxed-stop cassette and WPRE) | Cortical pyramidal neurons | (Madisen et al., submitted) | ||
| Ai32 (ChR2H134R-EYFP) | Knock-in to the Rosa26 locus | CAG promoter (with floxed-stop cassette and WPRE) | Cortical pyramidal neurons | (Madisen et al., submitted) | ||
| Ai35 (Arch-EGFP-ER2) | Knock-in to the Rosa26 locus | CAG promoter (with floxed-stop cassette and WPRE) | Cortical pyramidal neurons | (Madisen et al., submitted) | ||
| Ai39 (eNpHR3.0-EYFP) | Knock-in to the Rosa26 locus | CAG promoter (with floxed-stop cassette and WPRE) | Cortical pyramidal neurons | (Madisen et al., submitted) |
The first ChR2 transgenic mouse line was made using the Thy1 promoter, which had been shown to enable extremely high-level brain expression (Caroni, 1997; Feng et al., 2000), and it demonstrated for the first time the in vivo potential of ChR2 to investigate neural circuit properties (Arenkiel et al., 2007; Wang et al., 2007). The Thy1-ChR2-EYFP construct was randomly integrated into the mouse genome via pronuclear injection, and several transgenic lines with differential levels and patterns of expression were described. In the cortical pyramidal neurons of line 18 mice (Wang et al., 2007), which showed the highest CNS expression of ChR2, maximal peak photocurrents of 500–600 pA were reached with large-area-applied blue light pulses (~10 mW/mm2 of 5–10 ms duration). Action potentials could be induced by light as low as 0.2 mW/mm2, with ~6 ms average latency from light onset. Action potentials fired reliably following light pulses up to 30 Hz. These photoexcitation properties are similar to those of ChR2-positive neurons generated by viral transduction or in utero electorportion. In line 9 mice, a two-dimensional array light scanning method over the surface of a cell was used to map local synaptic inputs to ChR2-negative cells. Mapping data revealed that excitatory synaptic input maps to pyramidal neurons were qualitatively different from those to interneurons, with the former being much larger and more irregularly shaped. Furthermore, it was demonstrated for the first time that ChR2-positive cortical layer 5 cells can be activated in vivo by light on a millisecond scale (Arenkiel et al., 2007). Brief light pulses (3.5 ms), delivered through a 200-µm diameter optical fiber positioned directly above the surface of the brain, reliably evoked responses from individual neurons with an average of ~10 ms spike latency. Neurons followed spike trains at high fidelity at frequencies as high as 40 Hz.
In the olfactory bulb of line 18 Thy1-ChR2-EYFP mice (Arenkiel et al., 2007), only mitral cells have ChR2 expression, and in these cells photocurrents (mean of ~200 pA) and action potentials could be reliably induced by blue light in vitro. Illuminating the dorsal surface of the olfactory bulb could induce spiking in mitral cells. A circuit mapping study was conducted by stimulating mitral cells in the bulb and recording postsynaptic responses in piriform cortex. Large area (600 µm) light stimuli on the bulb drove the firing of piriform neurons much more effectively than smaller area (300 µm and 100 µm) light stimuli, supporting the model that multiple distant mitral cells or glomerular inputs are required for the activation of piriform cortical cells.
The 12-kb Omp promoter, another strong neuronal promoter, drives expression in all olfactory sensory neurons (OSNs), and several Omp-ChR2-EYFP transgenic mouse lines have been generated by pronuclear injection (Dhawale et al., 2010). Two transgenic lines had ChR2-EYFP expression restricted to the vomeronasal organ and the accessory olfactory bulb. A third line (termed ORC-M) that expressed ChR2-EYFP in the olfactory epithelium and the main olfactory bulb, as well as in the vomeronasal organ and accessory olfactory bulb, was used for study. Light stimulation in the glomerular layer activated ChR2 in the OSN axon terminals, which led to glutamate release and generated postsynaptic currents in the mitral cells. Light stimulation over the olfactory bulb surface induced a rapid and reliable increase of firing of the mitral/tufted cells through presynaptic activation (as opposed to the direct activation observed using cells from the Thy1-ChR2-EYFP mice above). Titration of light intensity (to lower than 2 mW/mm2) enabled mapping of a single glomerulus to each recorded single-unit mitral or tufted cell. Tetrad recordings coupled with light stimulation identified neighboring mitral/tufted cell pairs that are either sister cells (i.e., cells receiving inputs from the same glomerulus) or non-sister cells (i.e., cells receiving inputs from different glomeruli). It was found that sister mitral cells showed correlated changes in their firing rate, whereas non-sister cells did not. Intriguingly, however, odor presentation desynchronized sister cells, suggesting that the odor response properties of sister cells are not redundant and could be differentially affected by local circuitry.
The Vglut2-ChR2-YFP transgenic mouse line was generated using a BAC transgenic strategy in which human codon-optimized hChR2-YFP was inserted in frame at the start codon of Vglut2 (also known as Slc17a6) in the BAC (Hagglund et al., 2010). In these mice ChR2 is specifically expressed in Vglut2 expressing glutamatergic neurons, including those in the hindbrain and spinal cord, with variable intensities. Spinal cord ChR2-positive neurons responded to continuous blue light (~35 mW/mm2) with rapid depolarization of variable degrees (1–15 mV). The relatively small and variable depolarization might be related to the low and variable expression of ChR2 in these transgenic mice. Nonetheless, light stimulation of the glutamatergic neurons in the spinal cord or the hindbrain from the ventral side was sufficient to generate rhythmic locomotor-like activities, demonstrating the direct involvement of these glutamatergic neurons in intrinsic rhythm generation.
The Chat-ChR2-EYFP mouse line was also generated using the BAC transgenic strategy (Ren et al., 2011). It was used to study the cholinergic transmission in the habenulo-interpeduncular pathway. ChR2-EYFP fluorescence was found in the ventral two-thirds of the medial habenula (MHb) where CHAT+ cells are located, as well as the entire MHb-fr-IPN axon projection tract. Brief (5 ms) or continuous blue light pulses (20 mW/mm2) could evoke rapid action potential firing and large photocurrents (~500 pA peak amplitude) in MHb neurons. Light stimulation of the axonal terminals in the projection target area, interpeduncular nucleus (IPN), elicited postsynaptic responses in IPN neurons tested. Brief (5 ms) light pulses evoked fast EPSCs that were blocked only by glutamate antagonists. Prolonged light stimulation evoked an additional component, the slow inward currents that were not affected by glutamate antagonists but were reduced by nAchR blockers to about one-third of their original levels, thus revealing the dual transmission nature of the MHb cholinergic neurons.
The Mrgprd-ChR2-Venus knock-in mice were generated by inserting the mammalian codon-optimized ChR2(H134R)-Venus gene into the Mrgprd gene in frame at the start codon through homologous recombination (Wang and Zylka, 2009). Mrgprd molecularly marks ~75% of all IB4+ nonpeptidergic nociceptive neurons in the dorsal root ganglia (DRGs) and trigeminal ganglia, and these Mrgprd+ neurons exclusively innervate skin and terminate in lamina II (the substantia gelatinosa or SG) of dorsal spinal cord. Light stimulation evoked action potentials in 68% heterogyzous and 94% homozygous Mrgprd-ChR2-Venus+ DRG cells in dissociated culture. The latency between light onset and actional potential peak was ~20 ms, and the spike jitter (average standard deviation of the light-evoked spike latency from each neuron) was ~1.3 ms. In spinal cord slices, light-evoked excitatory postsynaptic currents (EPSCL) could be generated in ~50% of SG neurons. Using spike jitter and pre-post synaptic proximity as criteria, the connections between the Mrgprd-ChR2-Venus+ DRG cells and these light responsive SG neurons were classified into monosynaptic or polysynaptic. And it was found that monosynaptic connections made up of ~50% of the light responsive SG cells and included almost all known SG cell types except for the islet cells.
Conditional expression of ChR2 using the Tet system was achieved in the BTR mice (Chuhma et al., 2011), in which a bi-directional tetO (also known as TRE) promoter was used to drive ChR2-mCherry in one direction and HaloR-EGFP (HaloR being the same as NpHR) in the other direction. After pronuclear injection, 3 founder lines were obtain and crossed to a line of αCaMKII-tTA with striatal tTA expression restricted to medium spiny neurons (MSNs). Only one of the 3 crosses (αCaMKIIa-tTA::BTR6) had ChR2-mCherry expression, and was subsequently used in the study. Within the dorsal striatum, <10% MSNs were fluorescently labeled and they were randomly distributed in a mosaic pattern. While mCherry fluorescence was seen in the membrane, outlining the cell soma, and extended into the processes, EGFP accumulated in aggresomes in MSN cell bodies and was not seen in the plasma membrane, suggesting that ChR2, but not HaloR, would be functional. While yellow light illumination elicited marginal HaloR responses, blue light illumination evoked large ChR2 photocurrents (~350 pA) and depolarization (~30 mV). ChR2 activation was therefore used to map functional connections among different cell types in both specificity and strength (as measured in the size of IPSCs). Photostimulation of MSN presynaptic terminals expressing ChR2 elicits GABAA synaptic responses from local collaterals in the dorsal striatum (dStr), as well as in their projections to globus pallidus (GP) and substantia nigra (SN). It was found that MSN synaptic connections are quite specific. In the dStr, MSNs connect robustly to other MSNs and less robustly to tonically active neurons, but not to fast-spiking interneurons. In the GP, MSNs connect strongly to type B/C neurons and practically not to type A neurons. In the SN, MSNs make their strongest connections with SNr GABAergic neurons, but no connections with SNc dopamine neurons.
Cre dependent conditional expression of ChR2 was utilized in the creation of a floxed-STOP ChR2 mouse line (R26::ChR2-EGFP) (Katzel et al., 2011), in which a mammalian codon-optimized ChR2(H134R)-EGFP driven by the CMV early enhancer/chicken β-actin (CAG) promoter and a floxed-STOP cassette was targeted to the Rosa26 locus by homologous recombination. After Cre-mediated excision of the floxed-STOP cassette, recombination brings the ChR2-EGFP coding sequence in frame with an initiating ATG in the loxP site, resulting in translation of ChR2-EGFP with an 11-amino-acid N-terminal ‘loxP tag’ (MYAIRSYELAT). After crossing to a Gad2-ires-CreERT2 mouse line, ChR2-EGFP was expressed in all main subclasses of cortical GABAergic interneurons, but undetectable in αCaMKII-positive pyramidal neurons, following tamoxifen induction. The relatively low level expression of ChR2 required the use of homozygous mice that were fed with retinol, as well as longer stimulating laser light pulses (20 ms, 2 mW) to evoke spiking than those used to activate virally transduced or in utero transfected neurons. In these ChR2-positive cortical interneurons, under individual light pulses average peak photocurrents were ~190 pA, and average spike latency was ~16 ms. The light pulses were only able to elicit spiking in perisomatic areas, not in dendritic or axonal arborizations. Optical raster stimulation was used to map synaptic inputs from ChR2-positive interneurons to ChR2-negative pyramidal neurons in three cortical regions (M1, S1 and V1). It was found that the most common circuit motif is the lateral, intralaminar inhibition, supporting the view that inhibition is largely local, intralaminar and uniform across areas. However, rarer translaminar inhibitory inputs to subsets of layer 2/3 or layer 5 pyramidal neurons were also identified, predominantly in area V1.
The first functional Halo (NpHR) transgenic mice (orexin/Halo) were generated using the 3.2 kb human prepro-orexin promoter by pronuclear injection (Tsunematsu et al., 2011). Three founder lines showed sufficiently strong expression of Halo, and among these, line 5 showed the highest expression rate (94% of orexin neurons) and was used for the study. Interestingly, orexin neurons expressing Halo did not show blebbing or other features indicative of inappropriate trafficking. Maximum orange light illumination (586 nm, 4 mW) generated hyperpolarization of ~11 mV and photocurrents of ~6 pA in orexin neurons, and was able to inhibit either spontaneous or depolarizing current injection-induced action potential firing. Acute inhibition of orexin neurons in vivo using orange LED light through optical fibers results in time-of-day-dependent induction of slow-wave sleep and in reduced firing rate of dorsal raphe neurons in an efferent projection site.
We have created four new mouse lines with high-level and Cre-dependent expression of ChR2(H134R) (Nagel et al., 2005) fused to either tdTomato (Ai27; ChR2(H134R)-tdTomato) or EYFP (Ai32; ChR2(H134R)-EYFP), as well as a modified version of Arch (Ai35; Arch-EGFPER2) (Chow et al., 2010) or eNpHR3.0 (Ai39; eNpHR3.0-EYFP) (Gradinaru et al., 2010). These lines were generated using a transgenic expression strategy previously employed to generate a set of fluorescent reporter lines (Madisen et al., 2010), in which the expression cassette, which includes (from left to right) the CAG promoter, the floxed-STOP cassette, the transgene, and a WPRE sequence, was targeted to the Rosa26 locus by homologous recombination. After crossing to several Cre lines, including Emx1-Cre, Camk2a-CreERT2, Chat-Cre and Pvalb-ires-Cre, all optogenetic reporter genes were found to be strongly expressed, by in situ hybridization (ISH) and native fluorescence, in Cre-defined areas and cell types. Peak photocurrents reached nA range in both Ai27 and Ai32 ChR2 mice and 100–300 pA range in Ai35 Arch-ER2 and Ai39 eNpHR3.0 mice (unpublished data). The yet-to-published results demonstrate that robust, selective optical activation and silencing can be achieved in different neuronal cell types from different brain regions in all these mice, using a variety of photo-stimulation paradigms both in vitro on brain slices and in vivo in awake, behaving animals.
Transgenic expression of optical indicators
Genetically encoded calcium indicators (GECIs) offer unique opportunities to study calcium dynamics and cell signaling under a variety of physiologically relevant conditions. Since their inception, GECIs have undergone several rounds of optimization and are being increasingly used to monitor neuronal activities. In most applications, the GECIs have been produced in cells using viral approaches. Although several groups were successful in creating transgenic lines that expressed early-generation GECIs in non-neuronal tissues (Hara et al., 2004; Ji et al., 2004), the first mouse lines to express a functional GECI in the brain were two in which the indicators [inverse pericam (IP) and camgaroo-2 (Cg-2)] were transcribed from TRE-containing promoters following mating to the αCaMKII-tTA mouse line (Hasan et al., 2004). Two different transgenic lines designed to express GCaMP2 in the brain were also generated and have been used to monitor the activities of different cell populations, including cerebellar granule cells (Diez-Garcia et al., 2007; Diez-Garcia et al., 2005), and cells in the olfactory system (Chaigneau et al., 2007; He et al., 2008). A troponin C-based sensor, CerTN-L15, was also expressed under the Thy1 promoter in transgenic mice, and exhibited Ca2+ responses in cortical pyramidal neurons both in vitro and in vivo (Heim et al., 2007). Elucidation of the crystal structure of G-CaMP2 led to rationally designed changes in the molecule, resulting in GCaMP3, which demonstrates increased brightness, stability, and dynamic range (Tian et al., 2009) as compared to G-CaMP2. No transgenic lines have yet been published that describe expression of GCaMP3 in any cell type. However, we’ve recently generated a Cre-dependent GCaMP3 reporter mouse that utilizes the same Rosa26-based expression strategy we’ve successfully implemented for expressing other genetic tools. Preliminary data indicate that large calcium transients were observed with neuronal firing (Zariwala et al., unpublished data). Below we provide an overview of the published transgenic mouse lines expressing GECIs (see Table 2 for a summary).
Table 2.
Transgenic mouse lines expressing genetically encoded calcium indicators
| Name | Method of generation |
Promoter used |
Cell type examined | ΔF/F (or ΔR/R) | Reference |
|---|---|---|---|---|---|
| Ptetbi-Cg2/luc (lines MTH-Cg2-14 and MTH-Cg2-19) | Conventional transgenic, crossed to αCaMKII-tTA | tetO | Hippocampal or cortical pyramidal neurons | ~4% (wide-field) or ~10% (2-photon) in response to short trains of action potentials, 2–8% (wide-field) or 20–100% (2-photon) in response to synaptic stimulation |
(Hasan et al., 2004) |
| Ptetbi-IP/luc (line MTH-IP-12) | Conventional transgenic, crossed to αCaMKII-tTA | tetO | Hippocampal or cortical pyramidal neurons | −2% (wide-field) or −30% (2-photon) in response to synaptic stimulation | (Hasan et al., 2004) |
| Retina whole mount | −10% (2-photon) in response to light | ||||
| Olfactory bulb in vivo | −8% (wide-field) in response to odor | ||||
| pCAGGS-YC3.60pm | Conventional transgenic | CAGGS | Hippocampal CA1 region | 2–3% (wide-field) in response to synaptic stimulation | (Nagai et al., 2004) |
| Kv3.1-G-CaMP2 (line 846) | Conventional transgenic | Kv3.1 | Cerebellar granule cells | 5–25% (wide-field at 60×) or ~50% (2-photon) in response to 10 stimuli at 100 Hz | (Diez-Garcia et al., 2007; Diez-Garcia et al., 2005) |
| tetO-G-CaMP2 | Conventional transgenic, crossed to OMP-ires-tTA | tetO | VNO olfactory sensory neurons | 20% to >100% (2-photon) in response to pheromone stimuli | (He et al., 2008) |
| Thy1-CerTN-L15 (line C) | Conventional transgenic | Thy1.2 | Cortical layer 2/3 pyramidal neurons | ~4% (2-photon) per action potential, 30–60% (2-photon) in response to iontophoretic glutamate stimulations | (Heim et al., 2007) |
| Ai38 (GCaMP3) | Knock-in to the Rosa26 locus | CAG promoter (with floxed-stop cassette and WPRE) | Cortical layer 2/3 neurons | (Zariwala et al., in revision) |
Among the first transgenics to express a GECI in the brain were two series of mice that carried randomly integrated cassettes for Tet-inducible expression of camgaroo-2 (Cg-2) or inverse pericam (IP) (Hasan et al., 2004). Following mating to αCaMKII-tTA mice, many low-expressing lines exhibited punctate fluorescence inside cell bodies, while a few high-expressing lines showed more uniform cellular fluorescence. Functional Ca2+ responses in some of the highly-expressing lines were demonstrated in cells from several tissues. In hippocampal and cortical slices, short trains of stimuli evoked fluorescence changes (ΔF/F) in both Cg-2 and IP mice in the range of 2–8% by wide-field (WF) microscopy (with a CCD camera) and 10–100% by 2-photon (2P) microscopy. In retina light-evoked fluorescence changes were seen in whole-mount preps of IP-expressing mice (10% 2P), whereas no changes were detected in preps of Cg-2 mice. Finally, in vivo imaging of the olfactory bulb showed odor-evoked fluorescence changes in both Cg-2 and IP-expressing mice (1–8% WF).
An improved version of the indicator yellow cameleon, YC3.60, created by using a circularly permuted YFP (cpYFP), exhibited a larger dynamic range and SNR in biochemical assays than previous versions of the molecule (Nagai et al., 2004). A transgenic mouse line was made in which a modified CAG promoter (CAGGS) was used to drive expression of membrane-localized YC3.60. Upon tetanic stimulation of the Schaffer collateral/commissural pathway in these mice, a significant increase in FRET signal ([Ca2+]) was evoked in area CA1 (ΔF/F of 2–3% by WF microscopy), and oscillatory [Ca2+] in area DG (ΔF/F of 1–2% by WF microscopy). However, it was noted that the dynamic range of the indicator in cells of the CNS of these mice was greatly reduced from what had been observed in in vitro studies.
The Kv3.1 potassium channel promoter was used to direct expression of G-CaMP2 in a defined subpopulation of neurons of the transgenic mouse line 846 (Diez-Garcia et al., 2005). In the cerebellar cortex, G-CaMP2 was expressed exclusively in granule cells, where it reported presynaptic Ca2+ signals. In cerebellar slices, electrical stimulation in the molecular layer induced an increase in fluorescence in a beam-like area along the parallel fibers (ΔF/F of ~0.14% for a single stimulus, ~3% for 8 stimuli, and ~4% for 30 stimuli, WF microscopy). Stimulation at the granular layer induced both a local response and a beam-like response in the molecular layer. At high magnification (60×) in which brightest fluorescence was better localized, stimulations (10 pulses at 100 Hz) at either the molecular layer (antidromic activation) or the granular layer (orthodromic activation) both resulted in 5–25% ΔF/F in the target areas.
In subsequent studies on a subline of 846 (846HB) (Diez-Garcia et al., 2007), direct stimulation (10 pulses at 100 Hz) of parallel fibers in the molecular layer of cerebellar slices evoked a ~50% ΔF/F through 2-photon laser-scanning microscopy, which was larger than what was obtained with 1-photon laser-scanning microscopy (~30% ΔF/F). Ca2+ signals were also detected in the cerebellar molecular layer in vivo by both whole-field and 2-photon fluorescence imaging. Stimulations of parallel fibers in the molecular layer (10 pulses at 100 Hz) induced a ~3% whole-field fluorescence change that was clearly distinguishable from wild-type mice, as well as up to 50% ΔF/F by 2-photon imaging across responsive areas. Furthermore, using these transgenic mice, Ca2+ transients in the parallel fibers demonstrated presynatically-expressed long-term plasticity (both preLTP and preLTD) at the PF- Purkinje neuron synapses (Qiu and Knopfel, 2007, 2009).
The Kv3.1-G-CaMP2 mouse line (846) has also been used in studies of the olfactory system, where G-CaMP2 was shown to be expressed in the mitral cells, tufted cells, and some juxtaglomerular cells in the olfactory bulb (Chaigneau et al., 2007; Fletcher et al., 2009). Two-photon imaging detected odor induced Ca2+ responses in the glomeruli that were odor specific, concentration dependent, and that could be blocked by superfusion of glutamate receptor antagonists. Glomeruli Ca2+ signals reflected activation of multiple mitral cells synchronized during population bursts, and stimulation of individual external tufted (ET) cells could drive population bursts of mitral cells within the same glomerulus (De Saint Jan et al., 2009). These mice were employed to establish an odor-evoked sensory map with single glomerulus resolution, which reflected exclusively the activity of olfactory bulb neurons postsynaptic to sensory afferents (Fletcher et al., 2009). In these G-CaMP2-based postsynaptic odor maps, different odorants activated distinct but overlapping sets of glomeruli. Increasing odor concentration increased both the response amplitude of individual glomeruli as well as the total number of activated glomeruli. Furthermore, the G-CaMP2 response displayed a fast time course that enabled analysis of the temporal dynamics of odor maps over consecutive sniff cycles.
In another transgenic mouse line that employed G-CaMP2, the indicator was expressed from the Tet-inducible promoter (He et al., 2008). Following mating of this line with the OMP-ires-tTA line, G-CaMP2 expression was restricted to the olfactory sensory neurons in both the main olfactory epithelium and the vomeronasal organ (VNO). In VNO slices, diluted female or male urine samples evoked large Ca2+ transients (ΔF/F of 20% to >100%) in individual VNO neurons by 2-photon imaging. Diverse combinatorial activation patterns of VNO neurons were observed in response to gender, strain or individual specific pheromone stimuli.
Troponin C (TnC), the Ca2+ sensor protein in skeletal and cardiac muscle, was used as the basis to engineer a modified calcium sensor named CerTN-L15. Transgenic mice were generated to express CerTN-L15 under the Thy1 promoter (Heim et al., 2007). In transgenic line C, which had the highest level expression of the transgene, the indicator was widely expressed in many types of neurons, most prominently in the pyramidal neurons of the hippocampus and the neocortex. In cortical slices through 2-photon imaging, although fluorescence changes caused by single action potentials were not reliably detected, brief trains of multiple action potentials evoked clear changes in the ratio of Citrine/Cerulean fluorescence (ΔR/R), with a linear relationship between the number of action potentials and the ΔR/R (extrapolated ΔR/R = ~4% per action potential). Iontophoretic glutamate applications in layer 2/3 neurons of the visual cortex in vivo evoked cellular Ca2+ signals that were similar to those evoked in slices, as well as dendritic Ca2+ transients with ~49% ΔR/R. The TnC-based sensors may be advantageous compared to those based on calmodulin (CaM), in that CaM interacts extensively with other intracellular neuronal proteins, which could make the in vivo functionality of CaM-based Ca2+ sensors unpredictable.
Future directions
Tremendous progress has been made towards the goal of creating functionally relevant transgenic mice for optogenetic studies. The characterization of early-generation transgenic lines carrying light-activatable molecules has provided insight as to the prerequisites for an effective transgenic approach to applying these tools. Key among these are the requirement for targeted cells to be more light sensitive and to exhibit more rapid on/off kinetics. Increased light sensitivity of targeted cells can be achieved through a combination of reengineering current opsin molecules to have an increased response per light unit and improving current transgene expression strategies. Targeted cells that could be triggered by lower light conditions would be less vulnerable to both short and long-term photo damage. At the same time, since cells located in deeper tissue layers would be more reactive to low-light stimulation, any particular amount of applied light would have the potential to recruit a larger group of cells for the study. Increased light sensitivity is also often associated with a shorter latency of response (i.e., more rapid onset). For optical actuators, shorter latency could enable distinguishing direct light-induced activation or silencing from secondary effects mediated by synaptic transmission. For optical indicators, more rapid onset could facilitate faster tracking of spiking events.
Recently a number of new optogenetic variants with novel or superior light-response properties have emerged, including the activating opsin molecules CatCh (Kleinlogel et al., 2011), ChR2-ET/TC (Berndt et al., 2011), and C1V1 (Yizhar et al., 2011); the inhibitory opsin molecules ArchT (Han et al., 2011) and Halo57 (E. Boyden, personal communications); the genetically encoded calcium indicators GCaMP5 and GCaMP6 (L. Looger, personal communications); and the voltage sensitive fluorescent protein VSFP-Butterfly (T. Knopfel, personal communications). Continued efforts will be needed to assess the functionality of these new variants in a transgenic context. Also important is the development of methods aimed at increasing the overall level of transgene expression. Next generation transgenics may be based on stronger transcriptional promoters, whether ubiquitous or cell type-specific, or on novel genomic loci that enable highly stable and highly inducible expression of inserted exogenous genes. The latter would be especially beneficial for application of the tTA system in mice, where precise and non-leaky control of TRE-promoter activity has been difficult to achieve. TRE-promoter cassettes have often not been well expressed in mice and are prone to undergo chromatin silencing modifications over time. Targeting TRE-promoter-driven reporter genes to genomic loci less susceptible to epigenetic modification might increase the utility of the tTA approach for transgenesis [e.g., (Zeng et al., 2008)].
Elucidating the role individual components play in a complex neural circuit will require the ability to monitor and manipulate cell activity with extreme specificity. Expression strategies that rely on combinatorial or intersectional regulation should make possible the refined cell type-specific and region-specific transgene expression patterns required. Double reporter lines that demand multiple events to induce optogenetic molecule expression need to be generated, as will driver lines based on ‘drivers’ other than Cre recombinase. In some instances, the ability to limit optogenetic protein activity to particular cellular compartments (such as axons, dendrites, or soma) will be essential for understanding neuronal function. Such specificity can be attained by fusing known subcellular targeting domains with optogenetic molecules (Lewis et al., 2011; Lewis et al., 2009). Further, the use of sculpted light for photostimulation can also improve the specificity of cell activation, independent of the strategy for transgene expression (Andrasfalvy et al., 2010; Papagiakoumou et al., 2010).
Finally, a researcher’s dream is to be able to use a combined optogenetic approach to activate and silence different cell types at the same time while monitoring all cell activities. We can work on steps towards this goal. By generating a plethora of transgenic tools, people can pick and choose the combination to manipulate different cell types. With continued efforts in genetic engineering, protein engineering and optical/electronic engineering, we may not need to wait long to realize this dream.
Acknowledgements
This work was funded by the Allen Institute for Brain Science and NIH grants (MH085500, DA028298) to H.Z. The authors wish to thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support.
Contributor Information
Hongkui Zeng, Allen Institute for Brain Science, 551 N 34th Street, Seattle, WA 98103, USA, Tel: (206) 548-7104, Fax: (206) 548-7083, hongkuiz@alleninstitute.org.
Linda Madisen, Allen Institute for Brain Science, 551 N 34th Street, Seattle, WA 98103, USA, Tel: (206) 548-7069, Fax: (206) 548-7083, lindam@alleninstitute.org.
References
- Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, Oertner TG. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecondtimescale, genetically targeted optical control of neural activity. Nature neuroscience. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. Journal of neuroscience methods. 1997;71:3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]
- Chaigneau E, Tiret P, Lecoq J, Ducros M, Knopfel T, Charpak S. The relationship between blood flow and neuronal activity in the rodent olfactory bulb. J Neurosci. 2007;27:6452–6460. doi: 10.1523/JNEUROSCI.3141-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kelz MB, Zeng G, Sakai N, Steffen C, Shockett PE, Picciotto MR, Duman RS, Nestler EJ. Transgenic animals with inducible, targeted gene expression in brain. Molecular pharmacology. 1998;54:495–503. doi: 10.1124/mol.54.3.495. [DOI] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R, Rayport S. Functional connectome of the striatal medium spiny neuron. J Neurosci. 2011;31:1183–1192. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Saint Jan D, Hirnet D, Westbrook GL, Charpak S. External tufted cells drive the output of olfactory bulb glomeruli. J Neurosci. 2009;29:2043–2052. doi: 10.1523/JNEUROSCI.5317-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF. Nonredundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nature neuroscience. 2010;13:1404–1412. doi: 10.1038/nn.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Garcia J, Akemann W, Knopfel T. In vivo calcium imaging from genetically specified target cells in mouse cerebellum. NeuroImage. 2007;34:859–869. doi: 10.1016/j.neuroimage.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Diez-Garcia J, Matsushita S, Mutoh H, Nakai J, Ohkura M, Yokoyama J, Dimitrov D, Knopfel T. Activation of cerebellar parallel fibers monitored in transgenic mice expressing a fluorescent Ca2+ indicator protein. The European journal of neuroscience. 2005;22:627–635. doi: 10.1111/j.1460-9568.2005.04250.x. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fletcher ML, Masurkar AV, Xing J, Imamura F, Xiong W, Nagayama S, Mutoh H, Greer CA, Knopfel T, Chen WR. Optical imaging of postsynaptic odor representation in the glomerular layer of the mouse olfactory bulb. Journal of neurophysiology. 2009;102:817–830. doi: 10.1152/jn.00020.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain cell biology. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund M, Borgius L, Dougherty KJ, Kiehn O. Activation of groups of excitatory neurons in the mammalian spinal cord or hindbrain evokes locomotion. Nature neuroscience. 2010;13:246–252. doi: 10.1038/nn.2482. [DOI] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chow BY, Zhou HH, Klapoetke NC, Chuong A, Rajimehr R, Yang A, Baratta MV, Winkle J, Desimone R, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in systems neuroscience. 2011;5:1. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Bindokas V, Lopez JP, Kaihara K, Landa LR, Jr, Harbeck M, Roe MW. Imaging endoplasmic reticulum calcium with a fluorescent biosensor in transgenic mice. American journal of physiology. 2004;287:C932–938. doi: 10.1152/ajpcell.00151.2004. [DOI] [PubMed] [Google Scholar]
- Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, et al. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS biology. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ma L, Kim S, Nakai J, Yu CR. Encoding gender and individual information in the mouse vomeronasal organ. Science (New York, NY. 2008;320:535–538. doi: 10.1126/science.1154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nature methods. 2007;4:127–129. doi: 10.1038/nmeth1009. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Ji G, Feldman ME, Deng KY, Greene KS, Wilson J, Lee JC, Johnston RC, Rishniw M, Tallini Y, Zhang J, et al. Ca2+-sensing transgenic mice: postsynaptic signaling in smooth muscle. The Journal of biological chemistry. 2004;279:21461–21468. doi: 10.1074/jbc.M401084200. [DOI] [PubMed] [Google Scholar]
- Katzel D, Zemelman BV, Buetfering C, Wolfel M, Miesenbock G. The columnar and laminar organization of inhibitory connections to neocortical excitatory cells. Nature neuroscience. 2011;14:100–107. doi: 10.1038/nn.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C, Bamberg E. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nature neuroscience. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lewis TL, Jr, Mao T, Arnold DB. A role for myosin VI in the localization of axonal proteins. PLoS biology. 2011;9:e1001021. doi: 10.1371/journal.pbio.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TL, Jr, Mao T, Svoboda K, Arnold DB. Myosin-dependent targeting of transmembrane proteins to neuronal dendrites. Nature neuroscience. 2009;12:568–576. doi: 10.1038/nn.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26:5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Griesbeck O. Genetically encoded calcium indicators. Chemical reviews. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science (New York, NY. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monetti C, Nishino K, Biechele S, Zhang P, Baba T, Woltjen K, Nagy A. PhiC31 integrase facilitates genetic approaches combining multiple recombinases. Methods (San Diego, Calif. 2011;53:380–385. doi: 10.1016/j.ymeth.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global doublefluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic acids research. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Papagiakoumou E, Anselmi F, Begue A, de Sars V, Gluckstad J, Isacoff EY, Emiliani V. Scanless two-photon excitation of channelrhodopsin-2. Nature methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu DL, Knopfel T. An NMDA receptor/nitric oxide cascade in presynaptic parallel fiber-Purkinje neuron long-term potentiation. J Neurosci. 2007;27:3408–3415. doi: 10.1523/JNEUROSCI.4831-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu DL, Knopfel T. Presynaptically expressed long-term depression at cerebellar parallel fiber synapses. Pflugers Arch. 2009;457:865–875. doi: 10.1007/s00424-008-0555-9. [DOI] [PubMed] [Google Scholar]
- Ren J, Qin C, Hu F, Tan J, Qiu L, Zhao S, Feng G, Luo M. Habenula "cholinergic" neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron. 2011;69:445–452. doi: 10.1016/j.neuron.2010.12.038. [DOI] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nature neuroscience. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nature neuroscience. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Tasic B, Hippenmeyer S, Wang C, Gamboa M, Zong H, Chen-Tsai Y, Luo L. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7902–7907. doi: 10.1073/pnas.1019507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, Yamanaka A. Acute Optogenetic Silencing of Orexin/Hypocretin Neurons Induces Slow-Wave Sleep in Mice. J Neurosci. 2011;31:10529–10539. doi: 10.1523/JNEUROSCI.0784-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zylka MJ. Mrgprd-expressing polymodal nociceptive neurons innervate most known classes of substantia gelatinosa neurons. J Neurosci. 2009;29:13202–13209. doi: 10.1523/JNEUROSCI.3248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CR, Power J, Barnea G, O'Donnell S, Brown HE, Osborne J, Axel R, Gogos JA. Spontaneous neural activity is required for the establishment and maintenance of the olfactory sensory map. Neuron. 2004;42:553–566. doi: 10.1016/s0896-6273(04)00224-7. [DOI] [PubMed] [Google Scholar]
- Zeng H, Horie K, Madisen L, Pavlova MN, Gragerova G, Rohde AD, Schimpf BA, Liang Y, Ojala E, Kramer F, et al. An inducible and reversible mouse genetic rescue system. PLoS genetics. 2008;4:e1000069. doi: 10.1371/journal.pgen.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, Deisseroth K. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nature protocols. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, Deisseroth K, Augustine GJ, Feng G. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain cell biology. 2008;36:141–154. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]