Abstract
The 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine modification of cytosine is a known base modification produced in vitro by oxidative stress. However, the presence of this modification in vivo has not been established. In this study the introduction of this base modification into dinucleoside monophosphates was accomplished using the Fenton reaction. Subsequently, the modification was produced in short isotopically labeled oligomers. Labeled tetramers bearing the lesion were used as internal standards for LC-MS/MS determinations of the base modification in the DNA of white blood cells from healthy donors. The background level of the 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine modification of cytosine was found to be larger than the levels of the formamide and thymine glycol base modifications.
INTRODUCTION
Cells exhibit a surprisingly high level of oxidatively induced DNA damage. Presumably this damage is caused primarily by reactive oxygen species (ROS) generated in vivo (1). Superoxide radical and hydrogen peroxide are ROS generated in cells during synthesis of ATP. The level of the 8-oxo-7,8-dihydroguanine lesion in DNA is often taken as an indicator of oxidative stress. This indicator is used despite the fact that measurements of background levels of the 8-oxo-7,8-dihydroguanine in the DNA of white blood cells (WBC) disagree widely among different laboratories (2–7). The cited reports used either electrochemical detection or 32P postlabeling to measure the 8-oxo-7,8-dihydroguanine lesion.
Exposure of DNA in vitro to ROS produces a variety of base modifications in addition to the 8-oxo-7,8-dihydroguanine lesion. Being less subject to the vagaries associated with measurements of 8-oxo-7,8-dihydroguanine, other base modifications would likely be more reliable indicators of oxidative stress. Studies of the radiation chemistry of DNA and its constituents suggest many alternative modifications for study. For example, exposure of d(CpApTpG) to ionizing radiation in aqueous solution generates multiple products separable by chromatography, as shown in Fig. 1. Several of the products have been characterized by mass spectrometry and by proton NMR spectroscopy (8). The major base modifications identified included 8-oxo-7,8-dihydroguanine, the formamide remnant from the breakdown of either of the pyrimidine bases, thymine glycol derived from thymine, and an imidazolidine modification derived from cytosine. With the notable exception of the last mentioned, these modifications have also been detected in cells exposed to oxidative stress. The neglected cytosine modification, 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine, is the focus of this report. The isomers of this modification are off scale in Fig. 1. The structure of 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine derived from irradiated cytosine was determined by Hahn et al. (9). This base modification has also been characterized at the nucleoside level (10).
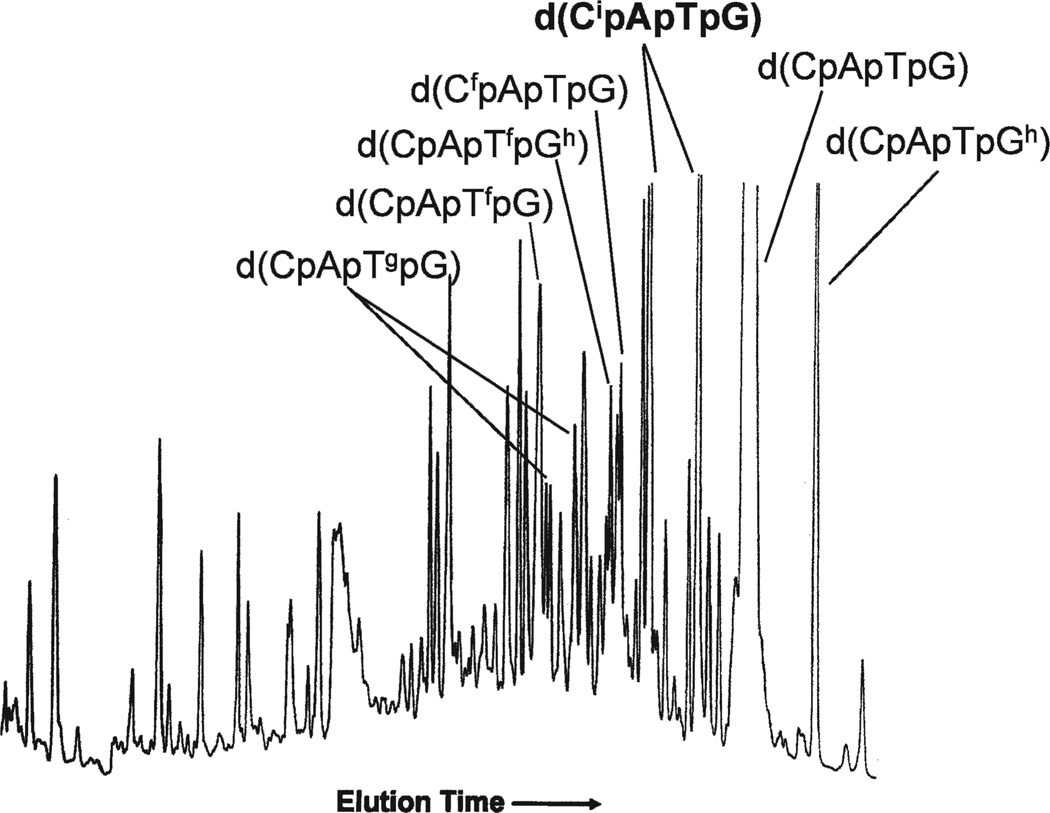
FIG. 1.
HPLC elution profile of d(CpApTpG) after exposure to X rays in oxygenated aqueous solution at a concentration of 0.028 mM. The dose was 240 Gy (8). The products of interest for this study are labeled d(CipApTpG) and are off scale.
In this study, the 1-carbamoyl-2-oxo-4.5-dihydroxyimidazolidine modification was characterized in dinucleoside monophosphates, for example d(CipA). An isotopically labeled tetramer containing the modification, d(GpCipA*pT), where A* stands for [15N]-labeled 2′-deoxyadenosine, was synthesized for use as an internal standard for mass spectrometry measurements. The imidazolidine modification was quantified in the DNA of WBC from human blood samples and was found to be comparable to the baseline levels of the formamide and thymine glycol modifications. The simultaneous measurement of the formamide, thymine glycol and imidazolidine base damages is proffered as a more reliable assessment of oxidative stress. For reasons enumerated below, DNA damage is measured in our assay in the form of modified dinucleoside monophosphates, as shown in Fig. 2 (11, 12). Modified dinucleoside monophosphates with 2′-deoxyadenosine as the 3′ nucleoside are preferred because overall these sequences chromatograph most advantageously.
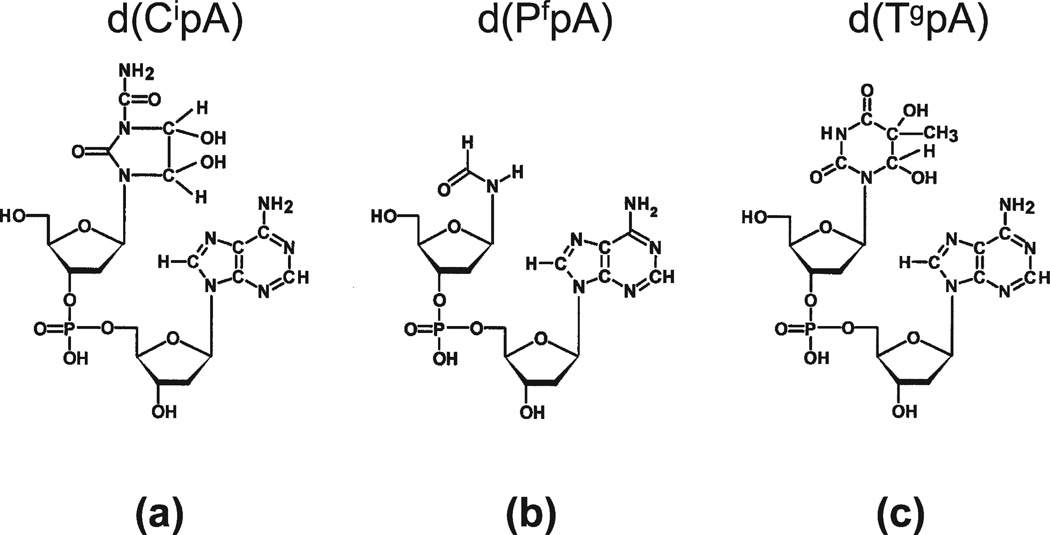
FIG. 2.
The imidazolidine, formamide and thymine glycol base modifications were measured in the DNA of WBC in the form of modified dinucleoside monophosphates, a, b and c, respectively.
MATERIALS AND METHODS
The modification of short oligomers to contain the imidazolidine lesion used the Fenton reaction. For example, a solution containing 500 µl 0.1 M Tris, pH 7.4, 120 µl 1 mM CuSO4, 30 µl H2O2, 30% and 2 mg d(CpA) (650 µl total volume) was maintained at room temperature for 24 h. Products were isolated by HPLC and characterized by mass spectrometry and by proton NMR spectroscopy.
The synthesis of isotopically labeled d(GpCipA*pT), where Ci stands for deoxycytosine bearing the imidazolidine modification and A* stands for [15N]-labeled 2′-deoxyadenosine, was a three-step process. Universally [15N]-labeled 2′-deoxyadenosine phosphoramidite was procured from Cambridge Isotopes Laboratories. The phosphoramidite was incorporated into the tetramer d(GpCpA*pT) by PrimeSyn Inc. The labeled tetramer was modified in our laboratory using the Fenton reaction. The imidazolidine modifications were isolated by HPLC analogous to Fig. 1. The labeled tetramer was enzymatically hydrolyzed and run on HPLC along with the modified dinucleoside monophosphate characterized in Fig. 3. The labeled and unlabeled dinucleoside monophosphates eluted identically. Internal standards are added to the DNA sample prior to enzymatic digest. In addition to d(GpCipA*pT), we employed internal standards of the form d(GpApApTfpA*) and d(GpCpApTgpA*) for quantifying the formamide and thymine glycol modifications (Fig. 2b and c, respectively) (12). The internal standards and the DNA sample are hydrolyzed together and the modifications were detected at the dimer level.
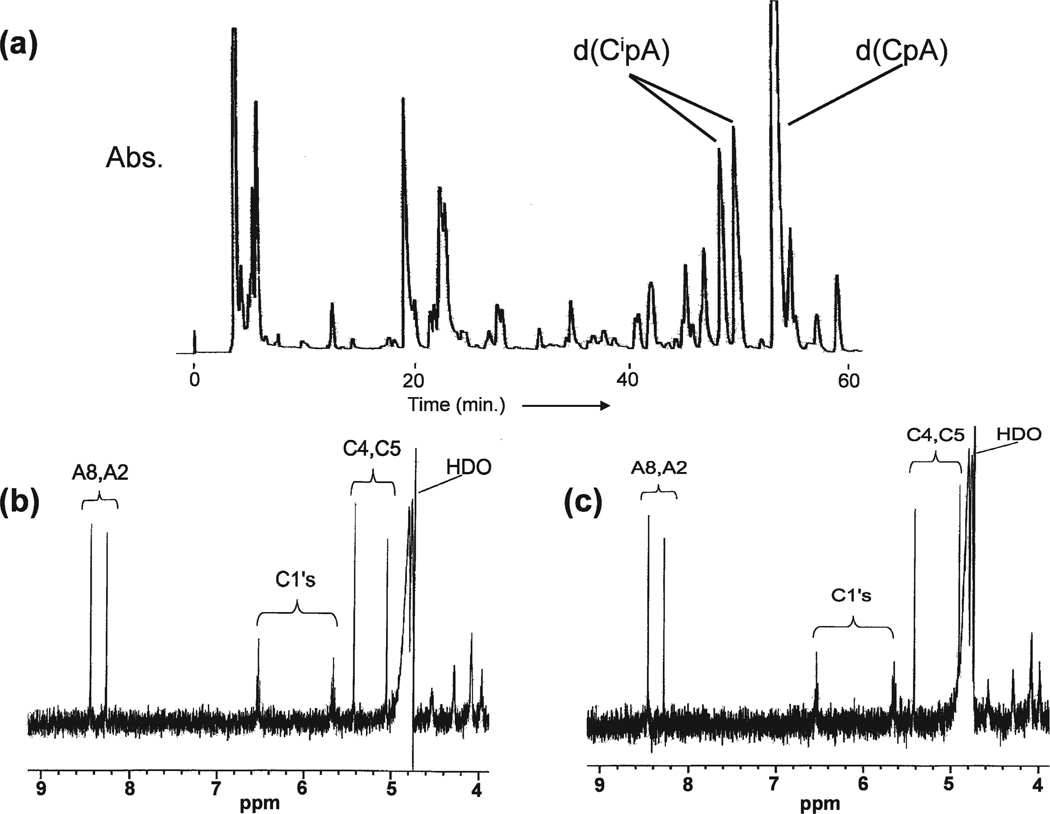
FIG. 3.
Panel a: HPLC elution profile of d(CpA) after treatment with Cu++ and H2O2. Peaks labeled d(CipA) contain imidazolidine base modifications. Panels b and c: Proton NMR spectra of early- and late-eluting stereoisomers, respectively, of d(CipA).
Blood samples were drawn with EDTA as anticoagulant under an IRB-approved protocol. The sample was centrifuged and the buffy coat was collected. DNA was extracted from the cells using a kit designed to minimize spurious oxidation reactions (ZeptoMetrix, Inc., Buffalo, NY). The kit uses chaotropic precipitation of the DNA together with desferol in the extraction procedure. One hundred micrograms of DNA was hydrolyzed and dephosphorylated using nuclease P1 and alkaline phosphatase. A solution containing 15 µl sodium acetate buffer (0.25 M, pH 5.2), 50 µl 3.0 mM Zn Cl2, 50 µl water, and either 1.0 or 3.0 U nuclease P1 (Sigma N8630) together with the DNA and isotopically labeled internal standards was incubated at 37°C for 2 h at pH 5.2. After addition of 25 µl of Tris-HCl (1 M, pH 9.0) and 70 U of alkaline phosphatase, the sample was incubated for an additional 2 h at pH 8.2.
RESULTS
The 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine modification has not been included previously in assays of oxidatively induced DNA damage in WBC. Characterization of this modification at the dimer level was a necessary preliminary step in this study. The dinucleoside monophosphate d(CpA) was treated with hydrogen peroxide in the presence of Cu++ (Fenton reaction) as described above. Separation of products by HPLC yielded the elution profile of Fig. 3a. Product peaks were isolated, desalted and analyzed by mass spectrometry. Peaks 7 and 8 had identical molecular weights, 589, corresponding to the negative ion weight of the product of interest, d(CipA) of Fig. 2a. The C4 and C5 carbon atoms of 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine are chiral, and the two products can be accounted stereoisomers. These products were further characterized by proton NMR spectroscopy. The low-field portions of the proton NMR spectra are shown in Fig. 3b and c. The chemical shifts associated with the C4-H and C5-H resonances distinguish between the two stereoisomers. The negligible spin-spin coupling between the C4 and C5 protons indicates that these are the trans stereoisomers. The modified dimers d(CipC), d(CipG) and d(CipT) were synthesized similarly.
Our assay takes advantage of the fact that base modifications typically inhibit the hydrolysis by nuclease P1 of the phosphoester bond 3′ to the modified nucleoside. Consequently, the nuclease P1 (plus alkaline phosphatase) hydrolysate tends to accumulate base modifications in the form of modified dinucleoside monophosphates as shown, for example, in Fig. 2. Detection by LC-MS/MS using electrospray ionization and operating in the negative ion mode is more sensitive for dinucleoside monophosphates than for nucleosides due to the presence of the phosphodiester bond. Furthermore, the fragmentation pattern of dinucleoside monophosphates reliably generates the 3′ fragment formed by breakage of the C5′-O bond of the 5′ nucleoside. The MS/MS values for detection of d(CipA), for example, are 589/330.
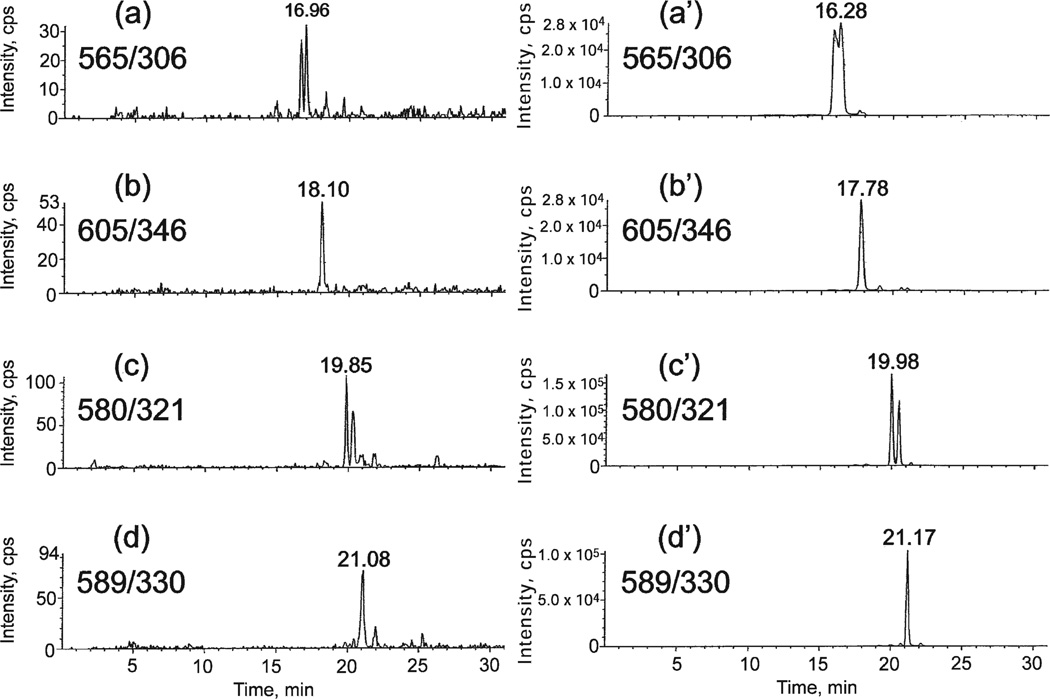
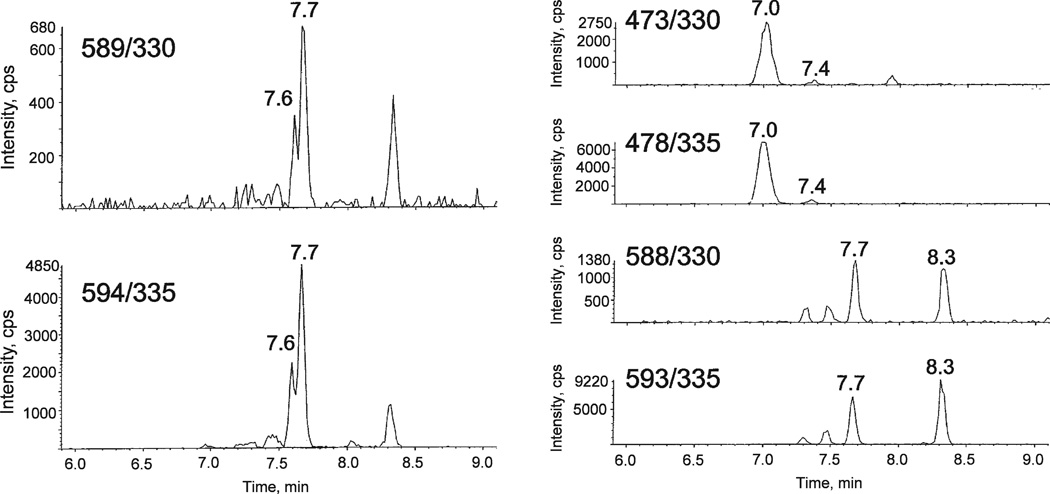
Two samples containing 100 µg of DNA derived from human cells in culture were prepared for LC-MS/MS analysis as described above. Synthetic d(CipC), d(CipG), d(CipT) and d(CipA) reference compounds were added to one of the samples. The spectrometer was programmed for MS/MS values: 565/306, 605/346, 580/321 and 589/330. The elution profiles of the two samples are compared in Fig. 4. In the left-hand set of elution profiles, the signal arises from the DNA of untreated cells; in the right-hand set of elution profiles, the signals arise predominantly from the added reference compounds The latter profile was substantially unchanged whether it was prepared from the early- or late-eluting components of each pair of isomers. Evidently, the imidazolidine modification is present in the DNA of untreated human cells. The sequence of elutions in Fig. 4 is as expected. The same sequence of elution is observed for this chromatography applied to unmodified dinucleoside monophosphates (14). The stereoisomers of d(CipG) and d(CipA) are not resolved in Fig. 4.
FIG. 4.
LC-MS/MS elution profiles for DNA samples with MS/MS values set to detect d(CipC), d(CipG), d(CipT) and d(CipA). Right-hand profiles were spiked with reference materials.
The imidazolidine modification was incorporated into a larger assay that also included the formamide and thymine glycol modifications. A typical set of LC-MS/MS profiles for the three modifications displayed in Fig. 2 from the DNA of untreated WBC from a healthy donor is shown in Fig. 5. A compilation of results for measurements of the three modifications in the DNA of WBC of healthy donors is given in Table 1. These measurements are a determination of the background levels of the three modifications of Fig. 2 in the DNA of healthy individuals. The measurements show that the level of the 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine modification is of the same order of magnitude as the formamide or thymine glycol modifications. The key value in Table 1, 7.5 fmol d(CipA)/µg of DNA, is equivalent to 7.7 dCi/106 nucleotides.
FIG. 5.
Typical LC-MS/MS elution profiles from a hydrolysate of DNA obtained from WBC. Elution profiles are shown for dinucleoside monophosphates containing the imidazolidine (left-hand profiles), formamide (upper right profiles), and thymine glycol (lower right profiles) modifications and for their respective internal standards. Internal standards were added to the DNA sample before enzymatic hydrolysis.
TABLE 1.
Background Levels of Imidazolidene, Formamide and Thymine Glycol in WBC of Healthy Donors
| d(CipA) (fmol/µg of DNA) |
d(TfpA (fmol/µg of DNA) |
d(TgpA) (fmol/µg of DNA) |
|
|---|---|---|---|
| Group 1, 3 U (4) | 8.3 ± 2.5 | 5.0 ± 0.8 | 4.4 ± 0.9 |
| Group 2, 1 U (4) | 6.7 ± 2.0 | 6.0 ± 1.0 | 3.2 ± 0.8 |
| Mean (8) | 7.5 ± 1.5 | 5.5 ± 0.6 | 3.8 ± 0.6 |
Notes. Mean values are given in terms of fm/µg of DNA ± SEM. The amount of endonuclease used in processing the two groups was either 1 U or 3 U (see the Materials and Methods).
The base modifications discussed herein occur as stereoisomers. The occurrence of isomers in the case of the imidazolidene modification has already been noted. It is generally preferable to quantify the product from DNA by referencing it to the sum of the isomers in the internal standard because of the possibility of interchange between isomers. In the case of the formamide modification, rotomers are present due to restricted rotation about the amide bond (15). The rotor states are unequally populated and are stable enough to yield distinct resonances in proton NMR spectra of nucleosides and oligomers containing this base modification (8, 16). Our measurements of the thymine glycol modification pertain to the cis stereoisomers.
The MS/MS values for d(CipA) differ from the values for d(TgpA) by a single mass unit. Furthermore, the later eluting isomer of d(CipA) and an earlier-eluting isomer of d(TgpA) tend to elute simultaneously. The natural abundances of isotopes 13C, 15N and 17O of d(TgpA) enhance the signal arising from d(CipA). Quantification of the imidazolidene modification was accomplished in this study by comparing the signals from the earlier-eluting components of the sample and internal standard signals.
DISCUSSION
Sequencing studies have shown that breast tumors and colorectal tumors accumulate of the order of 100 mutations during tumorigenesis (17, 18). Oxidative stress is a plausible global contributor to mutagenesis. Levels of oxidative DNA damage, inferred from the measurements of 8-oxo-7,8-dihydroguanine, are reported to be higher in patients with lung, leukemia, colorectal, bladder, breast and esophageal cancers than in controls (2–7), although the reported control levels vary widely. The level of the thymine glycol modification is higher in ovarian cancer patients than in controls (19). These data contribute to the interest in assessing and monitoring oxidatively induced DNA damage as an indicator of oxidative stress.
Hydroxyl radical is the ROS mainly responsible for generating oxidative stress. A further reservation concerning 8-oxo-7,8-dihydroguanine as an indicator of oxidative stress is that hydroxyl radical is relatively unreactive toward deoxyguanosine (20). On the other hand, the formation of 1-carbamoyl-4,5-dihydroxyimidazolidine is a classic example of a hydroxyl radical-induced damage product. The first step in the reaction of hydroxyl radical is to produce a free radical adduct, in this case 5(6)-hydroxy-6(5)-yl-cytosine. Under aerated conditions, addition of oxygen to the carbon atom bearing the unpaired electron generates the peroxyl radical. In the reducing environment of a cell, formation of 5(6)-hydroxy-6(5)-hydroperoxide is probable. The final product, 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine, follows upon rearrangement. The increase in molecular weight is 50, consistent with observed properties of the modification in question.
The main result of this study is to establish the 1-carbamoyl-2-oxo-4,5-dihydroxyimidazolidine modification of cytosine as a significant oxidatively induced modification of DNA. This conclusion is based on measurements of the level of the modification in the DNA from WBC of healthy donors. Typical basal levels of oxidative base damage in DNA are high, of the order of 1–10 per 106 nucleotides, although lower levels have been reported (2–7, 19, 21, 22). The levels of the imidazolidine modification are no exception. Our results show the abundance of the imidazolidine modification to be comparable to the formamide or thymine glycol modifications.
This study is another instance of a base modification identified initially in bare DNA exposed to ionizing radiation that was subsequently shown to be present at significant levels in human DNA. Oxidative stress generates a multiplicity of DNA modifications, most of which have not been quantified in the DNA from human donors. Oxidatively induced DNA damage is likely to be a risk factor for cancer because it is present in DNA as many types of damage, frequently at high levels. Levels of 1–10 base modifications per 106 nucleotides may be compared with, for example, smoking-induced DNA damage, which is typically an order of magnitude smaller (23–25).
The development of LC-MS/MS spectroscopy provides unique opportunities for DNA damage studies. Detection by LC-MS/MS is very sensitive, is highly selective, and has the potential for measuring multiple products simultaneously. It may be noted that previous assays of DNA damage in this laboratory used an HPLC step that removed unmodified nucleosides, constituting the bulk of the sample, from the hydrolyzed DNA before LC-MS/MS analysis (11, 12, 19). This labor-intensive step was omitted in the current protocol.
DNA damage analysis at the dimer level has advantages. Detection of modified dinucleoside monophosphates by LC-MS/MS operating in the negative ion mode is particularly sensitive due to the presence of the phosphodiester bond. Furthermore, it is often convenient to synthesize an isotopically labeled internal standard, needed for quantification of a lesion, by incorporating the isotopic label in the unmodified 3′ nucleoside.
ACKNOWLEDGMENTS
We are indebted to Mr. John Schaffer of Spectra Gases, Inc. and Dr. Janusz Kozak of Cambridge Isotopes Laboratories Inc. for making [15N]-labeled 2′-deoxynucleoside phosphoramidites available. We are also indebted to Dr. Surendra Chaturvedi of PrimeSyn Lab Inc. for manual synthesis of selected oligomers. The work is supported by grant CA139513 from the National Cancer Institute. Mass spectrometry facilities and the RPCI Data Bank and Biorepository (DBBR) are shared resources supported by NCI Grant CA 016056.
REFERENCES
- 1.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA. 1992;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akcay T, Saygili I, Andican G, Yalcin V. Increased formation of hydroxy-2′-deoxyguanosine in peripheral blood leukocytes in bladder cancer. Urol. Int. 2003;71:271–274. doi: 10.1159/000072677. [DOI] [PubMed] [Google Scholar]
- 3.Breton J, Sichel F, Pottier D, Prevost V. Measurement of 8-oxo-7,8-dihydro-2′-deoxyguanosine in peripheral blood mononuclear cells: optimization and application to samples from a case-control study on cancers of the oesophagus and cardia. Free Radic. Res. 2005;39:21–30. doi: 10.1080/10715760400023523. [DOI] [PubMed] [Google Scholar]
- 4.Gackowski D, Banaszkiewicz Z, Rozalski RA, Jawien A, Olinski R. Persistent oxidative stress in colorectal carcinoma patients. Int. J. Cancer. 2002;101:395–397. doi: 10.1002/ijc.10610. [DOI] [PubMed] [Google Scholar]
- 5.Oltra AM, Carbonell F, Tormos C, Iradi A, Saez GT. Antioxidant enzyme activities and the production of MDA and 8-oxo-dG in chronic lymphocytic leukemia. Free Radic. Biol. Med. 2001;30:1286–1292. doi: 10.1016/s0891-5849(01)00521-4. [DOI] [PubMed] [Google Scholar]
- 6.Soliman AS, Vulimiri SV, Kleiner HE, Shen J, Elissa S, Morad M, Taha H, Lukmanji F, Li D, Bondy ML. High levels of oxidative DNA damage in lymphocyte DNA of premenopausal breast cancer patients in Egypt. Int. J. Environ. Health Res. 2004;14:121–134. doi: 10.1080/0960312042000209534. [DOI] [PubMed] [Google Scholar]
- 7.Vulimiri SV, Wu X, Baer-Dubowska W, de Andrade M, Detry M, Spitz MR, DiGiovanni J. Analysis of aromatic DNA adducts and 7,8-dihydro-8-oxo-2′-deoxyguanosine in lymphocyte DNA from a case-control study of lung cancer involving minority populations. Mol. Carcinog. 2000;27:34–46. doi: 10.1002/(sici)1098-2744(200001)27:1<34::aid-mc6>3.3.co;2-7. [DOI] [PubMed] [Google Scholar]
- 8.Box HC, Dawidzik JB, Wallace JC, Hauer CR, Stack RF, Rajecki MJ, Budzinski EE. An overall perspective of X-ray damage to a DNA oligomer mediated by reactive oxygen species. Radiat. Res. 1999;152:575–582. [PubMed] [Google Scholar]
- 9.Hahn BS, Wang SY, Flippen JL, Karle IL. Radiation chemistry of nucleic acids. Isolation and characterization of glycols of 1-carbamoyl-imidazoidone as products of cytosine. J. Am. Chem. Soc. 1973;95:2711–2712. doi: 10.1021/ja00789a064. [DOI] [PubMed] [Google Scholar]
- 10.Wagner JR, Decarroz C, Berger M, Cadet J. Hydroxyl-radical-induced decomposition of 2′-deoxycytidine in aerated aqueous solutions. J. Am. Chem. Soc. 1999;121:4101–4110. [Google Scholar]
- 11.Greene KF, Budzinski EE, Iijima H, Dawidzik JB, DeFedericis H-C, Patrzyc HB, Evans MS, Bailey DT, Freund HG, Box HC. Assessment of DNA damage at the dimer level: Measurement of the formamide lesion. Radiat. Res. 2007;167:146–151. doi: 10.1667/rr0693.1. [DOI] [PubMed] [Google Scholar]
- 12.Bailey DT, DeFedericis H-C, Greene KF, Iijima H, Budzinski EE, Patrzyc HB, Dawidzik JB, Box HC. A novel approach to DNA damage assessments: measurement of the thymine glycol lesion. Radiat. Res. 2006;165:438–444. doi: 10.1667/rr3534.1. [DOI] [PubMed] [Google Scholar]
- 13.Box HC, Budzinski EE, Evans MS, French JB, Maccubbin AE. The differential lysis of phosphoester bonds by nuclease P1. Biochim. Biophys. Acta. 1993;1161:291–294. doi: 10.1016/0167-4838(93)90227-i. [DOI] [PubMed] [Google Scholar]
- 14.Dawidzik JB, Patrzyc HB, Iijima H, Budzinski EE, Higbee A, Cheng H-C, Box HC. DNA damage measured by liquid chromatography-mass spectrometry in mouse fibroblast cells exposed to oxidative stress. Biochim. Biophys. Acta. 2003;1621:211–217. doi: 10.1016/s0304-4165(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 15.Uesugi S, Shida T, Ikehara M, Koayashi Y, Kyogoku Y. Identification of degradation products of d(CpG) by a 1,10 phenanthroline-copper ion complex. J. Am. Chem. Soc. 1982;104:5494–5495. [Google Scholar]
- 16.Cadet J, Nardin R, Voituriez I, Remin M, Hruska FE. A 1H and 13C nmr study of the radiation-induced degradation products of 2′-deoxythymidine derivatives: N-(2′-deoxy-β-d-erythropentofuranosyl) formamide. Can. J. Chem. 1981;59:3313–3318. [Google Scholar]
- 17.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 18.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 19.Iijima H, Patrzyc HB, Budzinski EE, Freund HG, Dawidzik JB, Rodabaugh KJ, Box HC. A study of pyrimidine base damage in relation to oxidative stress and cancer. Br. J. Cancer. 2009;101:452–456. doi: 10.1038/sj.bjc.6605176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadet J, Douki T, Gasparutto D, Ravanat J-L. Oxidative damage to DNA: formation. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Mangal D, Vudathala D, Park J-H, Lee SH, Penning TM, Blair IA. Analysis of 7,8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 2009;212:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cadet J, Douki T, Ravanat JL. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 23.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Wang M, Villalta PW, Luo X, Feuer R, Jensen J, Hatsukami DK, Hecht SS. Quantitation of an acetaldehyde adduct in human leukocyte DNA and the effect of smoking cessation. Chem. Res. Toxicol. 2007;20:108–113. doi: 10.1021/tx060232x. [DOI] [PMC free article] [PubMed] [Google Scholar]