Abstract
Leucine-rich repeat kinase 2 (LRRK2) is linked to Parkinson’s disease and may represent an attractive therapeutic target. Here we report a 2,4-dianilino-5-chloro-pyrimidine, TAE684, a previously reported inhibitor of anaplastic lymphoma kinase (ALK), is also a potent inhibitor of LRRK2 kinase activity (IC50 of 7.8 nM against wild-type LRRK2, 6.1 nM against the G2019S mutant). TAE684 substantially inhibits Ser910 and Ser935 phosphorylation of both wild-type LRRK2 and G2019S mutant at a concentration of 0.1–0.3 µM in cells and in mouse spleen and kidney, but not in brain, following oral doses of 10 mg/kg.
Keywords: Parkinson’s disease, LRRK2, TAE684
Parkinson’s disease (PD) is a debilitating neurodegenerative disease that affects over one million Americans.1,2 Recent genetic studies have revealed an underlying genetic cause in at least 10% of all PD cases,3 which provides new opportunities for discovery of molecularly targeted therapeutics that may ameliorate neurodegeneration. Among the genes associated with PD, leucine-rich repeat kinase 2 (LRRK2) is unique because of a missense mutation, G2019S, is frequently found in not only familial but also sporadic Parkinson’s disease cases.4,5 The G2019S mutation enhances kinase activity, suggesting that small molecule LRRK2 kinase inhibitors may be able to block aberrant LRRK2-dependent signaling in Parkinson’s disease.6,7
LRRK2 kinase inhibitors are being actively pursued and recently first-generation ‘tool’ inhibitors that exhibit excellent potency and selectivity for LRRK2 such as LRRK2-IN-18 and CZC-251469 have been reported. However, neither compound is able to achieve good exposure in mouse brains which limits their utility in murine PD models and eventual translation in human clinical trials.8,9 Here we report that TAE684, a previously reported inhibitor of anaplastic lymphoma kinase (ALK),10 is also a potent LRRK2 inhibitor which posses a favorable pharmacokinetic profile in mice.
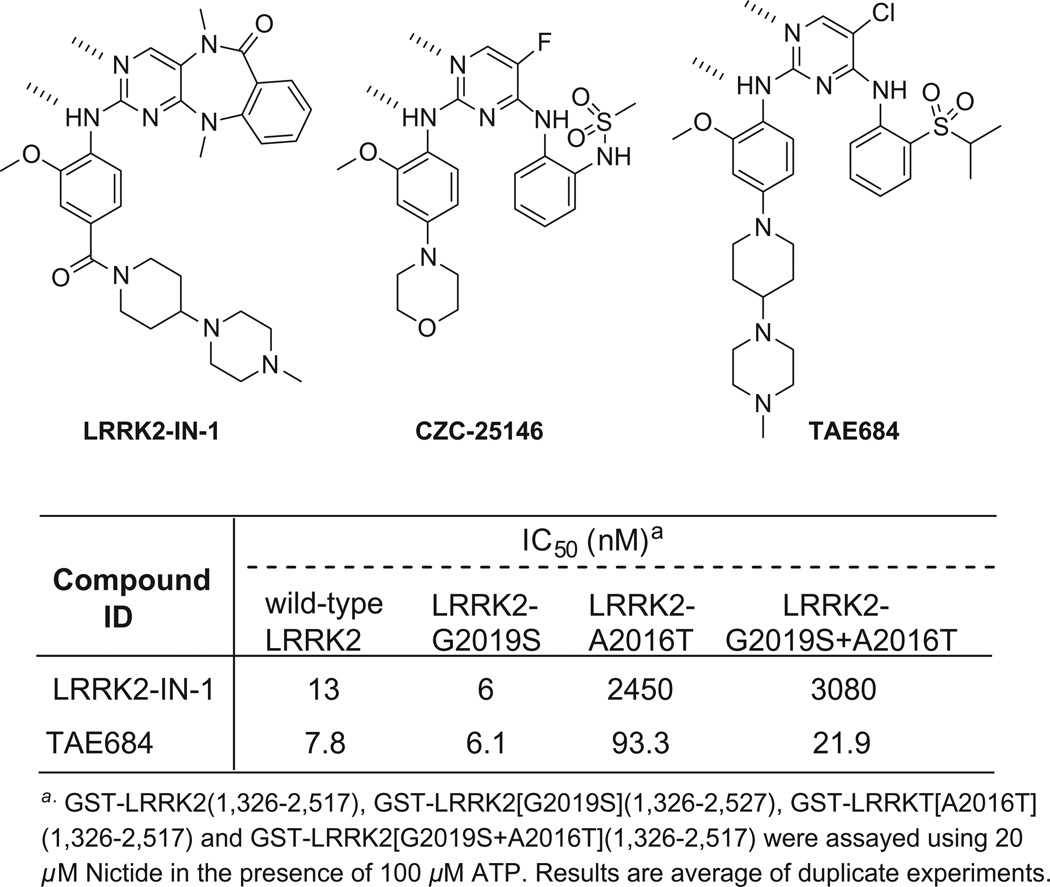
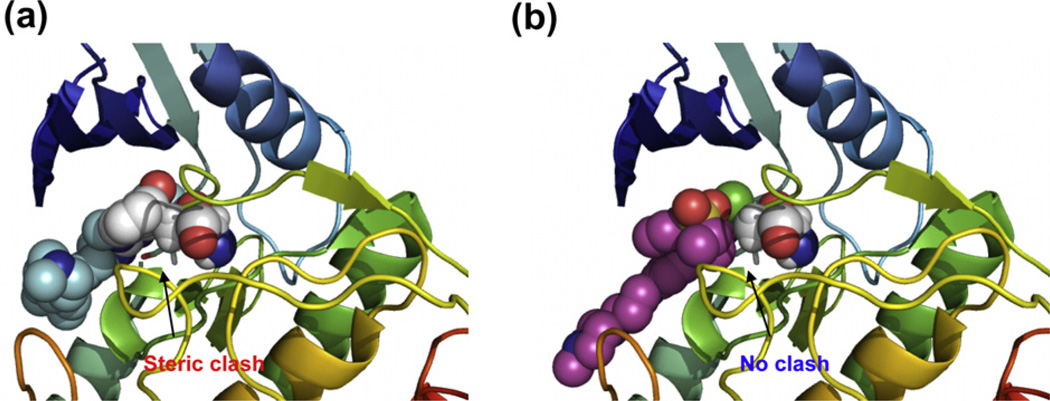
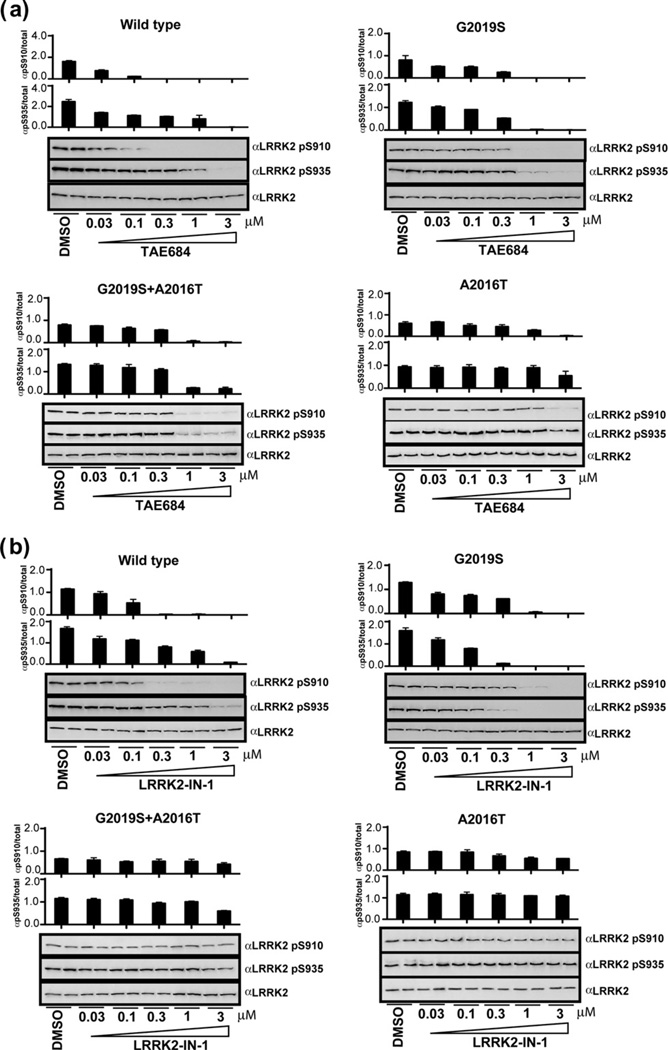
In an effort to discover new compound classes that are capable of potently inhibiting LRRK2 kinase activity and that also have the ability to traverse the blood–brain barrier (BBB), we screened our in-house kinase targeted compound library using an LRRK2 enzymatic assay. TAE684, 5-chloro-N4-(2-(isopropylsulfonyl)phenyl)-N2-(2-methoxy-4-(4-(4-methyl-piperazin-1-yl)piperidin-1-yl)phenyl)pyrimidine-2,4-diamine, a previously reported ALK kinase inhibitor,10 emerged as a potent LRRK2 inhibitor with biochemical IC50s of 7.8 and 6.1 nM against wild-type LRRK2 and LRRK2[G2019S], respectively (Fig. 1). While the biochemical potency of TAE684 for inhibition of wild-type and G2019S LRRK2 is similar to LRRK2-IN-1, TAE684 maintains significant inhibition of the A2016T mutation which induces dramatic resistance to LRRK2-IN-1 (Fig. 1). Although both LRRK2-IN-1 and TAE684 share the aminopyrimidine pharmacophore, a molecular model of TAE684 built based on a previously published crystallographic structure with ALK,11 suggests that isopropylsulfone moiety is able to avoid the steric clash that is likely between the anthranilic acid ring of LRRK2-IN-1 with the A2016T residue (Fig. 2a,b).8,12,13 We next examined the ability of TAE684 to inhibit LRRK2 in a cellular context in comparison to LRRK2-IN-1. As there are no validated direct phosphorylation substrates of LRRK2, we monitored phosphorylation of Ser910 and Ser935, two residues whose phosphorylation is known to be dependent upon LRRK2 kinase activity14 (Fig. 3). TAE684 induced a dose-dependent inhibition of Ser910 and Ser935 phosphorylation in both wild-type LRRK2 and LRRK2[G2019S] stably transfected into HEK293 cells (Fig. 3a). Significant dephosphorylation of Ser910 and Ser935 was observed at 0.1–0.3 µM of TAE684 for wild-type LRRK2 and at slightly higher doses for LRRK2[G2019S] (Fig. 3a), which is approximately 10-fold more potent compared to LRRK2-IN-1 (compare Fig. 3a to b). Consistent with the biochemical results, TAE684 also induced dephosphorylation of Ser910 and 935 at a concentration of 1–3 µM in the drug-resistant LRRK2[A2016T+G2019S] and LRRK2[A2016T] mutants (Fig. 3a), revealing that TAE684 has a different profile compared to LRRK2-IN-1 (compare Fig. 3a to b).
Figure 1.
LRRK2 inhibitors.
Figure 2.
Modeling study. (a) Molecular model of LRRK2-IN-1 with LRRK2[A2016]. Potential steric clash is indicated. (b) Molecular model of TAE684 with LRRK2[A2016T]. Small molecules are shown in space-filling mode and protein is depicted in ribbon format.
Figure 3.
Compound TAE684 inhibits LRRK2 in cells. (a) HEK 293 cells stably expressing wild-type GFP-LRRK2, GFP-LRRK2[G2019S], GFP-LRRK2[G2019S+A2016T], and GFP-LRRK2[A2016T] were treated with DMSO or increasing concentrations of TAE684 for 90 min. Cell lysates were subjected to immunoblotting for detection of LRRK2 phosphorylated at Ser910 and Ser935 and for total LRRK2. (b) As in (a) except LRRK2-IN-1 was used.
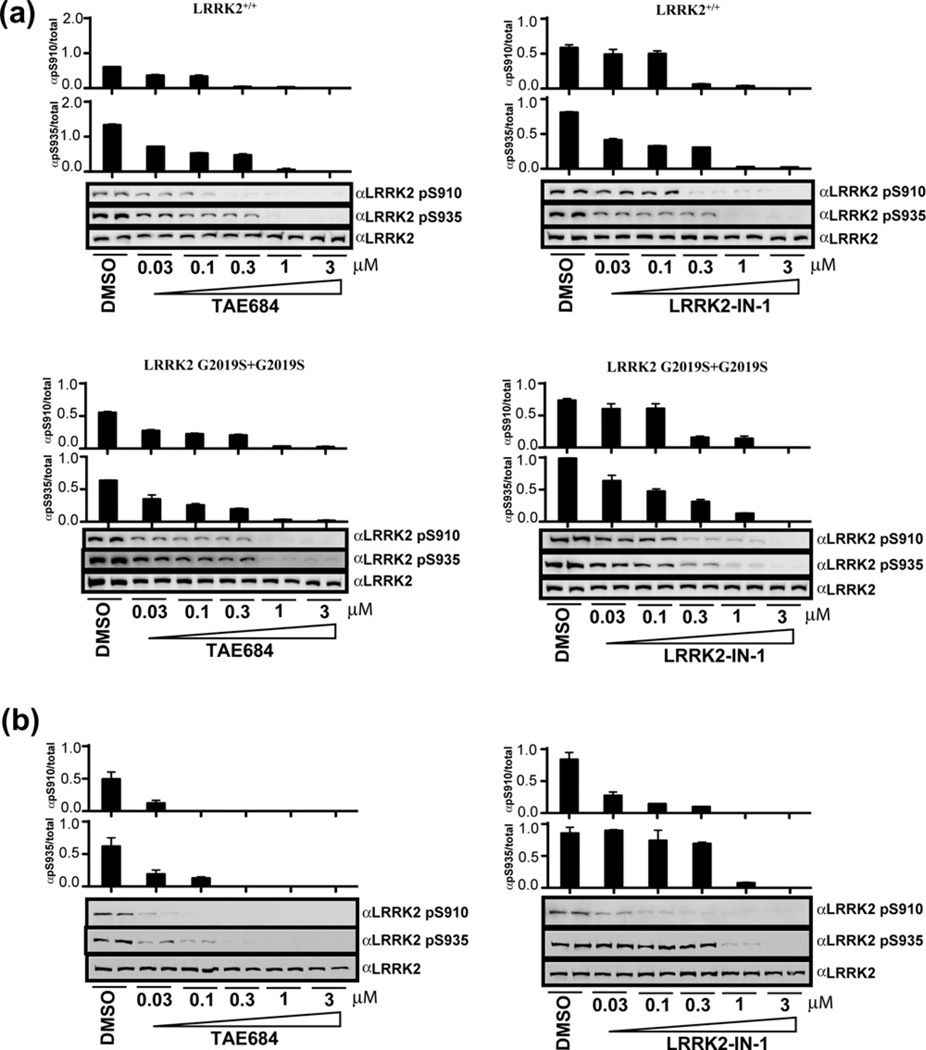
We next examined the effect of TAE684 on endogenously expressed LRRK2 in human lymphoblastoid cells derived from a control and Parkinson’s disease patient homozygous for the LRRK2[G2019S] mutation (Fig. 4a). We found that increasing doses of TAE684 led to similar dephosphorylation of endogenous LRRK2 at Ser910 and Ser935, as was observed in HEK293 cells stably expressing wild-type LRRK2 or LRRK2[G2019S] (compare Figs. 3a to 4a). Moreover, endogenous LRRK2 was also more sensitive to TAE684 than LRRK2-IN-1, which is consistent with the trend we observed in HEK293 cells. We also found that TAE684 induced similar dose-dependent Ser910 and Ser935 dephosphorylation of endogenous LRRK2 in mouse Swiss 3T3 cells (Fig. 4b).
Figure 4.
Compound TAE684 inhibits endogenously expressed LRRK2. (a) Endogenous LRRK2 from EBV immortalized human lymphoblastoid cells from a control subject and a Parkinson’s disease patient homozygous for the LRRK2[G2019S] mutation. After treatment of the cells with DMSO or the indicated concentration of TAE684 (or LRRK2-IN-1) for 90 min, cell lysates were subjected to immunoblot analysis with the purified indicated antibody for western analysis. Immunoblots were performed in duplicate, and results were representative of at least two independent experiments. (b) As in (a) except mouse Swiss 3T3 cells were used.
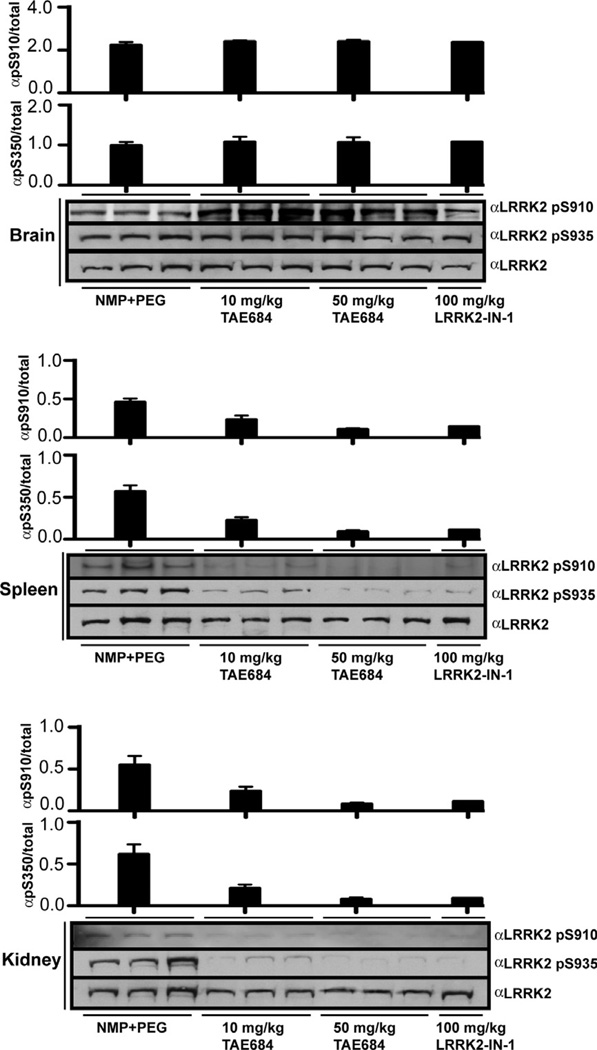
The mouse pharmacokinetic profile of TAE684 demonstrated excellent oral bioavailability (83% F), half-life of 11.3 h and excellent plasma exposure (6374 h*ng/mL, AUClast) (Table 1). More importantly, TAE684 exhibited brain to plasma ratio of 2, indicating superior BBB property (Table 1). Based on these excellent pharmacokinetic properties, pharmacodynamic experiments examining inhibition of LRRK2 Ser910/Ser935 phosphorylation were conducted after oral gavage administration with 10 mg/kg or 50 mg/kg of TAE684. We observed complete Ser910 and Ser935 dephosphorylation of LRRK2 in the kidney and spleen at dose of 50 mg/kg, and slightly lower effect at dose of 10 mg/kg. These also demonstrated improved potency relative to LRRK2-IN-1 (Fig. 5).8 Unfortunately, despite the significant exposure of TAE684 in the brain, no inhibition of LRRK2 Ser910 or Ser935 phosphorylation was observed in the brain (Fig. 5). We are currently investigating the reasons for this unexpected result.
Table 1.
Pharmacokinetic parameters of TAE684a
| Route | Matrix | AUClast (h*ng/mL) | T1/2 (h) | CL (mL/min/kg) | Vss (L/kg) | Brain/plasma (AUClast) ratio | %F |
|---|---|---|---|---|---|---|---|
| Iv | Plasma | 772 | 11.32 | 17.14 | 14.50 | 1.7 | 82.6 |
| Brain | 1318 | — | — | — | — | ||
| Po | Plasma | 6374 | — | — | — | 2.3 | — |
| Brain | 14,430 | — | — | — | — |
Experiments were done in male Swiss Albino Mice following single intravenous (iv, 1 mg/kg) and oral (po, 10 mg/kg) administration. AUC = area under the curve (measure of exposure), T1/2 = half life, CL = plasma clearance, Vss = volume of distribution, F = oral bioavailability.
Figure 5.
Pharmacodynamic analysis for TAE684. Pharmacodynamic study of TAE684 from brain, spleen and kidney following oral administration at the indicated doses. Tissues were collected and endogenous LRRK2 was resolved by SDS–PAGE and blotted with a phospho-specific antibody directed against Ser910, Ser935 and total LRRK2.
The kinase selectivity of TAE684 was assessed using standard radioactivity-base enzymatic assays against a panel of 124 kinases (Dundee profiling).15 At a concentration of 1 µM, TAE684 inhibited the kinase activities of CAMKKβ, CHK2, FGF-1R, NUAK1, PHKã1(PBK), and TSSK1 higher than 90% (detail results please see Supplementary data). Recently published Kinomescan binding data against a panel of 442 kinases revealed that TAE684 is a potent binder of many kinases with sub-100 nM Kds reported for: CAMKKβ, CHK2, FGF-1R, NUAK1, PHKã1(PBK), and TSSK1.16 These results show that TAE684 is a relatively broad-based kinase inhibitor and considerable less selective than LRRK2-IN-1 and CZC-25146.
In summary, we have discovered that TAE684 is a potent biochemical and cellular inhibitor of LRRK2 kinase activity. Detailed characterization of TAE684 using LRRK2-IN-1 as a bench mark revealed that TAE684 significantly inhibited phosphorylation of wild-type LRRK2 and LRRK2[G2019S] mutant at Ser910 and Ser935 at 0.1–0.3 µM in vivo, which is about 5–10-fold more potent than LRRK-IN-1. TAE684 is relatively insensitive to the A2016T mutation which suggests that this mutant will not be useful to validate whether the pharmacological effects of the compound are LRRK2-dependent. TAE684 achieves good exposure to mouse brain following oral administration but interestingly does not inhibit phosphorylation of Ser910 and Ser935 of LRRK2. Further characterization of clinical stage kinase inhibitors related to TAE684 may result in the identification of other compounds that might be relevant as pharmacological agents to investigate the impact of LRRK2 inhibition in animal models and eventually in humans.
Supplementary Material
Acknowledgments
We wish to thank staff at the National Centre for Protein Kinase Profiling (www.kinase-screen.mrc.ac.uk) for undertaking Dundee kinase specificity screening as well as Nicholas Dzamko for providing the LRRK2 rabbit monoclonal antibodies. We also thank Faycal Hentati Institut National de Neurologie, Tunis, Tunisia as well as Alastair D. Reith GlaxoSmithKline Stevenage U.K. for providing the human lymphoblastoid cells, SAI Advantium for performing pharmacokinetic studies, and the antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] coordinated by Hilary McLauchlan and James Hastie for generation of antibodies. This work was supported by NIH grant P41 GM079575-03 (N. Gray) the Medical Research Council (D. Alessi), the Michael J Fox foundation for Parkinson’s disease research (N. Gray & D. Alessi), the pharmaceutical companies supporting the DSTT (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA and Pfizer) (D. Alessi)
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2012.01.084. These data include MOL files and InChiKeys of the most important compounds described in this article.
References and notes
- 1.Gandhi PN, Chen SG, Wilson-Delfosse AL. J. Neurosci. Res. 2009;87:1283. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, Marshall FJ, Ravina BM, Schifitto G, Siderowf A, Tanner CM. Neurology. 2007;68:384. doi: 10.1212/01.wnl.0000247740.47667.03. [DOI] [PubMed] [Google Scholar]
- 3.Daniels V, Baekelandt V, Taymans JM. Neurosignals. 2011;19:1. doi: 10.1159/000324488. [DOI] [PubMed] [Google Scholar]
- 4.Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, Ferreira JJ, Tolosa E, Kay DM, Klein C, Williams DR, Marras C, Lang AE, Wszolek ZK, Berciano J, Schapira AH, Lynch T, Bhatia KP, Gasser T, Lees AJ, Wood NW. Lancet Neurol. 2008;7:583. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dächsel JC, Farrer MJ. Arch. Neurol. 2010;67:542. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
- 6.Greggio E, Cookson MR. ASN Neuro. 2009:1. doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Cookson MR. Expert Rev. Mol. Med. 2011;13:e20. doi: 10.1017/S146239941100192X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS. Nat. Chem. Biol. 2011;7:203. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsden N, Perrin J, Ren Z, Lee BD, Zinn N, Dawson VL, Tam D, Bova M, Lang M, Drewes G, Bantscheff M, Bard F, Dawson TM, Hopf C. ACS Chem. Biol. 2011;6:1021. doi: 10.1021/cb2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galkin AV, Melnick JS, Kim S, Hood TL, Li N, Li L, Xia G, Steensma R, Chopiuk G, Jiang J, Wan Y, Ding P, Liu Y, Sun F, Schultz PG, Gray NS, Warmuth M. Proc. Natl. Acad. Sci. U.S.A. 2007;104:270. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossi RT, Saccardo MB, Ardini E, Menichincheri M, Rusconi L, Magnaghi P, Orsini P, Avanzi N, Borgia AL, Nesi M, Bandiera T, Fogliatto G, Bertrand JA. Biochemistry. 2010;49:6813. doi: 10.1021/bi1005514. [DOI] [PubMed] [Google Scholar]
- 12.Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, Bamborough P, Reith AD, Alessi DR. Biochem. J. 2009;424:47. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.These images were produced using free version of Pymol software.
- 14.Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ. Biochem. J. 2010;430:405. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. Biochem. J. 2007;408:297. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Nat. Biotechnol. 2011;29:1046. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.