Abstract
Objective
Clinical questions are often long and complex and take many forms. We have built a clinical question answering system named AskHERMES to perform robust semantic analysis on complex clinical questions and output question-focused extractive summaries as answers.
Design
This paper describes the system architecture and a preliminary evaluation of AskHERMES, which implements innovative approaches in question analysis, summarization, and answer presentation. Five types of resources were indexed in this system: MEDLINE abstracts, PubMed Central full-text articles, eMedicine documents, clinical guidelines and Wikipedia articles.
Measurement
We compared the AskHERMES system with Google (Google and Google Scholar) and UpToDate and asked physicians to score the three systems by ease of use, quality of answer, time spent, and overall performance.
Results
AskHERMES allows physicians to enter a question in a natural way with minimal query formulation and allows physicians to efficiently navigate among all the answer sentences to quickly meet their information needs. In contrast, physicians need to formulate queries to search for information in Google and UpToDate. The development of the AskHERMES system is still at an early stage, and the knowledge resource is limited compared with Google or UpToDate. Nevertheless, the evaluation results show that AskHERMES’ performance is comparable to the other systems. In particular, when answering complex clinical questions, it demonstrates the potential to outperform both Google and UpToDate systems.
Conclusions
AskHERMES, available at http://www.AskHERMES.org, has the potential to help physicians practice evidence-based medicine and improve the quality of patient care.
Keywords: Clinical question answering, Question analysis, Passage retrieval, Summarization, Answer presentation
1. Introduction
Physicians generate up to six questions for every patient encounter [1–6], and these questions may be of a variety of types. Although it is important for physicians to meet their information needs, studies have shown that many of their questions go unanswered. For example, Ely and colleagues observed that physicians did not pursue answers to 45% of the 1062 questions posed in clinical settings, often because they doubted they could find good answers quickly, and for those they did pursue answers to, they failed to find answers to 41% of them [7]. As a result, more than 67% of the clinical questions posed by physicians remained unanswered.
One way to meet information needs is to refer to the published literature for related clinical evidence [8]. Although original research articles that are both scientifically rigorous and clinically relevant appear in high concentrations in only a few select journals (e.g., The New England Journal of Medicine, Annals of Internal Medicine, JAMA, and Archives of Internal Medicine), much clinical evidence appears in a wide range of other biomedical journals [9]. Even with the development of search engines for facilitating relevant biomedical literature searching, the needs of physicians still cannot be met properly, as an evaluation study showed that it took an average of more than 30 min for a healthcare provider to search for an answer from MEDLINE, which made “this type of information seeking is practical only ‘after hours’ and not in the clinical setting” [10].
Internet search engines (e.g., Google) provide another solution for physicians seeking answers to their questions [11–13]. However, the success of Internet searching often depends on skilled physicians [14], and Internet searches inevitably pose challenges in information content relatedness and quality [15–24]. Additionally, traditional search engines (e.g., Google) return long lists of articles rather than self-contained answers to specific questions, and many of these articles turn out to be irrelevant to specific questions due to the inevitable query ambiguity of open-domain search engines. In a recent study [25], for instance, PubMed appeared to perform better than Google Scholar at locating relevant and important literature articles to answer specific drug-related questions.
The importance of answering physicians’ questions related to patient care has motivated the development of many clinical resources (e.g. UpToDate, Thomson Reuters, eMedicine, National Guideline Clearinghouse) to provide high-quality summaries of clinically relevant information. These summaries, however, are written by domain experts who manually review the literature concerning specific medical topics. As such, these resources may be limited in scope and timeliness. An evaluation study showed that when provided with the 10 most commonly used clinical resources without time constraint physicians were able to answer only 70% of the 105 questions randomly selected from the 1062 questions collected by Ely and his associates [26]. Another study found that despite UptoDate being the top target site used by physicians, only 10.8% physicians use it to do research on rare diseases [27]. In fact, UpToDate was reported to be used infrequently in a number of evaluation studies [13,28–31]. In addition, as databases become more complex, it takes a correspondingly greater amount of time to search for an answer even in commercial clinical databases. For example, one evaluation study [32] has shown that it takes over four minutes to search for answers in UpToDate. Studies have found, however, that when a search takes longer than two minutes, it is likely to be abandoned [10,26].
Question answering (QA) systems have the potential to overcome these shortcomings. First, to maximize coverage and improve timeliness, they can automatically mine relevant knowledge from multiple sources and summarize the results to form answers based on important concepts embedded in the question. Secondly, to improve efficiency, they can provide succinct answers rather than entire documents, which can help users pinpoint useful information quickly. However, due to the difficulties that machines have understanding text, current QA approaches are mainly focused on answering factoid questions based on fact extraction or specific question types, such as definitional questions. Unfortunately, because of the potential for inaccurate mining results in general, such common sense factoid QA systems provide inferior usability compared to emerging manually managed fact databases, such as Wikipedia, Answers, Freebase, etc.
Our goal is to go beyond such factoid systems and develop a QA system with the ability to handle the kinds of complex questions that are commonly asked in the clinical domain through the use of a structured domain-specific ontology. Domain-specific knowledge can be used to enhance the capabilities of a system to automatically answer questions oriented to sophisticated problem-solving rather than mere fact discovery, which is vital for answering questions asked in the clinical domain. We hypothesize that domain knowledge can greatly enhance the machine learningbased computational model for information retrieval (IR), and text-mining technologies can be coupled with IR to present semantically inherent answer summarization. Our focus is on improving answer quality, especially in response to complex clinical questions, so that instead of providing a single fact or a list of documents, the system will decrease human effort by extracting the most pertinent information to a given question from the large amount of literature in the clinical domain.
Our fully automated system AskHERMES – Help physicians Extract and aRticulate Multimedia information from literature to answer their ad hoc clinical quEstionS [33–43] – automatically retrieves, extracts, analyzes, and integrates information from multiple sources that include the medical literature and other online information resources to formulate answers in response to ad hoc medical questions.
Fletcher [9] identified three basic skills necessary for physicians to manage their information needs: (1) find potentially relevant information, (2) judge the best from a much larger volume of less credible information, and (3) judge whether the best information retrieved provides sufficient evidence for making clinical decisions. AskHERMES addresses the first two components by finding and filtering clinical information. We previously found that AskHERMES outperforms several other systems (e.g., PubMed) for answering definitional questions [38,39]. Currently, AskHERMES attempts to answer all types of clinical questions, and this paper reports the development, implementation, and evaluation of the AskHERMES system.
2. Background
Question answering can be considered an advanced form of information retrieval. In the 1990s, the Text REtrieval Conference (TREC) supported research within the information retrieval community by providing the infrastructure necessary for large-scale evaluation of text retrieval methodologies. In 1999, TREC introduced a question answering (QA) track, and the earliest instantiations of the QA track focused on answering factoid questions (e.g., “How many calories are there in a Big Mac?”). Since 2003, TREC has addressed scenario questions (e.g., definitional questions such as “What is X?”) that require long and complex answers. TREC Genomics introduced passage retrieval for question answering in the genomics domain [44,45].
Development in question answering has mainly focused on improving the underlying answer extraction performance, especially against a standard set of questions, which has been, for example, the objective of the QA track at TREC [46]. However, improvements by such batch-run experiments may not translate into actual benefits for end users [47]. To date, few question answering systems have focused on designing effective interfaces. Many existing commercial search engines, such as Google, Yahoo and Bing, only return a long ranked list of relevant documents, an interface that is similarly used by PubMed in the biomedical domain. As discussed earlier, such interfaces are insufficient for providing succinct and relevant answers, which is an especially pertinent issue for physicians who have little time for wading through long lists of retrieved documents. Search results clustering, a visualization technique first introduced in the Scatter–Gather system [48], attempts to provide the user with essential information about the structure of topics in the retrieved results, and similar approach has been applied in medical literature search [49] and refined by several search sites, including Vivisimo, iBoogie, and the Carrot system. However, the output of these systems is still based on traditional retrieval results, and users must read through a ranked document list even for a query on a single topic.
Addressing clinical question answering has been an active effort of the biomedical community. Cimino et al. [50] tagged clinical questions semantically to make them generic (for example, “Does aspirin cause ulcers” became “Does <drug> cause <disease>”). Zweigenbaum [51,52] surveyed the feasibility of question answering in the biomedical domain. Rinaldi and colleagues [106] adapted an open-domain question answering system to answer genomic questions (e.g., “Where was spontaneous apoptosis observed?”). The EpoCare project (Evidence at Point of Care) proposed a framework to provide physicians the best available medical information from both literature and clinical databases [53,54]. Infobuttons [8,31,55–62] served as a medical portal to external information retrieval systems (e.g. PubMed) and databases (e.g., UpToDate). The CIQR (Context Initiated Question Response) project [63] focuses on the analysis of the types of questions asked by clinician when looking up references, which allows speech input in the clinical setting [64]. To incorporate patient specific information in seeking relevant and up-to-date evidence, the PERSIVAL (Personalized Retrieval and Summarization of Image, Video and Language Resource) system [65–67] was designed to provide personalized access to a distributed digital library.
Automatically analyzing clinical questions is an important step toward answering clinical questions. Physicians often ask complex and verbose questions comprising a wide variety of types. The typology of question types with representative examples collected in four studies [7,26,68,69] was shown in Table 1. There is a wealth of research proposing ways of categorizing such ad hoc questions. Ely and colleagues manually mapped 1396 clinical questions [70] to a set of 69 question types (e.g. “What is the cause of symptom X?” and “What is the dose of drug X?”) and 63 medical topics (e.g. drug or cardiology). Cimino and associates [50] predefined a set of generic question types (e.g., “What is treatment for disease?”) and then mapped ad hoc clinical questions to those types. Seol and associates [71] identified four major question types: treatment, diagnosis, etiology, and prognosis. Such typologies offer different solutions for automated systems to overcome the wide range of variability in the forms that clinical questions may take. Other researchers have applied the popular Population, Intervention, Comparison, and Outcome (PICO) framework as a way of dealing with the variability in clinical questions [53,54, 72–75].
Table 1.
A typology of question types, with representative examples, collected by Ely and associates in four studies. The left column represents the proportion of generic question types that the 4654 questions could be mapped to; questions beginning with the interrogatives “What”, “How”, “Do”, and “Can” account for 2231 (or 48%), 697 (or 15%), 320 (or 7%), and 187 (or 4%) of the questions, respectively. Representative examples are in the right column.
| General question type (and percentage) | Sample questions |
|---|---|
| “What …” (48%) | 1. What is the cause and treatment of this old man’s stomatitis? |
| 2. What should you do with someone who is not getting better from epicondylitis after physical therapy and nonsteroidal anti-inflammatory drugs have not worked? | |
| “How …” (15%) | 3. How long should you leave a patient on Coumadin and heparin? |
| “Do …” (7%) | 4. Do angiotensin II inhibitors work like regular angiotensin converting enzyme inhibitors to preserve kidney function in mild diabetes? |
| “Can …” (4%) | 5. Can Lorabid cause headaches? |
| Others (25%) | 6. I wonder if this patient could have a rotator cuff thing? |
Information retrieval component can be integrated with question analysis in the question answering system to retrieval relevant documents. There have been different models developed for information retrieval, including Boolean models [76], vector space models [77], ontology-based approaches [78], latent semantic indexing [79,80], and language models [81].
In addition, many existing systems turn to external knowledge to support deeper semantic analysis in question answering. SemRep [82,83] maps biomedical text to the Unified Medical Language System (UMLS) [84] concepts and represents concept relations with the UMLS semantic relationships (e.g., TREATS, Co-OCCURS_WITH, and OCCURS_IN). The SemRep summarization system condenses the concepts and their semantic relations to generate a short summary [83]. Essie is an information retrieval engine developed and used at the US National Library of Medicine (NLM) that incorporates knowledge-based query expansion and heuristic ranking [85]. CQA-1.0 [72] is designed as a clinical question answering system. Unlike AskHERMES that process ad hoc natural language question and incorporates mostly statistical and machine-learning approaches, CQA-1.0 requires a user to enter a question by the PICO framework (Patient, Intervention, Comparison, and Outcome), and provides semantic analysis at document and text level, identifying PICO elements and documents that are of clinical relevance. Sneiderman et al. [82] integrated three systems (SemRep, Essie, and CQA-1.0) to achieve the best information retrieval system (which outperformed each of the three systems) in response to clinical questions.
None of the aforementioned systems, however, are available online for testing. Although question answering is an active research field, most online QA systems as shown in Table 2 are not applicable to the clinical domain. The contributions of the AskHERMES system include:
Table 2.
A list of online non-clinical question answering systems.
| System | Domain | Characteristics |
|---|---|---|
| AnswerBus (http://www.answerbus.com/index.shtml) | Open domain | Returns relevant documents from WWW in response to an ad hoc question |
| Ask (http://www.ask.com/) | Open domain | Returns relevant paragraphs in response to a particular question |
| BrainBoost (http://www.answers.com/bb/) | Open domain | Returns sentences relevant to an ad hoc question |
| EAGLi (http://eagl.unige.ch/EAGLi/) | Genomics domain | Returns MEDLINE documents in response to an ad hoc genomics question |
| Start (http://start.csail.mit.edu/) | Open domain | Returns a short phrase in response to a factoid question |
| Why-Question (http://lands.let.ru.nl/cgi-bin/retrieve_wikidoc.pl/) | Open domain | Returns relevant paragraphs in Wikipedia in response to a why-type question |
| KnowItAll (http://www.cs.washington.edu/research/knowitall/) | Open domain | Returns a list of extracted relations in response of a predicative query |
| Wolfram Alpha (http://www.wolframalpha.com) | Open domain | Computational knowledge engine based on internal database |
A tailored machine learning model that automatically extracts information needs from complex clinical questions.
A dynamic model for hierarchically clustering sentences as answers and a new sentence ranking function.
A new answer presentation model in which answers, rather than document lists, are organized by question-orientated keywords.
Finally, AskHERMES is the only online system that attempts to automatically answer the full range of complex clinical questions.
3. Methods
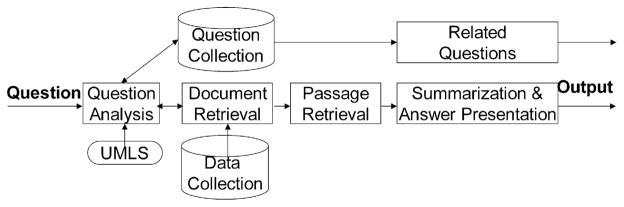
Fig. 1 shows the system architecture of AskHERMES, which takes as its input a natural language clinical question. Question Analysis automatically extracts information needs from the question and outputs a list of query terms. The UMLS knowledge resource is used for query term expansion. The Related Questions Extraction module returns a list of similar questions. Information Retrieval returns relevant documents that have been locally indexed. Information Extraction identifies relevant passages. Summarization & Answer Presentation aggregates answer passages, removes redundant information, automatically generates structured summaries, and presents the summaries to the user posing the question. In the following sections, we will provide a detailed description of each component.
Fig. 1.
The AskHERMES architecture.
3.1. Data sources and pre-processing
3.1.1. Data collection
At the time of evaluation, AskHERMES had indexed over 17 million MEDLINE abstracts (1966–2008); 2732 eMedicine documents (downloaded in 2008); 2254 clinical guidelines (downloaded in 2008); 167,000 full-text articles (downloaded from PubMed Central in 2008); and 735,200 Wikipedia documents. In total, there are 15,046,596 articles containing 3 million unique word tokens.
3.1.2. Pre-processing for retaining semantic content
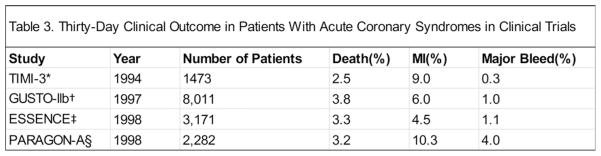
Most NLP approaches focus on mining narrative texts in different kinds of documents or articles, which, however, would lose semantic information embedded in tables and lists. It happens quite often particularly in clinically relevant articles such as those from eMedicine, clinical guidelines, and Wikipedia. For example, Fig. 2 shows a partial table in an article from eMedicine. The cells (e.g., “1994,” “year,” “2.5%” and “death”) in the table alone are of little meaning, but together, the content can help correctly answer questions like “What is the death rate for Acute Coronary Syndromes in 1994?” Similarly, grouping items from the list in Fig. 3 allows AskHERMES to answer such questions as “What is involved in Survival Skills for diabetes patients?” Therefore, we implemented manually curated rules to retain semantic information contained in each table and list. Specifically, for each table, all the textual information in each row or column (depending on the header location) together with the corresponding header and caption text would be formed as a separate passage to be indexed by the system; for each list, we treated it as a tree and each non-leaf branch node (including the root node) would generate a separate passage to be indexed, by collapsing all the nodes it contains and then combining the corresponding textual information with the caption text. In the meantime, we removed noisy short anchor texts (e.g., “Contact Us”) from the raw text before indexing.
Fig. 2.
An excerpt of a partial table appearing in an eMedicine article.
Fig. 3.
An excerpt of a list from a guideline in National Guideline Clearinghouse.
Another strategy we took in the pre-processing is merging all the section titles with any sentence within the corresponding section, with the assumption that section titles in the articles carry important semantic content which may not necessarily be explicitly described in narrative paragraphs. Despite the simplicity of this approach, it has improved the performance of AskHERMES. For example, by merging the title “cures for type 1 diabetes” with the sentence “simultaneous pancreas-kidney transplant is a promising solution, showing similar or improved survival rates over a kidney transplant alone”, the merged text can be recognized by AskHERMES as an answer to the question “What is the cure for type 1 diabetes?”, which would otherwise be missed because the sentence alone does not incorporate the word “cure”, and the title alone does not provide a description of a potential cure.
3.2. Question analysis
In the open domain, a common approach for question analysis is to map questions into a predefined question template (e.g., “What-type” and “How-type”) [86]. Such an approach has also been implemented into most of the existing online QA search engines (e.g., AnswerBus). Such template-driven approaches have significant limitations because they cannot handle the variation that is abundant in clinical questions. For example, three questions, shown below, belong to three different templates but require the same answer:
How should I treat polymenorrhea in a 14-year-old girl?
What is the treatment (or therapy) of polymenorrhea for a 14-year-old girl?
Who can tell me the treatment of polymenorrhea for a 14-year-old girl?
Furthermore, many clinical questions cannot be mapped to a specific template, as shown in the example below:
“The maximum dose of estradiol valerate is 20 milligrams every 2 weeks. We use 25 mg every month which seems to control her hot flashes. But is that adequate for osteoporosis and cardiovascular disease prevention?”
Therefore, a clinical QA system must have the ability to deal with a wide variety of complex questions, many of which cannot be answered by approaches depending on predefined templates. Accordingly, we have developed novel approaches to automatically extract information needs from complex questions [40]. Firstly, we classify a question into 12 general topics to facilitate information retrieval. Those topics include device, diagnosis, epidemiology, etiology, history, management, pharmacology, physical finding, procedure, prognosis, test and treatment & prevention, which have been used to annotate the 4654 clinical questions [40] by clinicians who recorded the questions. For example, the question above represents a medication, and we can therefore identify a pharmacological database (e.g., Thomson Reuters) as the best resource for potential answer extraction. Secondly, keywords that capture the most important content of the question are identified automatically. In the same question example, the keywords are “estradiol valerate” and “osteoporosis and cardiovascular disease prevention”. The keywords can be used as query terms for retrieving relevant documents as well as the anchor terms for answer extraction.
We developed supervised machine-learning approaches to automatically classify a question by general topics and to automatically identify keywords. For question classification, we explored several learning algorithms, showing support vector machines (SVMs) [87] achieved the best result. Since a question can be assigned to multiple topics, we developed a binary classifier (Yes or No) for each of the 12 topics. For keyword identification, we formulated it as sequence labeling problem using conditional random fields (CRFs) [88] model. In addition to basic lexical features(e.g. unigram, bigram) and syntactic features(e.g. parts-of-speech), we incorporated the lexical tool MMTx, an implementation of MetaMap [89], to map text to the UMLS concepts and semantic types as learning features for both tasks. Using an annotated collection of the 4654 clinical questions as the training and testing data, our results show an average of 76.5% F1-score for question classification and 58% F1-score for keyword extraction. Details of this work appear in [40].
3.3. Document retrieval
We integrated the latest version of the probabilistic relevance model BM25 [90] with the AskHERMES system for document retrieval, as it proved to be the best performing system [90,91] for tasks such as those at the recent Text REtrieval Conference (TREC) and held the advantage of simplicity, interpretation, and speed of computation. We empirically tuned the retrieval model in our system.
3.4. Passage retrieval
Previous work has shown that QA users prefer answers to be passages rather than sentences [92]. This is particularly true in our task, since much important content in terms of discourse relations (e.g., causal and temporal) is missing if groups of isolated sentences are extracted as answers. Much of the work done previously has defined a passage as a naturally occurring paragraph [93] or a fixed window [94]. However, candidate passages determined by such a definition will sometimes be too verbose to be of practical value for question answering. Therefore, we developed an approach that dynamically generates passage boundaries.
Specifically, we define a passage in AskHERMES as one or more adjacent sentences, in which every sentence incorporates one or more query terms from the question. Our approach is different from TextTiling [95], a popular method for multi-paragraph segmentation, in that the posed question plays an important role for passage recognition in our system.
As part of this work, we defined a novel scoring function for measuring the similarity between a sentence and the question (SS), which integrates both word-level and word-sequence-level similarity between a question and a sentence in the candidate answer passage, as shown in:
| (1) |
Sd denotes question-document similarity based on BM25 similarity, TFq is the total number of query terms that appear in the sentence, UTq is the unique number of query terms in the sentence, and LCS is the similarity between the sentence and the whole question based on the longest common subsequence (LCS) score [96].
LCS is an algorithm that identifies the longest subsequence common to all sequences in a set of sequences (typically just two) and is recognized as being very important for measuring similarity for many text processing applications, e.g. summarization evaluation (ROUGE score [97]). Incorporating the LCS score in the sentence score function can capture more detailed dependency information than mere bag-of-words or even bigrams. For example, given the question “How do I treat this man’s herpes zoster?” the candidate answers represented by sentences (1) and (2) below, have the same words and frequency that are matched against the extracted query terms (“treat”, “herpes”, and “zoster”), which means that an approach without LCS would rank the two answers the same. However, LCS assigns sentence (1) a value of 3 and sentence (2) a value of 2, giving sentence (1), which is the better answer for meeting the needs of the question, a higher ranking than sentence (2).
Corticosteroids have been used to treat herpes zoster for much longer than the antiviral drugs, but the effect of corticosteroids on PHN does not appear to be consistent.
A significant proportion of older subjects with herpes zoster develop post-herpetic neuralgia (PHN), a chronic condition that is difficult to treat.
Once the relevance score for each sentence is obtained, the score of a passage Sp is determined by the empirical metrics shown in:
| (2) |
where n is the number of sentences in this passage, max(Ss) is the maximum relevance score among all the sentences, and min(Ss) is the minimum score among all the sentences.
3.5. Summarization and answer presentation
We developed a new question-tailored summarization and answer presentation approach based on clustering technique. As stated earlier, clinical questions are typically long and verbose and frequently relate to multiple topics. Our automatic keyword extraction model effectively extracts content-rich keywords from ad hoc questions, and such keywords can then be used to hierarchically structure the summarized answers.
For example, in the question “How should I treat polymenorrhea in a 14-year-old girl?” the terms “treat”, “polymenorrhea”, “14-year-old” and “girl” are four important content terms, and an ideal answer would incorporate all four terms. However, in reality, most answer passages incorporate fewer content terms and sometimes contain only one of the four terms. We speculate that users would be able to identify the answer more efficiently if the answers could be grouped by the content terms.
Another benefit of this framework is that if physicians were interested in finding a general treatment for “polymenorrhea”, they would be able to examine the answer group containing “treat” and “polymenorrhea” without any age-related terms.
Responding to this motivation, we developed a novel summarization system based on structural clustering using content-bearing terms that provides a more user-friendly answer presentation interface to help physicians quickly and effectively browse answer clusters.
3.5.1. Topical clustering, ranking and hierarchical answer presentation
To extend the spirit of search results clustering, in this paper we propose an innovative hierarchical answer presentation interface in which all relevant passages are grouped into different topics based on two-layer clustering. We presented query-related answer passages that were structurally clustered based on content-bearing query terms rather than merely providing a ranked list of documents as output. Topic labels are assigned to each cluster in AskHERMES using query terms and expanded terms from the UMLS. Using a topic-labeled tree structure generated from first-layer clustering, physicians can easily locate information of interest before delving into more detail. In addition, second-layer clustering provides more refined categories for multi-faceted answers. Each leaf node is a small passage rather than a document, which facilitates browsing.
For query term-based clustering, we use the original query terms (Q) that appear in the question and the UMLS query expansion terms (QE). We first group together all the synonyms (a query term and its expanded terms are represented using the corresponding query term) and then generate root clusters, each of which contains different combinations of these synonym concepts. More formally, we assume that qi is the ith query term in Q and has Mi synonyms qei1, qei2, qei3, …, qeiMi. By “group together”, we mean that we use qi to stand for all the synonyms in the clustering, i.e. the ith concept. Passages containing different combinations of these concepts in the root node are divided into different clusters. Different variants from each synonym combination additionally lead to hierarchical subclusters.
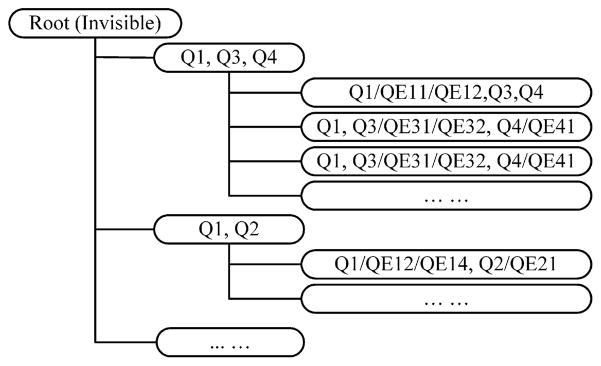
Fig. 4 further illustrates how the “bucket-based clustering” algorithm works, leading to the kind of hierarchical clustering structure shown in Fig. 5. All buckets/clusters can contain multiple attached passages.
Fig. 4.
Query term-based clustering algorithm.
Fig. 5.
Illustration of hierarchical clustering structure based on query terms.
All these hierarchical buckets are generated dynamically, which prevents a combination explosion, an issue especially relevant to complex questions. For a more compact answer presentation, we ignore root nodes to which no sentences are directly attached and promote their children branches one level up. As in Fig. 5, in cases where there is no sentence containing all three sets of the synonym terms in node “Q1, Q3, Q4”, only its children branches are displayed.
We rank clusters based on the query terms appearing in the cluster. We use the same ranking strategy as query weighting in Section 3.2 and sum up all the weights of the query term that occur as the ranking score of this cluster. We also rank the generated root buckets by summing up the IDF values of the query concept it covers. Since there are several synonyms for each query concept in the root buckets, we needed to find a way to choose an IDF value for each collapsed concept. We chose the minimum IDF value among synonyms based on the observation that some common words have rarely used synonyms with very high IDF values.
To further facilitate browsing the results, for the top p ranked clusters (not root clusters), if the number of passages in them exceeds q, we conduct a second layer of clustering based on the content of the passages along more refined semantic dimensions. In our current system, both p and q are empirically assigned to 5. For this task, we used Lingo [98], which uses a single vector decomposition approach to find the common labels for clusters and retrieves corresponding content (candidate answer passages in our system) for each cluster. This can generate readable labels for clusters and allow a passage to be put into multiple clusters instead of hard splitting.
3.5.2. Redundancy removal based on longest common substring
Because candidate answers are extracted from multiple sources, it is inevitable that they will contain some redundant information. To address this issue, we explored the longest common substring (LCSubstring) [99], which is an algorithm for identifying the longest string (or strings) that is a substring (or are substrings) of two or more strings. We applied LCSubstring in our system to remove redundancy among sentences as well as passages that were extracted as candidate answers. The difference between the longest common substring and the longest common subsequence (LCS) is that the former is required to be a continuous substring from the original strings, while the latter consists of all the common subsequences that share the same order but include intervals in between the original strings. Thus, the longest common substring has more constraints than the LCS, and we use it for redundancy removal in our system. To ensure that two units are similar enough to be considered duplicates, we set a threshold empirically. Although the longest common substring has been used for automatic summarization evaluation tasks such as ROUGE-L [100], and has been used for paraphrasing [101,102], it has not yet been reported for removing redundancy as a part of summarization.
4. System implementation
The AskHERMES system is built on the J2EE framework, in which JBoss is used for the application server and the JBoss Seam for building the user interface. JBoss has built-in EJB (Enterprise JavaBeans) caching and a reuse mechanismthat enables heavy load accessing. We also built a round-robin load-balancer in the front web server to distribute the accessing load among six backend servers. The six servers are running linux/solaris operating systems in which AskHERMES is deployed. Currently, AskHERMES system response time averages 20 s.
5. Results
In this section, we report our pilot evaluation of the AskHERMES system, which is compared with two frequently used state-of-the-art systems: the commercial Google search engine and the UpToDate clinical database system.
5.1. Evaluation design
To evaluate our AskHERMES system, we randomly selected 60 questions from the ClinicalQuestions collection [40] and asked three physicians (AB, JJC, and JE) for a manual evaluation of the output. For comparison, we also evaluated the Google search engine (using both Google and Google Scholar) and the UpToDate database system on the same set of questions. Our goal was to examine how well each of the three systems answer the questions and define the four metrics in the evaluation as follows:
Ease of Use (scale of 1 to 5).
Quality of Answer (scale of 1 to 5).
Time Spent (in s).
The Overall Performance (scale of 1 to 5).
For this pilot evaluation, each physician subject has been presented with a mutually exclusive set of 20 questions randomly selected from our question collection. For each question, each subject has been asked to identify answers from each of the three systems: AskHERMES, Google, and UpToDate, and then assign a score for each system on each evaluation metrics defined above.
5.2. Performance of AskHERMES in comparison with Google and UpToDate
Table 3 shows the results of three systems. These results show that AskHERMES’ Ease of Use score was very competitive with the same median evaluation score of 4 as both of the other systems, achieving an average score of 4.079 compared to the best score of 4.132 for UpToDate, which suggests that our clustering-based presentation interface is quite effective and beneficial in terms of user friendliness. This is also reflected in the fact that although AskHERMES has a slower response time than the other systems (a several second delay), the system’s ease of use is compensating for that lost time so that Time Spent scores are comparable.
Table 3.
Median evaluation score (with Interquartile range shown in parentheses) of Google, UpToDate and AskHERMES.
| UpToDate | AskHERMES | ||
|---|---|---|---|
| Ease of use | 4 (3, 5) | 4 (4, 5) | 4 (3.75, 5) |
| Quality of answer | 3 (1, 4.25) | 4 (1, 5) | 2.5 (1, 4) |
| Time spent (s) | 2.5 (2, 5) | 3 (2, 5) | 4 (2, 5) |
| Overall performance | 3 (1, 4) | 4 (1, 5) | 3 (1, 4) |
With regard to Quality of Answer, UpToDate attained the best score of 4, which is to be expected since it incorporates a rich domain-specific knowledge resource that is more clinically oriented. Although AskHERMES received the lowest score in this category, its difference with Google in this respect is not statistically significant (Wilcoxon signed test, p > 0.1).
Similarly, AskHERMES’ Overall Performance evaluation score was slightly lower than the UpToDate system, but as Table 4 shows, the pair-wise performance comparison of the three systems shows no statistically significant differences (p > 0.1) based on a two-sided Wilcoxon signed rank test. We also found that AskHERMES yielded the smallest standard deviation in the metrics for Quality of Answer (1.445) and Overall Performance (1.427) compared to Google (1.706/1.603) and UpToDate (1.653/1.636), demonstrating its robustness and ability to adapt.
Table 4.
Wilcoxon signed test (based on negative ranks) for overall performance comparison of the three systems (Z is the normal approximation value; p value indicates the significance level).
| Z | p value (2-tailed) | |
|---|---|---|
| Overall: Google – Overall: AskHERMES | −0.800 | .423 |
| Overall: Uptodate – Overall: AskHERMES | −1.604 | .109 |
| Overall: UptoDate – Overall: Google | −1.175 | .240 |
5.3. Impact of question length on the quality of answers
To gain further understanding of the quality of AskHERMES’ performance, we examined the questions that each system performed best on, as shown in Fig. 6. We found that each system has its own strengths for different kinds of questions; as Fig. 6 shows, AskHERMES performs best at answering complex questions within specific contexts, such as relationships, comparisons, and restrictions; Google performs best at answering short questions that can be answered in the open domain; and UpToDate performs best at answering short clinical questions on specific topics.
Fig. 6.
Sample questions that different systems perform best on.
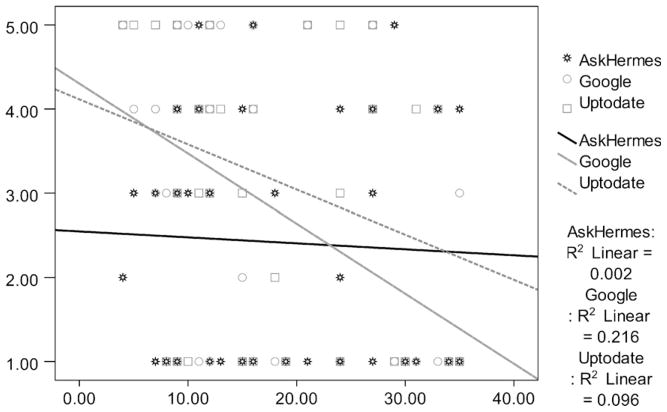
Fig. 7 shows the relationship between answer quality and the number of words in a given question, demonstrating that Google and UpToDate’s performance fluctuates for different questions while AskHERMES performs consistently across different questions. We observed that when questions contain more than 25–35 words, Google and UpToDate produce particularly poor answers compared to our system. Furthermore, we found that only when the word count of a question is less than 25 does UpToDate perform statistically better (p < 0.04) than AskHERMES for Quality of Answer. These results demonstrate that AskHERMES has a great potential for answering clinical questions, which are usually long and complex, despite its being a fully automatic system, in contrast to UpToDate, which relies on a great deal of manual effort.
Fig. 7.
Scatter graph of quality of answer and number of words in question.
6. Discussion
Currently, the development of AskHERMES is in an early stage, and the data resources it has indexed are very limited. Moreover, AskHERMES is an automatic QA system, while UpToDate uses domain experts to manually select only clinically related knowledge. Additionally, AskHERMES currently does not have access to all the full-text articles on a subject. As open-access full-text biomedical articles become increasingly available, we speculate that the performance of AskHERMES will be greatly improved.
Our pilot evaluation found that AskHERMES is competitive with other clinical information resources. The overall evaluation score of AskHERMES is not as high as UpToDate, but there were no statistically significant differences, according to Wilcoxon signed tests. Note that this is still a preliminary evaluation of our system, and the statistical test might be underpowered based on the current sample size, which therefore needs more investigation and validation on a larger scale evaluation with more independent samples in future work. Notably, we found that AskHERMES can actually perform better with longer and more complex clinical questions, which suggests that its integration of a question analysis component and domain-specific ontology offers AskHERMES the ability to understand and correctly recognize the information needs of physicians.
UpToDate is compiled by experts, while AskHERMES automatically assembles information from natural language texts. Frequently, the texts from which AskHERMES extracted information are not clinically relevant, and therefore, the non-relevancy leads to a lower performance. In addition, AskHERMES is built upon limited text resources, while UpToDate and Google Scholar have a much richer resource including full-text articles and e-books. The aforementioned factors all contribute to AskHERMES’ performance. We emphasize that despite the advantages of UpToDate and Google Scholar in their resources, the differences between AskHERMES and Google Scholar or UpToDate as shown in Table 4 were not statistically significant. Moreover, AskHERMES has achieved the same overall performance score (as shown in Table 3), suggesting that AskHERMES has the potential to outperform UpToDate and Google Scholar if rich resources are available.
On the other hand, AskHERMES holds a number of advantages over Google and UpToDate. First, the novel clustering-based summarization and presentation it offers have a clear advantage over the long document lists retrieved from Google, with the potential to save busy physicians’ time in retrieving potentially irrelevant documents. Second, AskHERMES is a fully automatic system, providing an unbeatable advantage over UpToDate in that it does not rely on time-consuming, labor-intensive human effort to maintain and update the system. Third, to use UpToDate, physicians are required to formulate their information needs clearly and succinctly and are required to know the workings of the UpToDate system, while AskHERMES requires little training, as the system has been developed to do this work itself.
Furthermore, there are some specific features of our system beyond the evaluation itself that can be summarized as follows:
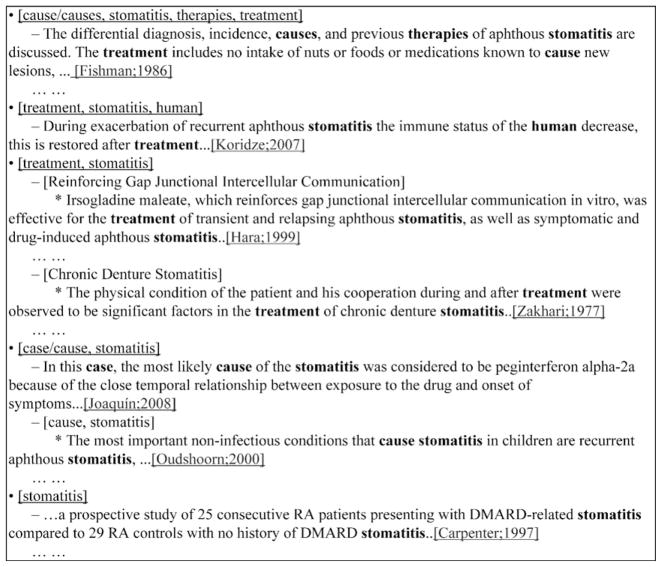
By using structural clustering based on content-bearing query terms, AskHERMES can deal with questions covering multiple focuses or topics. For example, in the question “What is the cause and treatment of this old man’s stomatitis?” there are two foci: “cause” and “treatment”, and it is very difficult to find a single sentence or a succinct passage that can cover both of them. AskHERMES can automatically separate “cause” and “treatment” based on query term-based clustering, as shown in Fig. 8.
LCS-based ranking and matching enables AskHERMES to immediately identify the best answer when the answer is similar enough to the question no matter how complex the question is. Fig. 9 shows the answer to the question “What is the difference between the Denver II and the regular Denver Developmental Screening Test?” where the highest-ranked sentence in our system’s output is the correct answer.
In order to help physicians easily obtain information from different points of view, the answer presentation interface in AskHERMES provides both clustered answers (illustrated in Fig. 8) and ranked answers (illustrated in Fig. 10). If a user’s question is simple enough or very specific, the user may find answers from ranked answers more quickly. Moreover, related questions (Fig. 10) retrieved from our question collection are also provided by our ‘interactive’ interface to assist physicians who may want to view answers to related questions.
Fig. 8.
AskHERMES’ answers to “What is the cause and treatment of this old man’s stomatitis?” (both focuses are covered in a succinct way).
Fig. 9.
AskHERMES’ answers to the question “What is the difference between the Denver II and the regular Denver Developmental Screening Test?”.
Fig. 10.
Illustration of AskHERMES’ interface for ranked answers and related questions on the query “what is the cause and treatment of this old man’s stomatitis?”.
In summary, clinical question answering is a very challenging task, and no current system can always perform well on the myriad questions that can be asked of it. AskHERMES provides a practical and competitive alternative to help physicians find answers.
7. Conclusions and future work
We present our online clinical question answering system, AskHERMES, which aims to help physicians quickly meet their information needs. The system relies on the use of supervised and unsupervised learning techniques in different components for exploring various linguistic features. AskHERMES is currently able to analyze and understand complex clinical questions of diverse types that cannot be answered by factoids or single sentences.
Our pilot evaluation shows that AskHERMES performs comparably to such state-of-the-art systems as Google and the UpToDate. In particular, our system demonstrates a better ability to answer long and complex clinical questions than other systems, showing robustness across questions of different word counts. In general, according to our preliminary results, there were no statistically significant differences between AskHERMES and the other two systems.
Since AskHERMES currently does not integrate the clinical evidence identification component that is manually entered by UpToDate, we plan to develop an automatic system to recognize clinical information to further enhance the answer quality of AskHERMES. Instead of document-based retrieval, we will investigate retrieving clinical information directly based on passage units for which we will also analyze more systematic ways of integrating keyword information. In addition, we are seeking more effective ways for improving the precision of query expansion, as explored in [103–105]. Finally, a future line of research is to conduct more extensive evaluations on the AskHERMES system, including intrinsic evaluation of clustering approach for summarization, extrinsic evaluation of the whole system using a larger data set as well as comparing with existing systems, such as Essie system (http://essie.nlm.nih.gov/), and Semantic Medline (http://skr3.nlm.nih.gov/SemMedDemo/).
Acknowledgments
The authors acknowledge support from the National Institute of Health (NIH), Grant Number 2R01LM009836. Any opinions, findings, or recommendations are those of the authors and do not necessarily reflect the views of the NIH.
References
- 1.Timpka T, Arborelius E. The GP’s dilemmas: a study of knowledge need and use during health care consultations. Methods Inform Med. 1990;29:23–9. [PubMed] [Google Scholar]
- 2.Bergus GR, Randall CS, Sinift SD, Rosenthal DM. Does the structure of clinical questions affect the outcome of curbside consultations with specialty colleagues? Arch Family Med. 2000;9:541–7. doi: 10.1001/archfami.9.6.541. [DOI] [PubMed] [Google Scholar]
- 3.Ely JW, Burch RJ, Vinson DC. The information needs of family physicians: case-specific clinical questions. J Family Pract. 1992;35:265–9. [PubMed] [Google Scholar]
- 4.Osheroff JA, Forsythe DE, Buchanan BG, Bankowitz RA, Blumenfeld BH, Miller RA. Physicians’ information needs: analysis of questions posed during clinical teaching. Ann Int Med. 1991;114:576–81. doi: 10.7326/0003-4819-114-7-576. [DOI] [PubMed] [Google Scholar]
- 5.Covell DG, Uman GC, Manning PR. Information needs in office practice: are they being met? Ann Int Med. 1985;103:596–9. doi: 10.7326/0003-4819-103-4-596. [DOI] [PubMed] [Google Scholar]
- 6.Smith R. What clinical information do doctors need? BMJ. 1996;313:1062–8. doi: 10.1136/bmj.313.7064.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005;12:217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ, Li J, Graham M, Currie LM, Allen M, Bakken S, et al. Use of online resources while using a clinical information system. AMIA ann symp proc. 2003:175–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher RH, Fletcher SW. Evidence-based approach to the medical literature. J Gen Int Med. 1997;12(Suppl 2):S5–S14. doi: 10.1046/j.1525-1497.12.s2.1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh WR, Crabtree MK, Hickam DH, Sacherek L, Friedman CP, Tidmarsh P, et al. Factors associated with success in searching MEDLINE and applying evidence to answer clinical questions. J Am Med Inform Assoc. 2002;9:283–93. doi: 10.1197/jamia.M0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen RJ. In search of evidence: family practitioners’ use of the Internet for clinical information. J Med Lib Assoc. 2002;90:370–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens MB, Von Thun AM. Military medical informatics: accessing information in the deployed environment. Military Med. 2009;174:259–64. doi: 10.7205/milmed-d-00-7108. [DOI] [PubMed] [Google Scholar]
- 13.Kitchin DR, Applegate KE. Learning radiology a survey investigating radiology resident use of textbooks, journals, and the internet. Acad Radiol. 2007;14:1113–20. doi: 10.1016/j.acra.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Tang H, Ng JH. Googling for a diagnosis – use of Google as a diagnostic aid: internet based study. BMJ. 2006;333:1143–5. doi: 10.1136/bmj.39003.640567.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell GP, Wilson P, Delamothe T. The quality of health information on the internet. BMJ. 2002;324:557–8. doi: 10.1136/bmj.324.7337.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jadad AR, Gagliardi A. Rating health information on the Internet: navigating to knowledge or to Babel? JAMA. 1998;279:611–4. doi: 10.1001/jama.279.8.611. [DOI] [PubMed] [Google Scholar]
- 17.Silberg WM, Lundberg GD, Musacchio RA. Assessing, controlling, and assuring the quality of medical information on the Internet: Caveant lector et viewor – let the reader and viewer beware. JAMA. 1997;277:1244–5. [PubMed] [Google Scholar]
- 18.Glennie E, Kirby A. The career of radiography: information on the web. J Diag Radiogr Imaging. 2006;6:25–33. [Google Scholar]
- 19.Childs S. Judging the quality of internet-based health information. Perform Meas Metrics. 2005;6:80–96. [Google Scholar]
- 20.Griffiths KM, Tang TT, Hawking D, Christensen H. Automated assessment of the quality of depression websites. J Med Int Res. 2005;7:e59. doi: 10.2196/jmir.7.5.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths KM, Christensen H. Quality of web based information on treatment of depression: cross sectional survey. BMJ. 2000;321:1511–5. doi: 10.1136/bmj.321.7275.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt JC. Commentary: measuring quality and impact of the World Wide Web. BMJ. 1997;314:1879–81. doi: 10.1136/bmj.314.7098.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benigeri M, Pluye P. Shortcomings of health information on the Internet. Health Promot Int. 2003;18:381–6. doi: 10.1093/heapro/dag409. [DOI] [PubMed] [Google Scholar]
- 24.Cline RJ, Haynes KM. Consumer health information seeking on the Internet: the state of the art. Health Educ Res. 2001;16:671–92. doi: 10.1093/her/16.6.671. [DOI] [PubMed] [Google Scholar]
- 25.Freeman MK, Lauderdale SA, Kendrach MG, Woolley TW. Google Scholar versus PubMed in locating primary literature to answer drug-related questions. Ann Pharmacother. 2009;43:478–84. doi: 10.1345/aph.1L223. [DOI] [PubMed] [Google Scholar]
- 26.Ely JW, Osheroff JA, Ebell MH, Bergus GR, Levy BT, Chambliss ML. Evans ER: analysis of questions asked by family doctors regarding patient care. BMJ. 1999;319:358–61. doi: 10.1136/bmj.319.7206.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Leo G, LeRouge C, Ceriani C, Niederman F. Websites most frequently used by physician for gathering medical information. AMIA Annu Symp Proc. 2006:902. [PMC free article] [PubMed] [Google Scholar]
- 28.McCord G, Smucker WD, Selius BA, Hannan S, Davidson E, Schrop SL, et al. Answering questions at the point of care: do residents practice EBM or manage information sources? Acad Med. 2007;82:298–303. doi: 10.1097/ACM.0b013e3180307fed. [DOI] [PubMed] [Google Scholar]
- 29.Goodyear-Smith F, Kerse N, Warren J, Arroll B. Evaluation of e-textbooks. DynaMed, MD Consult and UpToDate Aust Family Phys. 2008;37:878–82. [PubMed] [Google Scholar]
- 30.Phua J, Lim TK. How residents and interns utilise and perceive the personal digital assistant and UpToDate. BMC Med Educ. 2008;8:39. doi: 10.1186/1472-6920-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cimino JJ, Borotsov DV. Leading a horse to water: using automated reminders to increase use of online decision support. AMIA Annu Symp Proc. 2008:116–20. [PMC free article] [PubMed] [Google Scholar]
- 32.Hoogendam A, Stalenhoef AF, Robbe PF, Overbeke AJ. Answers to questions posed during daily patient care are more likely to be answered by UpToDate than PubMed. J Med Int Res. 2008;10:e29. doi: 10.2196/jmir.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Sable C, Zhu HR. Classifying medical questions based on an evidence taxonomy. Proceedings of the AAAI 2005 workshop on question answering in restricted domains; 2005. [Google Scholar]
- 34.Yu H, Sable C. Being Erlang Shen. Identifying answerable questions. Proceedings of the nineteenth international joint conference on artificial intelligence on knowledge and reasoning for answering questions; 2005. [Google Scholar]
- 35.Yu H. Towards answering biological questions with experimental evidence. Automatically identifying text that summarize image content in full-text articles. AMIA Annu Symp Proc. 2006:834–8. [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M, Cimino J, Zhu HR, Sable C, Shanker V, Ely J, et al. Beyond information retrieval–medical question answering. AMIA Annu Symp Proc. 2006:469–73. [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M, Wang W, Yu H. Exploring supervised and unsupervised methods to detect topics in biomedical text. BMC Bioinformatics. 2006;7:140. doi: 10.1186/1471-2105-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Kaufman K. A cognitive evaluation of four online search engines for answering definitional questions posed by physicians. Pacific Symp Biocomput. 2007;12:328–39. [PubMed] [Google Scholar]
- 39.Yu H, Lee M, Kaufman D, Ely J, Osheroff J, Hripcsak G, et al. Development, implementation, and a cognitive evaluation of a definitional question answering system for physicians. J Biomed Inform. 2007;40:236–51. doi: 10.1016/j.jbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Cao YG. Automatically extracting information needs from ad hoc clinical questions. AMIA Annu Symp Proc. 2008:96–100. [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H, Cao YG. Using the weighted keyword models to improve biomedical information retrieval. AMIA summit on translational bioinformatics; San Francisco, USA. 2009. [PMC free article] [PubMed] [Google Scholar]
- 42.Cao YG, Ely J, Antieau L, Yu H. Evaluation of the clinical question answering presentation. BioNLP. 2009 [Google Scholar]
- 43.Yu H, Cao YG. Using the weighted keyword models to improve clinical question answering. IEEE international conference on bioinformatics & biomedicine workshop NLP approaches for unmet information needs in health car; 2009. [Google Scholar]
- 44.Hersh W, Cohen A, Ruslen L, Roberts P. TREC 2007 genomics track overview. The TREC genomics track conference; 2007. [Google Scholar]
- 45.Hersh W, Cohen A, Roberts P, Rekapalli H. TREC 2006 genomics track overview. TREC genomics track conference; 2006. [Google Scholar]
- 46.Voorhees EM. The TREC-8 question answering track report. Proceedings of TREC; 1999. [Google Scholar]
- 47.Hersh W. The quality of information on the World Wide Web. J Am Coll Dent. 1999;66:43–5. [PubMed] [Google Scholar]
- 48.Hearst M, Pedersen J. Reexamining the cluster hypothesis: scatter/gather on retrieval results. The 19th annual international ACM conference on research and development in information retrieval (SIGIR-96); 1996. [Google Scholar]
- 49.Pratt W. Dynamic organization of search results using the UMLS. Proc AMIA Annu Fall Symp. 1997:480–4. [PMC free article] [PubMed] [Google Scholar]
- 50.Cimino JJ, Aguirre A, Johnson SB, Peng P. Generic queries for meeting clinical information needs. Bull Med Lib Assoc. 1993;81:195–206. [PMC free article] [PubMed] [Google Scholar]
- 51.Zweigenbaum P. Question answering in biomedicine. EACL workshop on natural language processing for question answering; Budapest. 2003. pp. 1–4. [Google Scholar]
- 52.Zweigenbaum P. Question-answering for biomedicine: methods and state of the art. MIE 2005 workshop; 2005. [Google Scholar]
- 53.Niu Y, Hirst G. Analysis of semantic classes in medical text for question answering. ACL 2004 workshop on question answering in restricted domains; 2004. [Google Scholar]
- 54.Niu Y, Hirst G, McArthur G, Rodriguez-Gianolli P. Answering clinical questions with role identification. ACL workshop on natural language processing in biomedicine; 2003. [Google Scholar]
- 55.Del Fiol G, Haug PJ, Cimino JJ, Narus SP, Norlin C, Mitchell JA. Effectiveness of topic-specific infobuttons: a randomized controlled trial. J Am Med Inform Assoc. 2008;15:752–9. doi: 10.1197/jamia.M2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins SA, Currie LM, Bakken S, Cimino JJ. Information needs, infobutton manager use, and satisfaction by clinician type: a case study. J Am Med Inform Assoc. 2008 doi: 10.1197/jamia.M2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cimino J. Infobuttons: anticipatory passive decision support. AMIA Annu Symp Proc. 2008:1203–4. [PubMed] [Google Scholar]
- 58.Cimino JJ. Use, usability, usefulness, and impact of an infobutton manager. AMIA Annu Symp Proc. 2006:151–5. [PMC free article] [PubMed] [Google Scholar]
- 59.Lei J, Chen ES, Stetson PD, McKnight LK, Mendonca EA, Cimino JJ. Development of infobuttons in a wireless environment. AMIA Annu Symp Proc. 2003:906. [PMC free article] [PubMed] [Google Scholar]
- 60.Cimino JJ, Li J. Sharing infobuttons to resolve clinicians’ information needs. AMIA Annu Symp Proc. 2003:815. [PMC free article] [PubMed] [Google Scholar]
- 61.Cimino JJ, Li J, Bakken S, Patel VL. Theoretical, empirical and practical approaches to resolving the unmet information needs of clinical information system users. Proc AMIA Symp. 2002:170–4. [PMC free article] [PubMed] [Google Scholar]
- 62.Cimino JJ, Elhanan G, Zeng Q. Supporting infobuttons with terminological knowledge. Proc AMIA Annu Fall Symp. 1997:528–32. [PMC free article] [PubMed] [Google Scholar]
- 63.Mendonça EA, Kaufman D, Johnson SB. Answering information needs in workflow. Proceedings of the 9th world congress on health information and libraries; 2005. [Google Scholar]
- 64.Chase HS, Kaufman DR, Johnson SB, Mendonca EA. Voice capture of medical residents’ clinical information needs during an inpatient rotation. J Am Med Inform Assoc. 2009;16:387–94. doi: 10.1197/jamia.M2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elhadad N, Kan M, Klavans JL, McKeown KR. Customization in a unified framework for summarizing medical literature. Artif Intell Med. 2005;33:179–98. doi: 10.1016/j.artmed.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 66.Elhadad N, McKeown K, Kaufman D, Jordan D. Facilitating physicians’ access to information via tailored text summarization. AMIA Annu Symp Proc. 2005:226–30. [PMC free article] [PubMed] [Google Scholar]
- 67.McKeown K, Chang SF, Cimino JJ, Feiner SK, Friedman C, Gravano L, et al. PERSIVAL, a system for personalized search and summarization over multimedia healthcare information. Proceedings of the 1st ACM/IEEE-CS joint conference on digital libraries; 2001. [Google Scholar]
- 68.Ely JW, Osheroff JA, Ferguson KJ, Chambliss ML, Vinson DC, Moore JL. Lifelong self-directed learning using a computer database of clinical questions. J Family Pract. 1997;45:382–8. [PubMed] [Google Scholar]
- 69.D’Alessandro DM, Kreiter CD, Peterson MW. An evaluation of information-seeking behaviors of general pediatricians. Pediatrics. 2004;113:64–9. doi: 10.1542/peds.113.1.64. [DOI] [PubMed] [Google Scholar]
- 70.Ely JW, Osheroff JA, Gorman PN, Ebell MH, Chambliss ML, Pifer EA, et al. A taxonomy of generic clinical questions: classification study. BMJ. 2000;321:429–32. doi: 10.1136/bmj.321.7258.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seol YH, Kaufman DR, Mendonca EA, Cimino JJ, Johnson SB. Scenario-based assessment of physicians’ information needs. Medinfo. 2004;11:306–10. [PubMed] [Google Scholar]
- 72.Demner-Fushman D, Lin J. Answering clinical questions with knowledgebased and statistical techniques. Comput Linguist. 2007;33:63–103. [Google Scholar]
- 73.Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annu Symp Proc. 2006:359–63. [PMC free article] [PubMed] [Google Scholar]
- 74.Demner-Fushman D, Lin J. Answer extraction, semantic clustering, and extractive summarization for clinical question answering. Proceedings of the 21st international conference on computational linguistics and 44th annual meeting of the association for computational linguistics (COLING/ACL); 2006. pp. 945–52. [Google Scholar]
- 75.Lin J, Demner-Fushman D. The role of knowledge in conceptual retrieval: a study in the domain of clinical medicine. Proceedings of the 29th annual international ACM SIGIR conference on research and development in information retrieval (SIGIR), Seattle, Washington; 2006. pp. 99–106. [Google Scholar]
- 76.Frakes WB, Baeza-Yates R. Information retrieval: data structure and algorithms. Prentice-Hall; 1992. [Google Scholar]
- 77.Salton G. A vector space model for information retrieval. CACM. 1975;18:613–20. [Google Scholar]
- 78.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deerwester SC, Dumais ST, Landauer TK, Furnas GW, Harshman RA. Indexing by latent semantic analysis. J Am Soc Inform Sci. 1990;41(6):391–407. [Google Scholar]
- 80.Hofmann T. Probabilistic latent semantic indexing. Proceedings of the 22nd annual international ACM SIGIR conference on research and development in information retrieval; California, USA. 1999. pp. 50–7. [Google Scholar]
- 81.Ponte J, Croft W. A language modeling approach to information retrieval. Proceedings of the 21st annual international ACM SIGIR conference on research and development in information retrieval; Melbourne, Australia. 1998. pp. 275–81. [Google Scholar]
- 82.Sneiderman CA, Demner-Fushman D, Fiszman M, Ide NC, Rindflesch TC. Knowledge-based methods to help clinicians find answers in MEDLINE. J Am Med Inform Assoc. 2007;14:772–80. doi: 10.1197/jamia.M2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proc AMIA Symp. 2002:722–6. [PMC free article] [PubMed] [Google Scholar]
- 84.Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Lib Assoc. 1993;81:170–7. [PMC free article] [PubMed] [Google Scholar]
- 85.Ide NC, Loane RF, Demner-Fushman D. Essie: a concept-based search engine for structured biomedical text. J Am Med Inform Assoc. 2007;14:253–63. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hovy E, Hermjakob U, Lin CY. The use of external knowledge in factoid QA. TREC; 2001; 2001. pp. 644–52. [Google Scholar]
- 87.Joachims T. Lecture notes in computer science. Berlin/Heidelberg: Springer; 1997. Text categorization with support vector machines: learning with many relevant features; pp. 137–42. [Google Scholar]
- 88.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Proceedings of the eighteenth international conference on machine learning (ICML); 2001. pp. 282–9. [Google Scholar]
- 89.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 90.Robertson S, Zaragoza H, Taylor M. ACM CIKM. 2004. Simple BM25 extension to multiple weighted fields. [Google Scholar]
- 91.Zaragoza H, Craswell N, Taylor M, Saria S, Robertson S. Microsoft Cambridge at TREC-13: web and HARD tracks. Proc of TREC; 2004; 2004. [Google Scholar]
- 92.Tellex S, Katz B, Lin J. Quantitative evaluation of passage retrieval algorithms for question answering. Proceedings of the 26th annual international ACM SIGIR conference on research and development in information retrieval; 2003. pp. 41–7. [Google Scholar]
- 93.Gobeill J, Ehrler F, Tbahriti I, Ruch P. Vocabulary-driven passage retrieval for question-answering in genomics. The sixteenth text retrieval conference, TREC; Gaithersburg, MD. 2007. [Google Scholar]
- 94.Liu X, Croft WB. Passage retrieval based on language models. Proc of CIKM; 2002. pp. 375–82. [Google Scholar]
- 95.Hearst MA. TextTiling: segmenting text into multi-paragraph subtopic passages. Comput Linguist. 1997;23:33–64. [Google Scholar]
- 96.Paterson M, Dancik V. Longest common subsequences. Proc of 19th MFCS, No. 841 in LNCS; 1994. pp. 127–42. [Google Scholar]
- 97.Feifan Liu, Yang Liu. Exploring correlation between ROUGE and human evaluation on meeting summaries. Audio, speech, and language processing. IEEE Trans. 2010;18:187–96. [Google Scholar]
- 98.Osinski S, Stefanowski J, Weiss D. Lingo: search results clustering algorithm based on singular value decomposition. Intell Inform Syst. 2004:359–68. [Google Scholar]
- 99.Hirschberg DS. Algorithms for the longest common subsequence problem. J ACM. 1977;24:664–75. [Google Scholar]
- 100.Lin C. ROUGE: a package for automatic evaluation of summaries. Proceedings of the ACL workshop: text summarization braches out; 2004; 2004. pp. 74–81. [Google Scholar]
- 101.Ng RT, Zhou X. Scalable discovery of hidden emails from large folders. ACM SIGKDD’05; 2005. pp. 544–9. [Google Scholar]
- 102.Zhang Y, Patrick J. Paraphrase identification by text canonicalization. Proceedings of the Australasian language technology workshop; 2005; 2005. [Google Scholar]
- 103.Abdou S, Savoy J. Searching in Medline: query expansion and manual indexing evaluation. Inform Process Manage. 2008;44:781–9. [Google Scholar]
- 104.Zighelnic L, Kurland O. Query-drift prevention for robust query expansion. Proceedings of the 31st annual international ACM SIGIR conference on research and development in information retrieval; 2008. pp. 825–6. [Google Scholar]
- 105.Stokes N, Li Y, Cavedon L, Zobel J. Exploring criteria for successful query expansion in the genomic domain. Inform Retrieval. 2009;12:17–50. [Google Scholar]
- 106.Rinaldi F, Dowdall J, Schneider G, Persidis A. Answering questions in the genomics domain. ACL-2004 workshop on question answering in restricted domains; Barcelona, Spain. 2007. [Google Scholar]